Abstract
The structure-activity models of the VEGFR2 kinase inhibitory activity of the derivatives of 2-anilino-5-aryloxazole have been investigated using Combinatorial Protocol in Multiple Linear Regression (CP-MLR) with nearly 500 topological descriptors which were calculated from DRAGON software. Among the descriptor classes considered collectively in the study, the inhibitory activity was, however, correlated with simple functional (FUN), topological (TOPO), atom centered fragments (ACF), molecular walk counts (MWC) and 2D-autocorrelation (2D-AUTO) descriptors. The developed models and participating descriptors in them have suggested that the substitutional modifications in the 2-anilino-5-aryloxazole moiety may have sufficient scope in optimization of prevailing inhibitory activity of these analogues.
Introduction
Mitogenic endogeneous proteins have important function in the formation and growth of solid tumors (angiogenesis) [Citation1]. Among these proteins vascular endothelial growth factor (VEGF) controls a key step in the angiogenesis. It is up regulated by tumor cells and induces mitogenic response on binding to the tyrosine kinase receptor VEGFR2 (KDR/Flk-1) of nearby endothelial cells [Citation2]. Hence, this pathway has attracted widespread interest in anticancer therapy [Citation3]. A variety of compounds such as anilinophthalazine [Citation4], anilinoquinazoline [Citation5], indolinone [Citation6] and isothiazole [Citation7] are known to inhibit the VEGFR2. Recently, Harris et al. [Citation8] have explored a new chemical class, 2-anilino-5-aryloxazoles (), as VEGFR2 inhibitors. The variation in the chemical space of these analogues is focused around the substituents of 2-anilino and 5-phenyl moieties of the structure. In order to investigate the scope of chemical space of 2-anilino-5-aryloxazoles as VEGFR2 inhibitors, a high dimensional quantitative structure-activity relationship (QSAR) study has been undertaken on these analogues to rationalize their activity profile. For this, it is necessary to characterize the molecules or their varying structural fragments from different perspectives. Among different methods, graph theoretical approaches provide large number of structural indices characteristic to the molecules and their functional units [Citation9–13]. Moreover, when dealing with a large number of descriptors, for the optimum utilization of information content of the generated data sets, it is necessary to adopt typical protocol(s) to identify the best models as well as information rich descriptors corresponding to the phenomenon under investigation. The Combinatorial Protocol in Multiple Linear Regression (CP-MLR) [Citation14–19] is an approach among many others to address the model evolution in high-dimensional QSAR studies. The aim of present communication is therefore, to establish QSAR between the reported VEGFR2 inhibitory activity of 2-anilino-5-aryloxazoles and the molecular descriptors calculated from DRAGON software [Citation12] using the CP-MLR analysis.
Materials and method
Data set
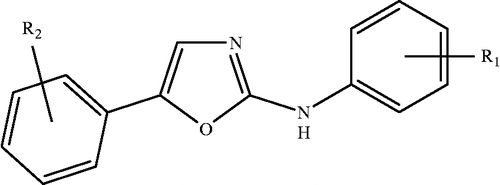
In this study 2-anilino-5-aryloxazole analogues () have been considered from the literature report [Citation8] along with their tyrosine kinase receptor, vascular endothelial growth factor receptor 2 (VEGFR2) (KDR/Flk-1) inhibitory activity in the form of logarithm of the inverse of inhibitory concentration (pIC50 where IC50 is in moles per liter against VEGFR2). The structures, for the varying R1 and R2 substituents of respective compounds, have been generated in ChemDraw [Citation20] using the standard procedure. These structures have been ported to DRAGON software for computing the parameters corresponding to 0D-, 1D-, and 2D-descriptor classes. A total number of 491 descriptors corresponding to these classes were generated. The descriptor classes along with their definitions and scope in addressing the structural features are given in . As the total number of descriptors involved in this study is very large, only the names of descriptor classes and the actual descriptor involved in the models have been addressed in the discussion. The QSAR model generation and validation have been done using the combinatorial protocol in multiple linear regression (CP-MLR) analysis [Citation14].
Table I. The observed, calculated and predicted VEGFR2 inhibition activity values of 2-anilino-5-aryloxazole derivatives ( for structures).
Table II. Descriptor classes used for the analysis of VEGFR2 inhibitory activity of derivatives of 2-anilino-5-aryloxazole and identified categories in modeling the activity.
