Abstract
A new compound, 9-dihydroxyl-2'-O-(Z)-cinnamoyl-7-methoxy-aloesin (1), and eight known compounds (2–9) were isolated from Aloe vera. Their structures were elucidated using 1D/2D nuclear magnetic resonance and mass spectra. Compound 9 exhibited reversible competitive inhibitory activity against the enzyme tyrosinase, with an IC50 value of 9.8 ± 0.9 µM. A molecular simulation revealed that compound 9 interacts via hydrogen bonding with residues His244, Thr261, and Val283 of tyrosinase. Additionally, compounds 3 and 7 were shown by half-leaf assays to exhibit inhibitory activity towards Pepper mild mottle virus.
Introduction
Aloe (Xanthorrhoeaceae), which consists of approximately 400 species, is a perennial plant that has been used as a traditional medicine for about 3000 yearsCitation1. The most well-known species is Aloe vera L., a stemless plant that grows to 100 cm. Its green leaves contain a clear, colorless and unsavory material; this gel is used in cosmetics, health drinks and beverages, and medicinesCitation1. These A. vera-containing substances consist of 99.5% water and 0.5% active ingredients, including vitamins, polysaccharides, phenolic compounds and organic acidsCitation2. This plant gel has many biological activities, including anti-inflammatoryCitation3,Citation4, anti-viralCitation4, anti-bacterialCitation4, anti-cancerCitation3,Citation4, anti-diabeticCitation4 and anti-allergy activitiesCitation4; it also provides protection against radiationCitation4. The main phytochemicals in this gel are derivatives of anthraquinone and C-glucosylanthrone: aloin A and B, emodin, desoxyaloin, aloinoside B and C, and elgonica dimer ACitation1,Citation5. Aloesin, aloe emodin and aloin A have been reported to have antioxidant activitiesCitation6, while aloinoside B and C are known to inhibit the activity of soluble epoxide hydrolase and phosphodiesterase-4DCitation5,Citation7.
Widely distributed in plants, insects and mammals, tyrosinase (EC 1.14.18.1) is a multifunctional oxidoreductase with two copper ions. It is a key enzyme in melanogenesis, in which monophenol is converted to o-diphenol and then converted to o-quinoneCitation8,Citation9. Melanin is important in protection from ultraviolet (UV) radiation from the sun in humans. Excessive production of melanin, however, causes several side effects, including age spots, ephelide, senile lentigines, melasma and frecklesCitation8–10. Moreover, this enzyme, which is found in insect cuticles, is responsible for the sclerotization of the insect exoskeletonCitation11. Accordingly, tyrosinase has therefore been regarded as the primary target for the treatment of skin pigmentation, the browning of fruits and vegetables, and the development of insecticides in agricultureCitation8–11.
The purpose of this study was to isolate and characterize the potential tyrosinase inhibitors from A. vera. One new compound 1, and eight known compounds 2–9, were purified from A. vera by silica gel and C-18 column chromatography and were assessed for their inhibitory activity against tyrosinase. Additionally, they were tested the inhibitory activity on PMMoV that is a major pepper-infecting virus that causes significant reductions in pepper yield throughout the worldCitation4.
Materials and methods
General experimental procedures
Column chromatography was performed using silica gel (Kieselgel 60, 70–230, and 230–400 mesh, Merck, Darmstadt, Germany) and YMC RP-18 resins (30–50 μm, Fuji Silysia Chemical Ltd., Kasugai, Aichi, Japan). Thin layer chromatography (TLC) was performed using pre-coated silica-gel 60 F254 and RP-18 F254S plates (both 0.25 mm, Merck, Darmstadt, Germany). Compounds were visualized by spraying with 10% aqueous H2SO4 solution and by heating for 2–3 min. NMR spectra were recorded using a JEOL ECA 600 and 400 spectrometeres (Tokyo, Japan), using DMSO-d6 and methanol-d4 as solvents. Mass spectra were measured by Bruker Daltonics MicroQ-TOF III mass spectrometer (Bruker Daltonics, 255748 Germany). Tyrosinase (T3824) and L-tyrosine (T3754) were purchased from Sigma-Aldrich (St. Louis, MO).
Plant material
Aloe vera was purchased from company of plant resources in Jeju island, Korea (Prndle Inc.), in May 2015. This specie was identified by Dr. J. H. Kim. A voucher specimen (A1) was deposited at the herbarium, Department of Horticultural and Crop Environment, National Institute of Horticultural and Herbal Science.
