Abstract
The enzyme deoxyuridine 5′-triphosphate nucleotidohydrolase (dUTPase) catalyses the hydrolysis of dUTP to dUMP and PPi thus controlling the incorporation of uracil into DNA genomes. In Campylobacter jejuni dUTPase exhibits structural properties of dimeric proteins characteristic of protozoa of the Kinetoplastidae family. In the present study we perform a kinetic analysis of Campylobacter dUTPase using the continuous spectrophotometric method and show that the enzyme is highly specific for deoxyuridine nucleotides. The Michaelis-Menten constant for dUTP was 0.66 μM while the kcat was 12.3 s− 1. dUDP was also efficiently hydrolysed although the specificity constant, kcat/Km, was five fold lower than for dUTP. The reaction product and the non hydrolysable analogue α,β imido dUDP are potent inhibitors of the enzyme while several analogues of dUMP with substituents at the 3′- and 5′-positions active against trimeric dUTPases, show poor inhibitory activity. Apparent structural and kinetic differences with other eukaryotic dUTPases suggest that the present enzyme might be exploited as a target for new drugs against campylobacteriosis.
Introduction
Deoxyuridine 5-triphosphate nucleotidohydrolase (dUTPase –EC 3.6.1.23) catalyzes the magnesium dependent hydrolysis of dUTP to dUMP and pyrophosphate [Citation1], providing the substrate for methylation of uracil by thymidylate synthase and preventing accidental incorporation of uracil into DNA by DNA-polymerase. Its widespread presence in a variety of organisms, including bacteriophages and certain retroviruses with relatively small genomes, suggests that the dUTPases are vital to DNA replication in all systems [Citation2–4]. dUTPases are present in a variety of different oligomeric forms: monomeric forms are found in herpes virus [Citation5]; homodimers in the Trypanosomatidae and certain bacteria [Citation6,Citation7]; and homotrimers in bacteria and mammals. The homodimeric enzymes lack sequence and structure conservation with the trimeric group of enzymes [Citation8,Citation9]. Campylobacter jejuni, is a microaerophilic, Gram-negative, flagellate, spiral bacterium closely related to the gastric pathogen Helicobacter pylori. It is the leading cause of bacterial food-borne diarrhoeal disease throughout the world. Not only is gastrointestinal infection with Campylobacter the leading cause of bacterial diarrhoeal disease worldwide but also the most common antecedent to the peripheral neuropathies Guillain Barre syndrome (GBS) and Miller Fisher syndrome (MFS) [Citation10,Citation11]. Campylobacter is a zoonotic pathogen of humans and livestock animals such as cows, pigs, sheep, and farmed poultry are reservoirs for the organism [Citation12].
The complete genome is available [Citation13]. Several genes were found which have homologues only in the eukaryotic domain; these include the deoxyuridine 5′-triphosphate nucleotidohydrolase (dut) gene (Cj1451), which is highly similar to those of Leishmania major and Trypanosoma cruzi and presents sequence motifs common to the dUTPase-dCTPase of T2 and T4 phages. Conversely, there is no orthologue of the E. coli dut gene which belongs to the conserved trimeric family of dUTPases and bears no resemblance with dUTPases of the Trypanosomatidae family. The crystal structure of the Campylobacter enzyme has been determined showing that, as other members of this family, dimeric dUTPases have a novel all-α fold and are unrelated to the all-β dUTPases of the majority of organisms including eukaryotes such as humans [Citation14]. A structure-guided analysis of the dimeric dUTPase family revealed its sequence relationship to the phage T4 dCTPase, phosphoribosyl-ATP pyrophosphatase HisE, NTP pyrophosphatase MazG, and several uncharacterized protein families, including the human protein XTP3TPA (RS21-C6) [Citation7]. The unification of these enzymes in one superfamily of all-α NTP pyrophosphatases is proposed, suggesting that dimeric dUTPases evolved from a tetrameric MazG-like ancestor by gene duplication.
In the present report we describe a detailed study of the kinetic properties of the enzyme, assessed by the stopped-flow spectrophotometric method. The bacterial protein exhibits properties typical of dimeric dUTPases lacking the ability to hydrolyze nucleotides other than uridine di and triphosphate.
