Abstract
The increase of antibiotic resistance amongst Mycobacterium tuberculosis strains has become one of the most pressing problems of modern medicine. Therefore, the search of antibiotics against M. tuberculosis with novel mechanisms of action is very important. We have identified inhibitors of M. tuberculosis leucyl-tRNA synthetase (LeuRS) among the derivatives of 5-phenylamino-2H-[1,2,4]triazin-3-one. The most active compounds 5-(5-chloro-2-hydroxy-phenylamino)-6-methyl-2H-[1,2,4]triazin-3-one and 5-(5-chloro-2-hydroxy-phenylamino)-2H-[1,2,4]triazin-3-one inhibit M. tuberculosis LeuRS with IC50 of 7.6 μМ and 7.2 μМ, respectively. It was established that the inhibitory activity of compounds against pathogenic LeuRS is 10-fold better, than for human enzyme.
Introduction
Mycobacterium tuberculosis infection remains a major cause of global mortality and morbidity. Despite the availability of effective treatment, tuberculosis drugs are getting less effective and associated with side effects limiting their use, especially in multidrug resistant (MDR) and extensive-drug-resistant (XDR) patients. Resistance to rifampicin and isoniazid is called MDR tuberculosis. Fluoroquinolones are key second-line antituberculosis drugs, usually used in the treatment of MDR tuberculosis. Therefore, fluoroquinolones resistance in M. tuberculosis increases chances of getting XDR tuberculosis. This situation is driving the search for development of novel antibiotics with different modes of action. Aminoacyl-tRNA synthetases (aaRSs) are promising antiinfective drug targets. These enzymes play pivotal role in protein synthesis and have wide evolutionary divergence in prokaryotes and eukaryotes which allow the development of selective aaRSs inhibitorsCitation1–6.
For instance, leucyl-tRNA synthetase (LeuRS) has been validated as an attractive therapeutic target. Earlier, it was shown that antifungal 5-fluoro-1,3-dihydro-1-hydroxy-2,1-benzoxaborole (AN2690 which has received FDA approval in July 2014 for the topical treatment of onychomycosis) inhibits yeast cytoplasmic LeuRSCitation7. The compound ZCL039, a benzoxaborole-based derivative of AN2690, was reported as potent antipneumococcal agent that inhibits Streptococcus pneumonia LeuRS activityCitation8. Ding et al. discovered effective Trypanosoma brucei LeuRS inhibitorsCitation9. Recently, we have found inhibitors of Mycobacterium tuberculosis LeuRS among the derivatives of N-Benzylidene-N′-thiazol-2-yl-hydrazineCitation10. The main aim of this research is to identify novel small molecule inhibitors of M. tuberculosis LeuRS.
Methods
Receptor preparation for flexible docking
Homology model of Mycobacterium tuberculosis LeuRS has been built using SWISS-MODEL workspaceCitation11. The crystal structure of Thermus thermophilus LeuRS (PDB ID: 2V0C)Citation7 was used as a template for homology modeling.
A receptor molecule has been minimized in water with GROMACS molecular dynamics simulation package (GROMACS force field, steepest descent algorithm, 1000 steps, em_tolerance = 100, em_step = 0.001). The partial atom charges of the receptor molecule were taken from the Amber force field. Active site spheres were calculated with DOCK sphgen software. The spheres with the positions outside of the active site of LeuRS were deleted manually. Connolly MS and Grid programs from DOCK package were used to generate receptor Connolly surface and energy grids. Surface and grid calculations were performed with parameter settings detailed in the referenceCitation12. The grid spacing was set to 0.3.
Ligand database processing
Calculations of ligand geometry were performed using YFF force field described in the referenceCitation13. Partial atomic charges of the ligands were calculated using Kirchhoff methodCitation14.
Flexible docking
DOCK program has been used for receptor-ligand flexible docking. DOCK input parameters have been set as described previouslyCitation12. In our virtual screening experiments “multiple anchors” parameter was set as following: the minimum of heavy atoms in the anchor was set to 6, the maximum number of orientations was set to 1000, and the “all atoms” model has been chosen. After docking calculations, we have performed complex procedure to select compounds for in vitro testing. At first, we have carried out filtration of compounds based on scores (less than −40 kcal/mol) and ability to form hydrogen bonds with amino acids in active site of LeuRS, such as Phe97, Tyr99, Glu103, His109, Tyr113, Asp137, Ser631, Gly678, Glu680, His681, Gln714, Ile717, Lys759, and Ile760. The evaluation of compounds ability to form hydrogen bonds was conducted using our in-house program y_hbonds, which based on analysis of the angles and distances between the corresponding donor and acceptor atoms. Taking into account these criteria, we have selected 6000 compounds for further visual inspection of ligand–enzyme complexes. Visual analysis is an important step which allows correct calculation errors concerning position of the ligand in the active site and physicochemical intermolecular contacts. During visual inspection, we have analyzed complementarity of ligand surface to receptor surface, correctness of torsion angles, stacking interactions, etc. Therefore, for biological investigations, we have selected 270 compounds. The structures were visualized using the Discovery Studio Visualizer 4.0Citation15.