Extraction and isolation
The powder product of A. vera (5 kg) was extracted with 95% MeOH (36 L) three times at room temperature during 7 days. The MeOH extract (247 g) condensed under reduced pressure was dissolved in 2.0 L distilled water. This was extracted three times with n-hexane, chloroform (CHCl3), ethyl acetate (EtOAc), and butanol (BuOH) to yield n-hexane (41 g), CHCl3 (10 g), EtOAc (25 g), and BuOH extracts (80 g), respectively. CHCl3 extract was chromatographed on silica gel column chromatography using gradient elution with CHCl3/MeOH (40:1 → 10:1) to yield seven fractions (C(0).1-C.7). C-4 was separated using C-18 column chromatography with gradient system of MeOH/H2O (1:1 → 5:1) to give compound 2 (450 mg). C(0).6 was subjected to C-18 column chromatography with isocratic system 50% MeOH to obtain compound 6 (13 mg). EtOAc extract was loaded on the silica gel column chromatography and eluted with CHCl3/MeOH (20:1 → 5:1) to yield ten fractions (E.1–E.10). E.5 was subject to C-18 column chromatography with gradient system of MeOH/H2O (0.5:1 → 3:1) to achieve six fractions (E.51–E.56). Compounds 5 (5.1 mg) and 1 (7.8 mg) were isolated from E.53 on C-18 column chromatography with isocratic system of 60% MeOH. E.55 was chromatographed using a C-18 column chromatography and also eluted with isocratic system of 60% MeOH to obtain compounds 8 (21.2 mg) and 9 (31.4 mg). E.7 was separated using C-18 column chromatography with gradient system of MeOH/H2O (1:1 → 5:1) to give five fractions (E.71–E.75). E.73 was subjected to silica gel column chromatography with isocratic system of CHC3/MeOH (9:1) to obtain compounds 3 (42.9 mg) and 4 (11.5 mg). Compound 7 (35.2 mg) was purified with C-18 column chromatography using the isocratic system of 70% MeOH from E.74 fraction.
Compound 2
Yellow powder; m.p. 224–225 °C, ESI-Ms m/z = 271 [M + H]+, (calcd C16H11O5+, 271). 1H NMR (400 MHz, CDCl3-d1) δ 7.82 (1H, t, J = 7.8 Hz, H-6), 7.73 (1H, d, J = 7.8 Hz, H-5), 7.71 (1H, s, H-4), 7.4 (1H, J = 7.8 Hz, H-7), 7.31 (1H,s, H-2), 5.62 (1H, t, J = 5.0 Hz, 3-OH), 4.64 (2H, d, J = 5.0 Hz, H-15).
Compound 3
Yellow powder; m.p. 147–149 °C, ESI-Ms m/z = 419 [M + H]+, (calcd C21H23O9+, 419), [α]25D: + 4.5° (C 0.1, MeOH). 1H NMR (400 MHz, MeOH-d4) δ 7.49 (1H, t, J = 7.8 Hz, H6), 7.07 (1H, H5), 7.05 (1H, s, H4), 6.88 (1H, s, 8, H2), 6.82(1H, d, J = 8.2 Hz, H7), 4.66 (1H, s, H15b), 4.65 (1H, s, H15a), 4.61 (1H, d, J = 0.9 Hz, H10), 3.56 (1H, dd, J 11.4, 1.6 Hz, H-6b'), 3.41 (1H, dd, J = 9.7, 2.4 Hz, H-6a'), 3.37 (1H, m, H-1'), 3.25 (1H, t, J = 8.7 Hz, H-3'), 3.01 (1H, t, J = 8.7 Hz, H-2'), 2.91 (1H, br s, H-5'), 2.90 (1H,br s, H-4').
Compound 4
Yellow powder; m.p. 141–145 °C, ESI-Ms m/z = 419 [M + H]+, (calcd C21H23O9+, 419), [α]25D: - 85° (C 0.1, MeOH). 1H NMR (400 MHz, MeOH-d4) δ 7.48 (1H, t, J = 7.8 Hz, H6), 7.08 (1H, overlapped peak, H5), 7.06 (1H, s, H4), 6.87 (1H, s, 8, H2), 6.85 (1H, overlapped peak, H7), 4.64 (2H, s, H15), 4.61 (1H, s, H10), 3.55 (1H, J =11.7 Hz H-6b'), 3.42–3.24 (4H, m, H-6a', 1', 3', 2'), 2.90 (1H, br s, H-5'), 2.89 (1H,br s, H-4').
