Abstract
The synthesis and stability of 4-methylumbelliferyl (1 → 3)-β-D-pentaglucoside 3 are described. The (1 → 3)-β-D-glucan isolated from the cell walls of Saccharomyces cerevisiae was recovered from the aqueous medium as water-insoluble particles by the spray drying (GS) method. The acid-solubilized (1 → 3)-β-D-oligoglucosides were prepared by partial acid hydrolysis of glucan. The peracetylated (1 → 3)-β-D-pentaglucoside 1 was obtained by isolation of peracetylated (1 → 3)-β-D-oligoglucoside mixture. The peracetylated 4-methylumbelliferyl (1 → 3)-β-D-pentaglucoside 2 was synthesized by treating compound 1 with the 4-methylumbelliferone and a Lewis acid (SnCl4) catalyst. NaOMe in dry methanol was used for the deacetylation of the blocked derivative, to give the target compound 3 in an overall yield of 35%. Activity assays with β-glucosidase indicated that compound 3 was much more stable than the corresponding pentasaccharide.
Introduction
Various types of such compounds (elicitor molecules) including oligosaccharides, (glyco)proteins, (glyco)peptides and lipids, have been shown to induce defence responses in plant cells and their involvement in the detection of (potential) pathogens in plant has been discussed [Citation1]. Oligosaccharides derived from fungal and plant cell wall polysaccharides are one class of well characterized elicitors that, in some cases, can induce defence responses at a very low concentration e.g. nM. In view of their well-defined chemical nature and the presence of highly sensitive perception systems in plants, some of these elicitors have provided good model systems to study how plant cells recognize such chemical signals and transduce them for activation of the defence machinery. These studies include structure-activity relationships of the elicitor molecules, characterization of the corresponding receptors and analysis of signal transduction cascades and elicitor-responsive genes.
However, the elicitor-active oligosaccharides can be hydrolyzed by endo- or exo-hydrolases from higher plants, and give elicitor-inactive oligosaccharide fragments Citation2, Citation3. Therefore, improving the stability of the elicitor-active oligosaccharides is the key to develop the oligosaccharide elicitors.
(1 → 3)-β-D-Oligoglucosides are potent defense elicitors, both in other dicots (tomato and bean) and in monocots (wheat and rice) Citation4, Citation5. With the aim of improving the stability of elicitor-active oligosaccharides and studying the degradation velocity by β-glucosidase on them, herein, we present a very facile and convergent synthesis of 4-methylumbelliferyl (1 → 3)-β-D-pentaglucoside 3. It is the analogue of the pentasaccharide, where the 4-methylumbelliferyl function group has been introduced at the reducing end of the glycone.
Experimental
General
Saccharomyces cerevisiae cell walls were purchased from the Anqi Company (Yichang, China). β-Glucosidase from Aspergillus niger was purchased from Fluka Chemical Company. (1 → 3)-β-D-Pentaglucoside was purchased from Sigma-Aldrich Chemical Company. IR spectra were recorded with an FT-IR apparatus, and wavenumbers are reported in cm− 1. 1H and 13C NMR spectra were recorded with Bruker ARX 400 spectrometers (400 MHz for 1H, 75 MHz for 13C) at 25°C for solns in D2O as indicated. Mass spectra were recorded with a VG PLATFORM mass spectrometer using the ESI mode. Kieselgel 60F254 (E. Merck) was used for TLC.
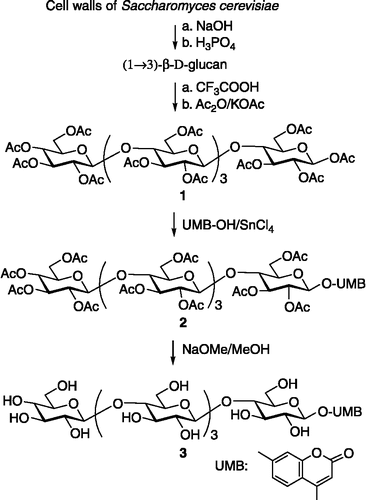
Preparation and isolation of the (1→3) - β -D-glucan
The water-insoluble (1 → 3)-β-D-glucan was obtained from Saccharomyces cerevisiae cell walls by extraction with 6% NaOH at 60°C for 4 h. Distilled water was added to the dispersion, and the insoluble part after stirring for 30 min was collected by centrifugation. The sediment was suspended in 3% NaOH and heated at 90°C for 2 h. The insoluble material was recovered by centrifugation, washed three times with distilled water, and subsequently extracted twice with 4% phosphoric acid at room temperature for 2 h. The insoluble residue, representing the cell-wall (1 → 3)-β-D-glucan was separated by centrifugation, resuspended in distilled water, and decanted with water until neutral. The aqueous suspension was taken for the recovery of the particulate glucan using the technique of spraying drying (GS). The value found was 13.5% for GS. The sample was characterized by FT-IR spectroscopy.