Purification of Mycobacterium tuberculosis LeuRS
The plasmid pET28a encoding the gene for M. tuberculosis LeuRS (the plasmid was the kind gift of Stephen Cusack and Andres Palencia) was used to express the protein in E. coli strain Rosetta (DE3). A single colony was used to inoculate LB medium containing 50 mg/L kanamycin and 36 mg/L chloramphenicol, which was then incubated overnight at 37 °C. A culture was induced to a final concentration of 1 mM IPTG and was incubated for 7 h at 23 °C. The cells were harvested by centrifugation.
The cells pellet was resuspended in 20–25 ml of buffer containing 20 mM Tris-HCl (pH 8.0), 100 mM NaCl, 5 mM MgCl2, 1 mM β-mercaptoethanol, and 1 tablet of Complete EDTA-free protease cocktail inhibitors and was sonicated for 10 min (in total) with 30 s of active cycle and 60 s of cool down. The lysate was cleared by centrifugation for 30 min at 16 000 rpm. The imidazole was added to supernatant to reach 15 mM in final and was mixed with 5 ml of Ni-NTA in binding buffer A (20 mM Tris-HCl (pH 8.0), 300 mM NaCl, 5 mM MgCl2, 15 mM imidazole, 1 mM β-mercaptoethanol, 0.1 mM PMSF). The mixture was stirred for 1 h at 4 °C and was applied into the empty column. The column was washed by 15 ml of buffer B (20 mM Tris-HCl (pH 8.0), 0.3 M NaCl, 5 mM MgCl2, 15 mM imidazole, 1 mM β-mercaptoethanol, 0.1 mM PMSF). Elution of protein was carried by buffer E (20 mM Tris-HCl (pH 8.0), 0.3 M NaCl, 5 mM MgCl2, 0.2 M imidazole, 5 mM β-mercaptoethanol, 0.1 mM PMSF). The homogeneity of the protein was tested using 12.5% SDS-PAGE. The fractions were pooled and dialyzed for 2 h with 20 mM Tris-HCl (pH 8.0), 100 mM NaCl, 5 mM MgCl2, and 5 mM mercaptoethanol, and kept overnight with 20 mM Tris-HCl (pH 7.4), 100 mM NaCl, 5 mM MgCl2, and 5 mM mercaptoethanol. The purified protein was concentrated using a Centricon-30 (Amicon, Miami, FL).
Purification of human cytoplasmic LeuRS
The plasmid pET16 encoding human cytoplasmic LeuRS, C-terminal histidine tag (the plasmid was the kind gift of Dr. Michael Alley), was transformed in E. coli strain Rosetta DE3. A single colony was inoculated into 10 ml of 2xYT medium containing 69 mg/L chloramphenicol, 100 mg/L ampicillin, and incubated at 37 °C overnight. About 10 ml of overnight culture was transferred to 1 l of 2xYT medium and grown at 37 °C until OD600 = 0.6. A culture was then induced with 0.4 mM IPTG and further grown at 20 °C for 5 hrs. The cells were pelleted by centrifugation at 5000 rpm for 15 min at 4 °C. The cells pellet was lysed with 20 ml buffer A (20 mM Na-p (pH 7.9), containing 300 mM NaCl, 5 mM imidazole, 5% glycerol, 1 mM β-mercaptoethanol, and 1 tablet of Complete EDTA-free protease cocktail inhibitors). The suspended cells were sonicated at 40% power for 3 min using an ultrasonic processor XL-Sonicator and centrifuged at 14 000 rpm for 30 min at 4 °C. The supernatant was combined with 5 ml of Ni-NTA agarose (pre-equilibrated with Buffer A). The gel and lysate were mixed for 1 h at 4 °C, and loaded on column. The column was then washed with 5 column volumes of Buffer A and eluted with 250 mM of imidazole in Buffer A. The eluted fractions containing desired protein were pooled and dialyzed against Buffer B (100 mM K-p (pH 6.8), 10 mM mercaptoethanol, and 20% glycerol). Protein was concentrated using a Centricone-30 (Amicon).