Cp-mlr
The CP-MLR is a ‘filter’-based variable selection procedure for the development of statistical models in high dimensional QSAR studies [Citation14–19]. It involves a combinatorial strategy with approximately placed ‘filters’ interfaced with MLR and extracts diverse models having unique combination of descriptors from the dataset. The filters set the thresholds for the descriptors in terms of inter-parameter correlation cutoff limits in subset regressions (filter-1), t-values of the regression coefficients (filter-2), internal explanatory power (filter-3; square root of adjusted multiple correlation coefficient of regression equation, r-bar), and the external consistency (filter-4; Q2 i.e. cross-validated R2 from the leave-one-out procedure). Throughout this study, the thresholds for the filters-1, 2 and 4 were assigned as 0.3, 2.0, and 0.3 ≤ Q2 ≤ 1.0, respectively. In the initial attempt, the base line models were generated by selecting a value of 0.71 for filter-3. In order to collect the descriptors with higher information content, the initial threshold of filter-3 was successively incremented with increasing the number of descriptors (per model) by considering the r-bar value of the preceding optimum model as the new threshold for the next generation.
Descriptor classification protocol
The three-stage descriptor classification protocol [Citation18] is implemented with two-descriptor combinations (baseline models), as they are the simplest to understand and to explain the activity. In the first stage of the classification protocol, the correlations of the activity with two descriptor combinations from the individual descriptor classes (DCs) of the dataset were used to sort them into four categories. They are primary contributors (category I: a DC forms a model with its constituent descriptors), collective contributors (category II: a DC unable to form a model with its constituent descriptors, but forms model(s) in combination with a descriptor from another such DC), secondary contributors (category III: a DC forms the model(s) only in combination with category I) and noncontributors (category IV: a DC unable to form a model(s) in any manner like that of category I, II, and III). The sorted DCs were collated in the second stage to identify all the 3-descriptor models across the categories. In the last stage, the individual descriptors emerged in all three-descriptor models were pooled to discover the higher models for quantification of the activity.
All the identified models have been put to the randomization test [Citation16,Citation21] by repeated randomization of the activity to discover the chance correlations, if any, associated with them. For this every model has been subjected to 100 simulation runs with scrambled activity. The scrambled activity models with regression statistics better than or equal to that of the original activity model have been counted to express the percent chance correlation of the model under scrutiny. The model development procedure has been finally validated by creating divergent training and test sets from complete data set.
Results and discussion
Initially, the VEGFR2 inhibitory activity of 35 analogues of 2-anilino-5-aryloxazole was investigated with a variety of 0D-, 1D- and 2D-descriptors obtained from DRAGON software. Several of these descriptors recognized from ten different classes, have shown significant correlations and are identified as the primary contributors (category I) in modeling the inhibitory activity of these compounds. lists various models (Equations 1–17) derived in such descriptors from each class along with their statistical parameters.
Table III. The QSAR models emerged in primary descriptors* from ten different classes.
Among the one and two descriptor models, the inhibition activity of the compounds have been best explained by CONS, TOPO, BCUT and 2DAUTO descriptor classes. The CONS descriptors appeared in model 2, favors the flexibility in molecular structure (RBN, number of rotatable bonds) in addition to the preference of five membered rings (nR05, number of five membered rings) in a structure for enhanced activity. The TOPO class descriptors, TIC2 (total information content index of neighborhood symmetry of order-2) and CIC5 (complementary information content of neighborhood symmetry of order-5) in model 4, favor the enhancement of these information contents for improving the activity. The BCUT descriptor BELe6 (lowest eigenvalue 6 of Burden matrix/weighted by atomic Sanderson electronegativities) alone have explained 74 per cent of variance in the activity (model 7). The 2DAUTO descriptors in model 11, ATS7m (Broto-Moreau autocorrelation of lag 7/weighted by atomic masses) and GATS2p (Geary autocorrelation of lag 2/weighted by atomic polarizabilities) suggest the importance of lags 7 and 2 weighted by respective properties.