Partial acid hydrolysis of glucan
To prepare hydrolysates, 50-100 mg of glucan were suspended at a concentration of 5 mg/mL in 2 mol/L TFA, heated at 85°C for 110 min in 25-mL reaction flasks. The cooled samples were centrifuged at 160 × g for 10 min at 25°C. Supernatant fractions from 10 reaction flasks were pooled, and the residual TFA was removed by rotoevaporation at 35-40°C in a silanized round-bottom flask. The sample was then lyophilized.
Synthesis of 4-methylumbelliferyl (1→3)-β-D-pentaglucoside 3
To boiling Ac2O (20 mL) in a three-necked flask, 200 mg of KOAc was added. Then, 200 mg of (1 → 3)-β-D-oligoglucoside mixture was gradually added under vigorous stirring. The solution was kept for 1 h at 140°C and then cooled to room temperature. The crude product directly was purified on a silical gel 60 column (eluant: CHCl3/MeOH 4:1), and the peracetylated (1 → 3)-β-D- pentaglucoside 1 was obtained in 45% yield. The peracetate 1 (350 mg) and 60 mg of 4-methylumbelliferone were added to 21 mL of dry 1,2-dichloroethane at 45°C, followed by the addition of 58 mg of SnCl4. The mixture was stirred for 5 h. After standard processing work-up, 210 mg of peracetylated 4-methylumbelliferyl (1 → 3)-β-D-pentaglucoside 2 was obtained. In succession, compound 2 was suspended in anhyd MeOH to a concentration of 100 mg/mL and deacetylated with an equal volume of 1 mol/L NaOMe at room temperature for 60 min with continuous mixing. The same was then neutralized with 1 mol/L HCl and filtered. The filtrate was evaporated to dryness under reduced pressure at 45°C. The target compound 3 was obtained in an overall yield of 35%. 1H NMR(D2O, 400 MHz): δ 5.24 (1H, d, J1,2 3.5 Hz, H-1), 4.62-4.54 (5H, m, H-1V, IV, III, II), 3.92-3.85 (5H, m), 3.75-3.33 (19H, m), 3.25-3.19 (6H, m), UMB: 6.24, 7.70, 7.03, 7.05 (4H, m, H-3, H-5, H-6, H-8), Me (3H, d, 2.39); 13C NMR (D2O, 75 MHz): δ 103.8, 103.65 (C-1I, 1V), 103.4 (3C) (C-1II, 1III, 1IV), 85.4 (C-3I), 85.2 (C-3II), 85.0 (2C) (C-3III, 3IV), 76.8 (C-5V), 76.45, 76.4 (5C) (C-3V, 5I, 5II, 5III, 5IV), 74.3 (C-2V), 74.1 (3C) (C-2II, 2III, 2IV), 73.6 (C-2I), 70.4 (C-4V), 69.0, 68.9 (4C) (C-4I, 4II, 4III, 4IV), 61.55 (5C) (C-6I, 6II, 6III, 6IV, 6V), UMB: 161.0 (C-2), 112.2 (C-3), 154.3 (C-4), 127.1 (C-5), 114.1 (C-6), 160.3 (C-7), 103.7 (C-8), 154.8 (C-1a), 114.8 (C-4a), Me (18.7); ESI-MS: m/z 1009 [M + Na]+; Anal. Calcd for C40H58O28: C, 48.68; H, 5.92. Found: C, 48.77; H, 6.09.
Assays for enzyme activity
Assays were terminated by heating to 100°C, and the release of reducing sugars was shown to be linear with respect to time and amount of enzyme used. The β-glucosidase (40 μg/mL), BSA (0.8 mg/mL), glycerol 10% (v/v), the substrate p-nitrophenyl β-D-glucoside (20 mmol/L), and 3 (or (1 → 3)-β-D-pentaglucoside, 20 mmol/L) were incubated at 37°C in 40 mmol/L sodium acetate buffer at pH4.0. The residual activity was determined at 100-min intervals.