Materials and methods
Overexpression of recombinant dUTPase of C. jejuni
The coding sequence for C. jejuni dUTPase was cloned in pET-11c to give pETCJDUT which was used to transform E. coli BL21 (DE3) cells. Transformed cells were cultured at 37°C in LB medium and expression of the enzyme was induced by addition of IPTG (Boehringer) at a final concentration of 1 mM for 3 h. Recombinant dUTPase accounts for 40–50% of soluble protein extract.
For enzyme purification a pellet of dUTPase overexpressing cells was resuspended in a solution consisting of buffer A (Tris-HCl 50 mM, EDTA 2 mM, MgCl2 2 mM) plus protease inhibitors (leupeptin 0.02 mg/mL, aprotinin 0.05 mg/mL, phenanthroline 10 mM, trypsin inhibitor 0.05 mg/mL, benzamidine 1 mM, PMSF 52μM). The soluble crude extract was obtained by sonication and centrifugation at 11000xg. The purification protocol developed consisted in three chromatographic steps (), hydroxyapatite, anion exchange chromatography in DEAE-cellulose, and mono-Q chromatography as previously described for dimeric enzymes [Citation15,Citation16]. This protocol gives 20 mg of protein per liter of culture and the protein is more than 95% free of impurities as judged by SDS-PAGE analysis with Coomassie blue staining of overloaded gels.
Table I. Kinetic parameters for dUTP and dUDP hydrolysis by Campylobacter jejuni dUTPase.
Measurement of dUTPase activity
Nucleotide hydrolysis was monitored by mixing enzyme and substrate with a rapid kinetic accessory (Hi-Tech Scientific) attached to a spectrophotometer (Cary 50) and connected to a computer for data acquisition and storage. Protons, released through the hydrolysis of nucleotides, were neutralised by a pH indicator in weak buffered medium with similar pKa and monitored spectrophotometrically at the absorbance peak of the basic form of the indicator. The indicator/buffer pair was cresol red/BICINE (pH 8, 573 nm) and the ratio between the indicator and the buffer concentration was 50:2000 (μM). The measurements were performed at 25°C, and the solutions containing 25 mM MgCl2 and 1 mg mL− 1 BSA were previously degassed. Absorbance changes were kept within 0.1 units. The final enzyme concentration was 0.03 μM. Indicator absorbance changes corresponding to complete hydrolysis of nucleotides were recorded in the computer, and the kinetic parameters (Vmax, Kmapp) were calculated by fitting the data to the integrated Michaelis-Menten Equation 17. Data points in the region of equilibrium and the immediate start were omitted. To obtain the real Km for substrate hydrolysis and the Ki for product inhibition, the obtained Kmapps were reploted versus [S0]. Units for Vmax were μmol min− 1.
Inhibitors
The preparation of inhibitors was described previously [Citation18].
Results and discussion
Metal ion requirement
dUTPases depend on the presence of divalent metals in the medium for catalysis. The metal participates in the coordination of the phosphates in the substrate, allowing for a correct orientation and recognition of the substrate in the active site. C. jejuni dUTPase was tested for metal requirement measuring activity at different concentrations of magnesium. The enzyme showed a strict requirement for the cation and the Vmax value increased with increasing concentrations of Mg2 + from 0.01 unit mg− 1 at 0.5 mM to 0.35 units mg− 1 at 50 mM. No changes in the apparent Km value were observed even when the Mg2+ concentration was raised up to 300 mM.
Hydrolysis of purine and pyrimidine nucleotides
The hydrolysis of different nucleotides by C. jejuni dUTPase was studied. The enzyme assay was carried out using as substrates dUTP, dUDP, UTP, dTTP, dCTP, dATP, and dGTP (nucleotides were purchased from Pharmacia except for dUDP and dUTP which were from Jena Bioscience) at a final concentration of 50 μM and 30 nM of enzyme. All nucleotides tested were inefficiently hydrolysed compared to the substrates dUTP and dUDP. Only in the case of dTTP an extremely slow reaction appeared to occur. The determination of the kinetic constants for this pseudo-substrate required very high concentrations of both enzyme and substrate, and the application of the integrated rate equation was unfeasible due to lack of ability to reach a situation of substrate saturation. Kinetic parameters for dUTP and dUDP are summarised in . Values obtained for dUTP hydrolysis are similar to those described for trimeric, dimeric and monomeric dUTPases while the unique capacity for dUDP hydrolysis is shared only with trypanosomatid enzymes (). In addition, the bacterial enzyme is incapable of hydrolysis of nucleotides where the base is not uracil and therefore exhibits features that limit binding of other nucleotides and are responsible for strict substrate specificity.