Aminoacylation assay
The standard aminoacylation assays were performed in 100 mM Tris-HCl (pH 8.0), 10 mM MgCl2, 2 mM DTT, 5 mM ATP, 10 mM KCl, 90 μM [14C]-L-leucine (238 mCi/mmol), 4 mg/ml E. coli tRNA, and 25 nM M. tuberculosis LeuRS. The reaction was incubated at 37 °C; aliquots were quenched by 10% trichloroacetic acid; and the level of aminoacylation of tRNA was determined by scintillation counting.
For the inhibitory studies in aminoacylation reaction, 20 μl solution containing 25 nM M. tuberculosis LeuRS, 100 mM Tris-HCl (pH 8.0), 10 mM MgCl2, 90 μM [14C]-L-leucine (238 mCi/mmol), 2 mM DTT, 4 mg/ml E. coli tRNA, and appropriate concentrations of inhibitor (dissolved in DMSO) was incubated for 5 min at 37 °C. Reactions were initiated by the addition of ATP to final concentration of 2 mM. At least triplicates were averaged to generate an IC50 value using Origin 7.0.
Results and discussion
The new LeuRS inhibitors were identified using receptor-based virtual screening of the commercially available compound library containing more than 100 000 diverse small organic compoundsCitation16. Among the best-scored compounds, one derivative of phenylamino-triazine was selected by the docking engine. In vitro tests showed that the compound 5-(2-Hydroxy-5-methyl-phenylamino)-6-methyl-2H-[1,2,4]triazin-3-one decreases aminoacylation activity of M. tuberculosis LeuRS. The residual activity of M. tuberculosis LeuRS in the presence of compound at 100 μM was 46%.
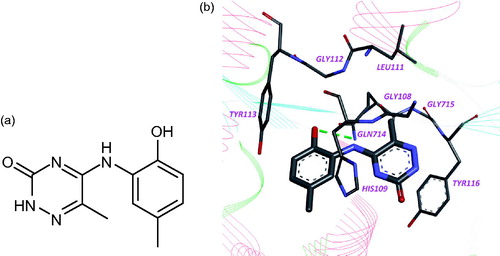
Accordingly to computer simulation, this compound interacts with amino acid residues Gly108, His109, Leu111, Gly112, Tyr113, Gln714, Gly715, Tyr716, and Ile717 in leucyl-binding region of LeuRS active site and forms hydrogen bond with amino group of Gln714 ().
Figure 1. Chemical structure (a) and binding mode of compound 1 in the active site of LeuRS M. tuberculosis (b). Hydrogen bond is indicated by green dotted line.
In order to find more effective inhibitors of LeuRS M. tuberculosis, we have selected 63 analogs of compound 1 from the library for evaluation of their inhibitory activity toward LeuRS M. tuberculosis. The chemical structure of compounds and residual activity of M. tuberculosis LeuRS in the presence of compound at 100 μM are presented in .
Table 1. Chemical structure of compounds and residual activity of M. tuberculosis LeuRS in the presence of compound at 100 μM.
As a result, it was found two compounds – 5-(5-Chloro-2-hydroxy-phenylamino)-6-methyl-2H-[1,2,4]triazin-3-one (compound 2) and 5-(5-Chloro-2-hydroxy-phenylamino)-2H-[1,2,4]triazin-3-one (compound 3) inhibiting M. tuberculosis LeuRS with IC50 values equal to 7.6 and 7.2 μМ, respectively. At the same time, these compounds demonstrate significantly less inhibition efficiency against human cytoplasmic LeuRS (IC50 = 68.9 and 76.5 μМ) ().
Table 2. Compounds inhibitory activity toward M. tuberculosis LeuRS and human cytoplasmic LeuRS.
The inhibitory activity of compounds against pathogenic LeuRS is 10-fold better, than for human enzyme. Therefore, the inhibitors from this chemical class seem to be selective toward M. tuberculosis LeuRS.
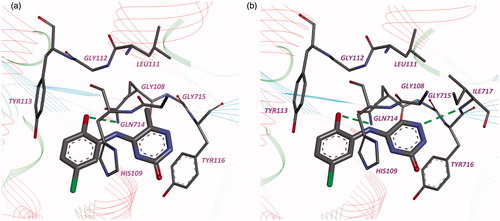
Accordingly to computer simulation, the compounds 2 and 3 have similar binding modes with compound 1 in leucyl-binding region of LeuRS active site ().