Compound 6
White amorphous substance; m.p. 170–172 °C. ESI-MS m/z = 345 [M + H]+, (calcd C15H17O7+, 345), [α]25D: -95° (C 0.1, MeOH). 1H-NMR(400 MHz, CD3OD) δ 6.29 (1H, d, J = 2.3 Hz, H-7'), 6.25 (1H, d, J = 2.3 Hz, H-5'), 6.19–6.18 (1H, m, H-7/-5), 4.73–4.69 (1H, m, H-3), 3.06 (1H, dd, J = 2.3 Hz, H-1'b), 2.95 (1H, dd, J = 2.3 Hz, H-1'a), 2.53 (3H, H-8'). 12C NMR (100 MHz, CD3OD) δ 206.9 (C-8'), 171.4 (C-1), 166.5 (C-6), 165.7 (C-8), 161.5 (C-4'), 160.3 (C-6'), 143.3 (C-4a), 139.5 (C-2'), 121.9 (C-3'), 111.6 (C-7'), 108.2 (C-5), 102.6 (C-5'), 102.3 (C-7), 101.6 (C-8a), 81.5 (C-3), 39.7 (C-1'), 33.6 (C-4, 8').
Compound 8
Yellowish amorphous substance; m.p. 141–143 °C. ESI-MS m/z = 541 [M + H]+, (calcd C29H33O10+, 541), [α]25D: -100° (C 0.1, MeOH). 1H-NMR (MHz, CD3OD) δ 7.36(3H, m, H-3", 5", 9"), 6.79 (3H, m, H-6, 6", 8"), 6.23 (1H, s, H-2), 6.07 (1H, d, J = 16.0 Hz, H-2"), 5.62 (1H, t, J = 9.7 Hz, H-2'), 5.14 (1H, d, J = 10.2 Hz, H-1'), 3.98–3.43 (5H, m, H-3'-6'), 3.92 (3H, s, -OMe), 2.74 (3H, s, 5-CH3), 2.34 (3H, s, CO-CH3). 13C-NMR (MHz, CD3OD) δ 204.6 (C-10), 182.2 (C-4), 168.1 (C-1"), 163.1 (C-7), 162.2 (C-7), 161.4 (C-7"), 159.7 (C-1a), 146.6 (C-3"), 144.7 (C-5), 131.2 (C-5",9"), 127.1 (C-4"), 116.9 (C-6",8"), 114.8 (C-4a), 113.6 (C-2"), 112.9 (C-2"), 112.1 (C-8a), 82.9 (C-5'), 77.9 (C-3'), 74.1 (C-2'), 72.6 (C-4'), 72.2 (C-1'), 63.4 (C-6'), 57.2 (C-OMe), 48.3 (CO-CH3), 23.7 (5-CH3).
Compound 9
Yellowish amorphous substance; m.p. 143–145 °C. ESI-MS m/z = 555 [M + H]+, (calcd C29H31O11+, 555.1) 1H-NMR (MHz, CD3OD) δ 7.36(3H, m, H-3", 5", 9"), 6.79 (3H, m, H-6, 6", 8"), 6.23 (1H, s, H-2), 6.07 (1H, d, J = 16.0 Hz, H-2"), 5.62 (1H, t, J = 9.7 Hz, H-2'), 5.14 (1H, d, J = 10.2 Hz, H-1'), 3.98–3.43 (5H, m, H-3'-6'), 3.92 (3H, s, -OMe), 2.74 (3H, s, 5-CH3), 2.34 (3H, s, CO-CH3). 13C-NMR (MHz, CD3OD) δ 204.6 (C-10), 182.2 (C-4), 168.1 (C-1"), 163.1 (C-7), 162.2 (C-7), 161.4 (C-7"), 159.7 (C-1a), 146.6 (C-3"), 144.7 (C-5), 131.2 (C-5",9"), 127.1 (C-4"), 116.9 (C-6",8"), 114.8 (C-4a), 113.6 (C-2"), 112.9 (C-2"), 112.1 (C-8a), 82.9 (C-5'), 77.9 (C-3'), 74.1 (C-2'), 72.6 (C-4'), 72.2 (C-1'), 63.4 (C-6'), 57.2 (C-OMe), 48.3 (CO-CH3), 23.7 (5-CH3).
Tyrosinase assay
Enzyme assay was performed as described previously with minor modificationsCitation12.
For the IC50 value 130 μL of tyrosinase (about 46 units/mL) solvated in 0.05 mM phosphate buffer (pH 6.8) and 20 μL of 1–0.032 mM concentrations of the inhibitors were mixed in 96 well plate, and then 50 μL of 1.5 mM L-tyrosine in buffer was added in the mixture. To test the enzyme kinetic study, 130 μL of tyrosinase and 20 μL inhibitor (6.2–25 µM) were also mixed, and then 50 μL 0.6–12 mM L-tyrosine was added in 96 well plate. The mixture was recorded at UV-Vis 475 nm for 20 min. The inhibitory ratio was calculated according to the following equation: Inhibitory activity rate (%) = 100-[(S20-S0/C20-C0] × 100where C20 and S20 were the intensity of control and inhibitor after 20 min, S0 and C0 is the intensity of control and inhibitor at zero min.