Table II. Kinetic parameters for dUTP hydrolysis accomplished by monomeric, dimeric and trimeric dUTPases.
Kinetic characterization and inhibition
Recombinant enzyme was purified using a combination of hydroxyapatite, anion exchange chromatography in DEAE-cellulose, and mono-Q chromatography as previously for the dimeric enzymes from the Trypanosomatidae family [Citation15]. Data of the multiple-turnover hydrolysis of dUTP by C. jejuni dUTPase was fitted to the integrated Michaelis-Menten equation as shown in , obtaining this way values for Kmapp and Vmax. Both kinetic parameters remained constant with changes in the concentration of buffer/pH indicator. However, while the Vmax value did not depend on variations in substrate concentration (dUTP), there was an increase in Kmapp values with increasing dUTP concentrations, indicating the existence of product inhibition. After replotting the different values of Kmapp versus dUTP concentration, the real Km and the Kip value of product inhibition were calculated ().
Figure 1. Complete hydrolysis of dUTP by dUTPase under multiple-turnover conditions. The inset shows the linear transformation of the data between the arrows, according to the integrated Michaelis-Menten equation adapted for enzymes with product inhibition, and the corresponding regression line. The reaction was recorded at 573 nm after reacting 30 nm dUTPase and 50 μM dUTP in the presence of 25 mM MgCl2, 2M BICINE, and 50 μM cresol red at pH 8.
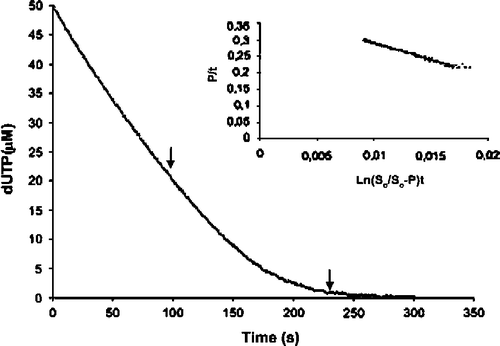
Figure 2. Inhibition by dUMP of dUTP hydrolysis. The Ki value for product (dUMP) inhibition was calculated from the Kmapp values obtained at different dUMP concentrations (5 to 100 μM). The Ki for dUMP was 11.4 μM and the real Km 0.66 μM.
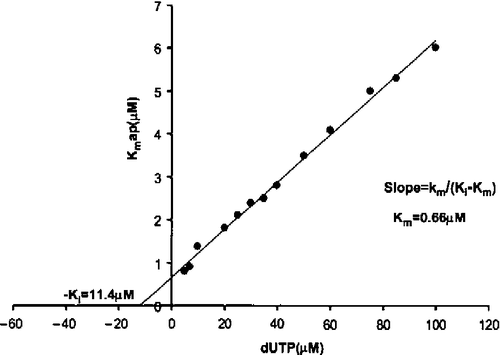
Using dUTP concentrations between 5 and 100 μM, the real Km value obtained was 0.66 μM. On the other hand, Vmax was 0.35 units mg− 1, remaining constant at the different dUTP concentrations tested. kcat was estimated to be 12.3+0.5 s− 1, assuming two active sites per dimer of dUTPase. The specificity constant, kcat/Km for dUTP was thus 18.6 × 106 M− 1 s− 1.
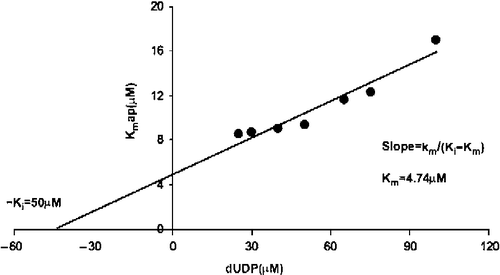
Analysis of the hydrolysis data of dUDP as described above for dUTP gave a real Km value of 4.74 μM, and a product inhibition constant Kip of 50 μM (). The kcat value was 16.6 s− 1 and the specificity constant, Kcat/Km, 3.5 x 106 s− 1M− 1.
Figure 3. Inhibition by dUMP of dUDP hydrolysis. The Ki value for product (dUMP) inhibition was calculated with the Kmapp value obtained at different dUMP concentrations (5 to 100 μM). The Ki for dUMP was 50 μM and the real Km 4.74 μM.