Figure 2. Binding modes of compounds 2 (a) and 3 (b) in the active site of LeuRS M. tuberculosis. Hydrogen bonds are shown by the green dotted lines.
The inhibitory activity of compounds against pathogenic LeuRS is 10-fold better, than for eukaryotic enzyme. Therefore, the inhibitors from this chemical class seem to be selective toward M. tuberculosis LeuRS and can be interesting compounds for further biological research.
Declaration of interest
There is no declaration of interest for this work.
This work was supported by the Science and Technology Center in Ukraine (contract No. 5218), the National Agency of Science, Innovation and Informatization of Ministry of Education and Science of Ukraine (contract No. DP/337-2013).
Acknowledgements
We thank Dr. Stephen Cusack and Dr. Andres Palencia (EMBL Grenoble Outstation, France) for the gift of clone for expression and purification of M. tuberculosis LeuRS. We thank Dr. Michael Alley (Anacor Pharmaceuticals, USA) for the gift of plasmid for expression and purification of human cytoplasmic LeuRS.
References
- Hurdle JG, O'Neill AJ, Chopra I. Prospects for aminoacyl-tRNA synthetase inhibitors as new antimicrobial agents. Antimicrob Agents Chemother 2005;49:4821–33
- Vondenhoff GH, Van Aerschot A. Aminoacyl-tRNA synthetase inhibitors as potential antibiotics. Eur J Med Chem 2011;46:5227–36
- Dewan V, Reader J, Forsyth KM. Role of aminoacyl-tRNA synthetases in infectious diseases and targets for therapeutic development. Top Curr Chem 2014;344:293–329
- Datt M, Sharma A. Novel and unique domains in aminoacyl-tRNA synthetases from human fungal pathogens Aspergillus niger, Candida albicans and Cryptococcus neoformans. BMC Genomics 2014;15:1069
- Fang ZP, Wang M, Ruan ZR, et al. Coexistence of bacterial leucyl-tRNA synthetases with archaeal tRNA binding domains that distinguish tRNA(Leu) in the archaeal mode. Nucleic Acids Res 2014;42:5109–24
- Pham JS, Dawson KL, Jackson KE, et al. Aminoacyl-tRNA synthetases as drug targets in eukaryotic parasites. Int J Parasitol Drugs Drug Resist 2013;4:1–13
- Rock FL, Mao W, Yaremchuk A, et al. An antifungal agent inhibits an aminoacyl-tRNA synthetase by trapping tRNA in the editing site. Science 2007;316:1759–61
- Hu QH, Liu RJ, Fang ZP, et al. Discovery of a potent benzoxaborole-based anti-pneumococcal agent targeting leucyl-tRNA synthetase. Sci Rep 2013;3:2475:1–8
- Ding D, Meng Q, Gao G, et al. Design, synthesis, and structure-activity relationship of Trypanosoma brucei leucyl-tRNA synthetase inhibitors as antitrypanosomal agents. J Med Chem 2011;54:1276–87
- Gudzera OI, Golub AG, Bdzhola VG, et al. Discovery of potent anti-tuberculosis agents targeting leucyl-tRNA synthetase. Bioorg Med Chem 2016;24:1023–31
- Homology model of LeuRS Mycobacterium tuberculosis was built using SWISS-MODEL workspace. Available from: http://swissmodel.expasy.org/workspace/index.php?func=modelling_simple1 [last accessed 25 May 2016]
- Bursulaya BD, Totrov M, Abagyan R, Brooks CL III. Comparative study of several algorithms for flexible ligand docking. J Comput Aided Mol Des 2003;17:755–63
- Yakovenko OY, Oliferenko AA, Golub AG, et al. The new method of distribution integrals evaluations for high throughput virtual screening. Ukr Bioorg Acta 2007;1:52–62
- Yakovenko OY, Oliferenko AA, Bdzhola VG, et al. Kirchhoff atomic charges fitted to multipole moments: implementation for a virtual screening system. J Comput Chem 2008;29:1332–43
- The structures were visualized using Discovery Studio Visualizer 4.0. Available from: http://accelrys.com/products/discovery-studio/visualization/discovery-studio-visualizer-download-instructions-40.html [last accessed 24 May 2016]
- We used a database of commercially available compounds. Otava Ltd. Available from: https://otavachemicals.com/products/compound-libraries-for-hts/screening-collection-for-prompt-delivery [last accessed 24 May 2016]