Results and discussion
Structural characterization of Saccharomyces cerevisiae glucan
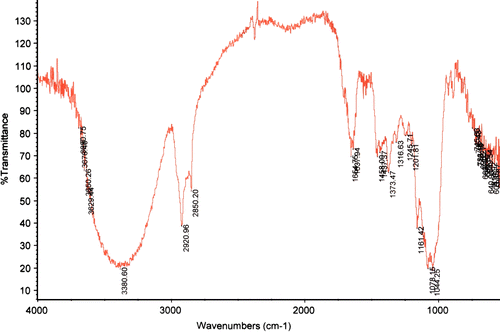
The FT-IR spectra of the glucan sample () showed the typical spectral pattern of (1 → 3)-β-D-glucan, that is, it contained absorption bands arising from the v(CC) and the v(COC) stretching vibrations at 1159 cm− 1, two partially overlapped bands at 1078 and 1041 cm− 1 attributable to ring and (C–OH) side group stretching, a band at 892 cm− 1 assigned to the β-glycosidic (C1–H) deformation mode, and the highest intensity of the v(OH) band at lower frequency (3425 cm− 1). The presence of amide I and amide II bands at 1642 and 1530 cm− 1 accords with the residual protein content (1.6%) of the glucan.
Preparation of peracetylated (1→3)-β-D-pentaglucoside 1
The acid-solubilized (1 → 3)-β-D-oligoglucoside mixture was prepared by partial acid hydrolysis of glucan in TFA (trifluoroacctic acid). The (1 → 3)-β-D-oligoglucoside mixture was acetylated with potassium acetate-acetic anhydride to maximize the yield of peracetylated (1 → 3)-β-D-oligoglucoside mixture. The peracetylated (1 → 3)-β-D-oligoglucoside mixture was isolated on a silical gel 60 column to give compound 1 in the yield of 45%.
Synthesis of 4-methylumbelliferyl (1→3)-β-D-pentaglucoside 3
Compound 1 was treated with 4-methylumbelliferone and stannic chloride as the Lewis acid catalyst to give peracetylated 4-methylumbelliferyl (1 → 3)-β-D-pentaglucoside 2. NaOMe in dry methanol at room temperature was used for the deacetylation of the blocked derivative, to give the corresponding target compound 3 in an overall yield of 35% ().
Binding β-glucosidase assay
We assayed the degradation velocity of β-glucosidase from Aspergillus niger on (1 → 3)-β-D-pentaglucoside and compound 3. The β-glucosidase activity was measured by determining the amount of reducing sugar equivalents released on incubation of the enzyme with the substrate p-nitrophenyl β-D-glucoside under the condition stated [Citation6]. The inhibition constants (k1) found for (1 → 3)-β-D-pentaglucoside and compound 3 were 80.6 mmol/L and 30.4 mmol/L, respectively, so that, compound 3 is much more stable than the pentasaccharide. This indicates that the 4-methylumbelliferyl at the reducing terminus of oligosaccharide increases its stability.
Acknowledgements
This work are supported by the Doctor Start-up Foundation of Chongqing Normal University (No. 07XLB025) and Chongqing Education Commission Foundation (No. KJ080810), China.
Declaration of interest: The author reports no conflicts of interest. The author alone is responsible for the content and writing of the paper.
References
- MG Hahn. (1996). Microbial elicitors and their receptors in plants. Annu Rev Phytopathol 34:387–412.
- GL Huang, MX Liu, XY Mei, and Y Wang. (2005). Synthesis, immunological activities, and scavenging ability toward superoxide anion of (1 → 3)-β-D-pentaglucoside and its epoxyalkyl derivatives. Bioorg Med Chem 13 (12):3873–3877.
- GL Huang, MX Liu, XY Mei, and YC Cao. (2004). Synthesis, protein-binding ability and phytoalexin-elicitor activity of epoxyalkyl (1 → 3)-β-D-oligoglucosides. Glycoconj J 20 (7–8):427–433.
- O Klarzynski, B Plesse, JM Joubert, JC Yvin, M Kopp, B Kloareg, and B Fritig. (2000). Linear β-1,3 glucans are elicitors of defense responses in tobacco. Plant Physiol 124 (3):1027–1038.
- GL Huang, XY Mei, MX Liu, and TC Liu. (2004). Synthesis, (1 → 3)-β-D-glucanase-binding ability and phytoalexin-elicitor activity of (R)-2,3-epoxypropyl (1 → 3)-β-D-pentaglucoside. Bioorg Med Chem Lett 14 (24):6027–6029.
- GL Huang, and XY Mei. (2007). Production and chitinase-binding ability of lipo-chitopentaose nodulation factor. J Enz Inhib Med Chem 22 (2):247–249.