The significance of various emerged models, listed in , may be ascertained through the statistical parameters, the correlation coefficient r, the standard error of the estimate s, and the Fisher's ratio F. Additionally, the cross-validated index Q2, obtained from leave-one-out (LOO) procedure, may assist in identifying the robustness of these models. In a comparative study, where a large number of QSAR models are generated from the descriptors belonging to different categories, the other important statistics such as the Kubinyi function, FIT [Citation22,Citation23] and the Akaike's information criterion, AIC [Citation24,Citation25] are very important in explaining the best predictive model equation. Even in stepwise development of a QSAR equation, these statistical parameters may play crucial role in ascertaining the overall significance of final model. The FIT function is closely related to the F-statistic but proved to be a useful parameter for the assessment of the quality of the models. The disadvantage of the F value is its sensitivity to changes in the number of independent variables, k in the equation that describes the model. The F value is more sensitive if k is small, whereas it is less sensitive if k is large. The FIT function, on the other hand, is less sensitive to a lower number k but is more sensitive to a larger number k. The best model would yield the highest value for this function. The AIC takes into account the statistical goodness of fit and the number of parameters that have to be estimated to achieve that degree of fit. The model that produces the lower AIC value should be considered potentially the most useful. The physical interpretation of resultant descriptors is given briefly in the footnotes under .
In search of statistical significant models, the equations in three descriptors were further derived and identified relevant descriptors were categorized under two different pools. Firstly, 25 descriptors () have emerged from the category analysis. These primary contributors were then subjected to CP-MLR. The resulting best model, in three descriptors, is shown in Equation (18)
In above equation, nR05, a CONS class descriptor, is accounting for the number of 5-membered rings in the structure under consideration. The positive regression coefficient of this descriptor recommends 5-membered rings in a structure. Likewise, ATS5e, which is Broto-Moreau autocorrelation of topological structure with path length (lag) 5 in the graph weighted by atomic Sanderson electronegativities, belonging to 2DAUTO class, indicated that the higher path lengths rich in electronic content would be favorable for the improvement of inhibition activity. The FUN class descriptor, nCaH, is indicative of the number of unsubstituted aromatic C (sp2) in a molecule. The associated negative regression coefficient of this parameter, therefore, demands for higher number of substituted aromatic carbons.
To explore models superior to the model in Equation (18), a second pool of descriptors was formulated from all the 10 classes considered collectively. This pool now comprising of 481 descriptors, was able to generate 16 models through the CP-MLR analysis. The 18 descriptors participated in these models along with their average regression coefficients, and total incidences are listed in . In these 16 models, Equation (18) was obtained once again in addition to a slightly superior Equation (19)
Table IV. Descriptors identified for modeling the VEGFR2 inhibitory activity along with the average regression coefficient, standard deviation and the total incidence.
The statistical parameter r is now able to account for 83% of variance in the observed activity and Q2 pointed to the robustness of this model. The newly appeared descriptors in above Equation, BEHe7 and MATS4v, belong to BCUT and 2DAUTO classes respectively. The former descriptor representing the highest eigenvalue n.7 of Burden matrix/weighted by atomic Sanderson electronegativities contributes positively to improve the activity while the later one being the Moran autocorrelation of topological structure with lag 4 in the graph weighted by atomic van der Waals volumes causes detrimental effect to the activity. Equation (19) was further subjected to randomization process, where 100 simulations were carried out but none of the identified models in these simulations has shown any chance correlation. The above equation was, therefore, used to calculate pIC50 values which were in close agreement with observed ones for all the compounds except the congener 17. The residual [pIC50(observed) − pIC50(calculated)] obtained for congener 17 was more than two times the standard deviation. This data point was, therefore, eliminated from the data set and the derived new model is as in Equation (20)
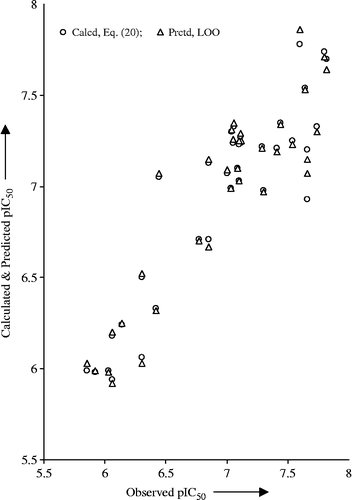
All the statistical parameters of Equation (20) have improved over to that of Equation (19). This, in turn, reflected upon the superiority of newly derived model. The outlier behavior of compound 17 is not immediately apparent. The calculated pIC50 values, using Equation (20), and predicted from LOO analysis are in close agreement with the observed ones (). The plot of observed versus calculated and predicted pIC50 values is given in to demonstrate the goodness of fit and to show systematic variations between them in the present congeneric series. From Equation (20), it has appeared that the more number of 5-membered rings and the highest eigenvalue n.7 of Burden matrix/weighted by atomic Sanderson electronegativities contributes positively to improve the activity while the Moran autocorrelation of topological structure with lag 4 in the graph weighted by atomic van der Waals volumes results in detrimental effect to it.