The inhibition of Campylobacter dUTPase by α-β-imido-dUDP was analysed. For each inhibitor concentration, the apparent Kmapp value was obtained using the integrated Michaelis-Menten equation. Km values are a linear function of the inhibitor concentration in competitive systems and:
A replot of Kmapp versus [I] has intercepts of
![]() (1+[S0]/Kiproduct)/(1 − Km/Kiproduct)(on the Kmapp axis) [Citation17]. A Ki value of 1.93 μM was obtained while no inhibition of the human enzyme was observed at 1 mM inhibitor indicative of a selectivity index greater than 500 for the bacterial enzyme and thus suggesting a basis for specific inhibitor design ().
(1+[S0]/Kiproduct)/(1 − Km/Kiproduct)(on the Kmapp axis) [Citation17]. A Ki value of 1.93 μM was obtained while no inhibition of the human enzyme was observed at 1 mM inhibitor indicative of a selectivity index greater than 500 for the bacterial enzyme and thus suggesting a basis for specific inhibitor design ().
Table III. Inhibition constants Ki (μM) for compounds against Campylobacter jejuni, P. falciparum and human dUTPases.
In addition, a series of analogues of dUMP with substituents at the 3′- and 5′-positions, together with variation in the heteroatom at the 5′-position were tested. Some of these compounds have been previously shown to inhibit efficiently the trimeric Plasmodium falciparum dUTPase while they were mostly inactive against the human enzyme [Citation18]. The structural basis for selective inhibition has been determined and is achieved by way of interactions between the trityl group and the side chains of residues Phe46 and Ile117 of the Plasmodium enzyme [Citation19]. This emphasises the strong structural and catalytic differences between dimeric and trimeric dUTPases which belong to different protein families and present different inhibition profiles [Citation18]. Further efforts are required for the identification of inhibitors of dimeric enzymes which undergo extensive secondary structure rearrangement with nucleotide binding. In the apo form, the active site is open; upon substrate binding, the enzyme undergoes a large conformational change and closes around the substrate. Thus the nature of the substrate binding site is distinctly different from that in the trimeric (including human) dUTPases [Citation14,Citation6].
In summary, the enzyme of C. jejuni has a high capacity of discrimination among the current nucleoside triphosphates present in cells. Kinetic properties and substrate specificity further support that the enzyme is related in function to dimeric enzymes of the Trypanosomatidae family. Likewise, differences in inhibition constants with trimeric enzymes such as human and Plasmodium suggest that the design of specific inhibitors of the Campylobacter enzyme may be exploited for the identification of new drugs.
Acknowledgements
We would like thank Medivir AB for contributing to the synthesis of the tritylated analogues. We would like to acknowledge the European Union LIFE programme (ICA4-CT-2001-00305), the Plan Nacional de Investigación (SAF2004-03828), the Plan Andaluz de Investigación (Cod. CVI-199) and the FIS Network RD06/0021 for funding. Antonio E. Vidal is a Postdoctoral scientist of the Ramon & Cajal programme.
References
- LE Bertani, A Haggmark, and P Reichard. (1961). Synthesis of pyrimidine deoxyribonucleoside diphosphates with enzymes from Escherichia coli. J Biol Chem 236:PC67–PC68.
- AM Baldo, and MA McClure. (1999). Evolution and horizontal transfer of dUTPase-encoding genes in viruses and their hosts. J Virol 73 (9):7710–7721.
- G Gribaudo, L Riera, P Caposio, F Maley, and S Landolfo. (2003). Human cytomegalovirus requires cellular deoxycytidylate deaminase for replication in quiescent cells. J Gen Virol 84 ((Pt 6)):1437–1441.
- AV Kaliman. (1996). Identification of the bacteriophage T5 dUTPase by protein sequence comparisons. DNA Seq 6 (6):347–350.
- N Tarbouriech, M Buisson, JM Seigneurin, S Cusack, and WP Burmeister. (2005). The monomeric dUTPase from Epstein-Barr virus mimics trimeric dUTPases. Structure 13 (9):1299–1310.
- M Harkiolaki, EJ Dodson, V Bernier-Villamor, JP Turkenburg, D Gonzalez-Pacanowska, and KS Wilson. (2004). The crystal structure of Trypanosoma cruzi dUTPase reveals a novel dUTP/dUDP binding fold. Structure 12 (1):41–53.
- OV Moroz, AG Murzin, KS Makarova, EV Koonin, KS Wilson, and MY Galperin. (2005). Dimeric dUTPases, HisE, and MazG belong to a new superfamily of all-alpha NTP pyrophosphohydrolases with potential “house-cleaning” functions. J Mol Biol 347 (2):243–255.