Equation (20) was further validated through three test sets, each containing 9 compounds out of the 34 active ones listed in . Of the three test sets, two were generated in the SYSTAT [Citation26] using the single linkage hierarchical cluster procedure involving the Euclidean distances with regard to the descriptors and to the activity values. In either case, the selection of the test set from the cluster tree was done in such a way as to keep the test compounds at a maximum possible distance from each other. The third test set of the compounds corresponds to the random selection procedure. The three test sets, selected in this manner, represent different cross-sections of all the compounds in present series. The remaining 25 data points in each of the three training sets were then used to derive new models. These models were next used to predict activities of compounds in test sets and of compound 10 having uncertain activity value and compound 17, the outlier congener. The residuals of the predictions and the corresponding predictive r2, s, Q2 and F-values have been given in . The predictions for compounds corresponding to three test sets are within the reasonable limits of their actual values.
Table V. Predicted residual activity of different test sets (9 compounds each) of the compounds listed in .
In conclusion, the present study has provided structure-activity relationship of the VEGFR2 kinase inhibitory activity of 2-anilino-5-aryloxazole analogues in terms of structural requirements. The inhibitory activity has, therefore become the function of the cumulative effect of different structural features which were identified in terms of individual descriptors. In order to improve the inhibitory activity of a compound, the descriptors, nR05 and BEHe7 have advocated, respectively, the presence of 5-membered rings in the structural frame work and the electronic content associated to eigenvalue n.7 of the Burden matrix while the descriptor MATS4v emphasized the requirement of the path length 4, weighted by atomic van der Waals volumes. The derived models and participating descriptors in them have suggested that the substituent of 2-anilino-5-aryloxazole moiety have sufficient scope for further modification. Thus, our study may provide a ground for modeling aspects of 2-anilino-5-aryloxazoles as the inhibitors of vascular endothelial growth factor receptor 2 (VEGFR2) kinase.
Acknowledgements
We express our sincere gratitude to our Institutions for providing the necessary facilities to complete this work.
References
- J Folkman. (2003). Fundamental concepts of the angiogenic process. Curr Mol Med 3:643–651.
- T Veikkola, M Karkkainen, L Claesson-Welsh, and K Alitalo. (2000). Regulation of angiogenesis via vascular endothelial growth factor receptors. Cancer Res 60:203–212.
- J Glade-Bender, JJ Kandel, and DJ Yamashiro. (2003). VEGF blocking therapy in the treatment of cancer. Expert Opin Biol Ther 3:263–276.
- G Bold, K-H Altman, J Frei, M Lang, PW Manley, P Traxler, B Weitfeld, J Bruggen, E Buschdunger, R Cozens, S Ferrari, P Furet, F Hoffman, G Martiny-Baron, J Mestan, J Rosel, M Sills, D Stover, F Acemoglu, E Boss, R Emmenegger, L Lasser, E Masso, R Roth, C Schlachter, W Vetterli, D Wyxx, and JM Wood. (2000). New anilinophthalazines as potent and orally active well absorbed inhibitors of the VEGF receptor tyrosine kinase useful as antagonists of tumor-driven angiogenesis. J Med Chem 43:2310–2323.
- LF Hennequin, ES Stokes, AP Thomas, C Johnstone, PA Ple, DJ Ogilvie, M Dukes, SR Wedge, J Kendrew, and JO Curwen. (2002). Novel 4-anilinoquinazolines with C-7 basic side chains: Design and structure-activity relationship of a series of potent orally active, VEGF receptor tyrosine kinase inhibitors. J Med Chem 45:1300–1312.
- DB Mendel, AD Laird, X Xin, SG Louie, JG Christensen, G Li, RE Schreck, TJ Abrams, TJ Ngai, LB Lee, LJ Murray, J Carver, E Chan, KG Moss, JO Haznedar, J Sukbuntherng, RA Blake, L Sun, C Tang, T Miller, S Shirazian, G McMahon, and JM Cherrington. (2003). In vivo antitumor activity of SU11248, a novel tyrosine kinase inhibitor targeting vascular endothelial growth factor and platelet derived growth factor receptors: Determination of a pharmacokinetic/pharmacodynamic relationship. Clin Cancer Res 9:327–337.
- JS Beebe, JP Jani, E Knauth, P Goodwin, CE Higdon, AM Rossi, E Emerson, M Finkelstein, E Floyd, S Harriman, J Atherton, S Hillerman, C Soderstrom, K Kou, T Grant, MC Noe, B Foster, F Rastinejad, MA Marx, T Schaeffer, PM Whalen, and WG Roberts. (2003). Pharmacological characterization of CP-547,632, a novel vascular endothelial growth factor receptor-2 tyrosine kinase inhibitor for cancer therapy. Cancer Res 63:7301–7309.
- PA Harris, M Cheung, RN HunterIII, ML Brown, JM Veal, RT Nolte, L Wang, W Liu, RM Crosby, JH Johnson, AH Epperly, R Kumar, DK Luttrell, and JA Stafford. (2005). Discovery and evaluation of 2-anilino-5-aryloxazoles as a novel class of VEGFR2 kinase inhibitors. J Med Chem 48:1610–1619.
- SC Basak, DK Harriss, and VR Magnuson. POLLY. Duluth, MN: University of Minnesota; (1988).
- Z, Molconn ver. 2.07, eduSoft Lc, a Virginia Corporation, Ashland, VA 23005 USA.www.edusoft-lc.com.
- (a) AR Katritzky, V Lobnov, and M Karelson. CODESSA (Comprehensive descriptors for structural and statistical analysis). Gainesville, FL: University of Florida; (1994) (b) AR Katritzky, S Perumal, R Petrukhin, E Kleinpeter. CODESSA-based theoretical QSPR model for hydantoin HPLC-RT lipophilicities. J Chem Inf Comput Sci 2001; 41:569–574.
- DRAGON software version 3.0-2003. By R Todeschini, V Consonni, A Mauri, M Pavan. Milano, Italy. http//disat.unimib.it/chm/Dragon.htm..
- MP Gonzalez, and AM Helguera. (2003). TOPS-MODE verces Dragon descriptors to predict permeability coefficients through low-density polymer. J Comput-Aided Mol Des 17:665–672.
- YS Prabhakar. (2003). A combinatorial approach to the variable selection in multiple linear regression: Analysis of Selwood et al. data set–A case study. QSAR Comb Sci 22:583–595.
- MK Gupta, and YS Prabhakar. (2006). Topological descriptors in modeling the antimalarial activity of 4-(3′,5′-disubstituted aniline)quinolines. J Chem Inf Model 46:93–102.
- YS Prabhakar, VR Solomon, RK Rawal, MK Gupta, and SB Katti. (2004). CP-MLR/PLS directed structure-activity modeling of the HIV-1 RT inhibitory activity of 2,3-diaryl-1,3-thiazolidin-4-ones. QSAR Comb Sci 23:234–244.
- YS Prabhakar. A combinatorial protocol in multiple linear regression to model gas chromatographic response factor of organophosphonate esters. Internet Electron J Mol Des2004; 3: 150–162, http://www.biochempress.com..
- MK Gupta, R Sagar, AK Shaw, and YS Prabhakar. (2005). CP-MLR directed QSAR studies on the antimycobacterial activity of functionalized alkenols–Topological descriptors in modeling the activity. Bioorg Med Chem 13:343–351.
- M Saquib, MK Gupta, R Sagar, YS Prabhakar, AK Shaw, R Kumar, PR Maulik, AN Gaikwad, S Sinha, AK Srivastava, V Chaturvedi, R Srivastava, and BS.C-3 Srivastava. (2007). Alkyl/arylalkyl-2,3-dideoxy hex-2-enopyranosides as antitubercular agents: Synthesis, biological evaluation and QSAR study. J Med Chem 50:2942–2950.
- Chemdraw ultra 6.0 and Chem3D ultra, Cambridge Soft Corporation, Cambridge, USA..
- SS So, and M Karplus. (1997). Three-dimensional quantitative structure-activity relationship from molecular similarity matrices and genetic neural networks. 1. Methods and validations. J Med Chem 40:4347–4359.
- H Kubinyi. (1994). Variable selection in QSAR studies. I. An evolutionary algorithm. Quant Struct–Act Relat 13:285–294.
- H Kubinyi. (1994). Variable selection in QSAR studies. II. A highly efficient combination of systematic search and evolution. Quant Struct–Act Relat 13:393–401.
- H Akaike. Information theory and an extension of the minimum likelihood principle. In: BN Petrov, and F Csaki. Second international symposium on information theory. Akademiai Kiado: Budapest; (1973) p.267–281.
- H Akaike. (1974). A new look at the statistical identification model. IEEE Trans Autom Control AC-19:716–723.
- SYSTAT, Version 7.0; SPSS Inc., 444 North Michigan Avenue, Chicago, IL, 60611..