- A Camacho, R Arrebola, J Pena-Diaz, LM Ruiz-Perez, and D Gonzalez-Pacanowska. (1997). Description of a novel eukaryotic deoxyuridine 5′-triphosphate nucleotidohydrolase in Leishmania major. Biochem J 325:441–447.
- V Bernier-Villamor, A Camacho, F Hidalgo-Zarco, J Perez, LM Ruiz-Perez, and D Gonzalez-Pacanowska. (2002). Characterization of deoxyuridine 5′-triphosphate nucleotidohydrolase from Trypanosoma cruzi. FEBS Lett 526 (1–3):147–150.
- JB Winer. (2001). Guillain Barre syndrome. Mol Pathol 54 (6):381–385.
- HJ Willison, and GM O'Hanlon. (1999). The immunopathogenesis of Miller Fisher syndrome. J Neuroimmunol 100 (1-2):3–12.
- JE Moore, L O'Riordan, DR Wareing, R Doyle, J Lanser, T Stanley, M Matsuda, T Matsui, and PG Murphy. (2003). Phenotypic and genotypic relationship between Campylobacter spp isolated from humans and chickens in Northern Ireland—a comparison of three phenotyping and two genotyping schemes. Int J Hyg Environ Health 206 (3):211–216.
- J Parkhill, BW Wren, K Mungall, JM Ketley, C Churcher, D Basham, T Chillingworth, RM Davies, T Feltwell, S Holroyd, and et al (2000). The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403 (6770):665–668.
- OV Moroz, M Harkiolaki, MY Galperin, AA Vagin, D Gonzalez-Pacanowska, and KS Wilson. (2004). The crystal structure of a complex of Campylobacter jejuni dUTPase with substrate analogue sheds light on the mechanism and suggests the “basic module” for dimeric d(C/U)TPases. J Mol Biol 342 (5):1583–1597.
- F Hidalgo-Zarco, AG Camacho, V Bernier-Villamor, J Nord, LM Ruiz-Perez, and D Gonzalez-Pacanowska. (2001). Kinetic properties and inhibition of the dimeric dUTPase-dUDPase from Leishmania major. Protein Sci 10 (7):1426–1433.
- A Camacho, F Hidalgo-Zarco, V Bernier-Villamor, LM Ruiz-Perez, and D Gonzalez-Pacanowska. (2000). Properties of Leishmania major dUTP nucleotidohydrolase, a distinct nucleotide-hydrolysing enzyme in kinetoplastids. Biochem J 346 (Pt 1):163–168.
- I Segel. Enzyme kinetics: behaviour and analysis of rapid equilibrium and steady-state enzyme systems USA, Wiley-Interscience. Ed., (1975).
- C Nguyen, G Kasinathan, I Leal-Cortijo, A Musso-Buendia, M Kaiser, R Brun, LM Ruiz-Perez, NG Johansson, D Gonzalez-Pacanowska, and IH Gilbert. (2005). Deoxyuridine triphosphate nucleotidohydrolase as a potential antiparasitic drug target. J Med Chem 48 (19):5942–5954.
- JL Whittingham, I Leal, C Nguyen, G Kasinathan, E Bell, AF Jones, C Berry, A Benito, JP Turkenburg, and EJ Dodson. (2005). and others. dUTPase as a platform for antimalarial drug design: structural basis for the selectivity of a class of nucleoside inhibitors. Structure 13 (2):329–338.
- G Larsson, PO Nyman, and JO Kvassman. (1996). Kinetic characterization of dUTPase from Escherichia coli. J Biol Chem 271 (39):24010–24016.
- J Nord, G Larsson, JO Kvassman, AM Rosengren, and PO Nyman. (1997). dUTPase from the retrovirus equine infectious anemia virus: specificity, turnover and inhibition. FEBS Lett 414 (2):271–274.
- O Bjornberg, and PO Nyman. (1996). The dUTPases from herpes simplex virus type 1 and mouse mammary tumour virus are less specific than the Escherichia coli enzyme. J Gen Virol 77 ((Pt 12)):3107–3111.
- AC Bergman, PO Nyman, and G Larsson. (1998). Kinetic properties and stereospecificity of the monomeric dUTPase from herpes simplex virus type 1. FEBS Lett 441 (2):327–330.