Molecular docking simulation
Molecular docking was performed as described previously using Autodock 4.2 program (La Jolla, CA)Citation13,Citation14. The 3D structure of the compound 9 was built and minimized energy by MM2 using the Chem3D Pro. The flexible bonds of the ligand were assigned with AutoDockTools. The protein structure (PDB ID:2Y9X) was obtained to RCSB, after which substrates and the chains (B-H) were removed by Chimera. All the hydrogen atoms and gasteiger charges in single chain (A) were added. Simulation studies were performed using the Autodock 4.2 version. Briefly, competitive inhibitor (9) was established the grid of number of points (X: 60, Y: 60, Z: 60) at 0.375 Å space in activity site by Autodock 4.2, and Molecular docking was performed by using the Lamarckian Genetic Algorithm (Runs 50 and the maximum number of evaluations was set as long). The result of 9 was prepared using Chimera (San Jose, CA) and LigPlot (Cambridge, UK).
Half leaf assay
Half-leaf assay on Nicotiana glutinosa (N. glutinosa) showing hypersensitive reaction (HR, called local lesions) to PMMoV was chosen to evaluate antiviral activity of compounds 1–9 from A. vera. They were pretreated on one half leaf of 6-leaf-stage N. glutinosa and a final extraction solvent without compounds was simultaneously pretreated on the other half leaf of N. glutinosa. Then, each half leaf pretreated with compounds or solvent was inoculated mechanically with 10 μg/ml PMMoV virions diluted in 10 mM potassium phosphate buffer (pH 7.0). N. glutinosa plants inoculated with PMMoV were kept in a wet chamber for 3 days after inoculation to count numbers of local lesions. The number of local lesions produced was calculated for a percentage inhibition. These experiments were independently repeated three times.
Results and discussion
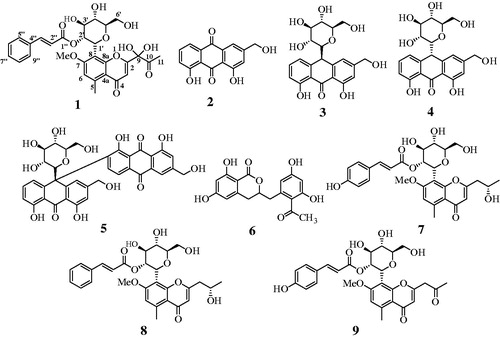
The A. vera plant was first extracted with 95% methanol, and the extract was then fractionated into n-hexane, CHCl3, EtOAc and BuOH fractions. The CHCl3 and EtOAc fractions were subjected to column chromatography over silica gel and C-18 to isolate one new compound 1, along with known compounds 2–9, which were identified as 9-dihydroxyl-2'-O-(Z)-cinnamoyl-7-methoxy-aloesin (1), aloe-emodin (2)Citation5, aloin A (3)Citation5, aloin B (4)Citation5, elgonica dimer A (5)Citation5, feralolide (6)Citation15, isoaloeresin D (7)Citation16, aloeresin E (8),Citation16 and, 7-O-methylaloeresin A (9)Citation17 (, Figure S8?15). The structures were identified by 1D/2D nuclear magnetic resonance (NMR), high-resolution electrospray ionization mass spectrometry (HRESIMS), and by comparison with previous reports.
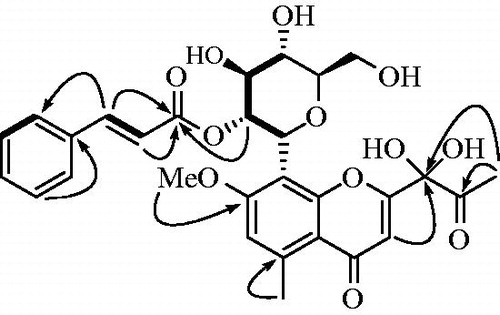
Compound 1 was isolated as a dark-brown powder and determined to have the molecular formula C29H30O12 according to its HRESIMS spectrum with a quasimolecular ion peak at m/z 571.1810 [M + H]+ (calculated for C29H31O12 571.1810). The UV spectrum displayed absorptions at 190, 216, 253, and 282 nm (Figure S1A and B). The 1H NMR data indicated the presence of seven aromatic [δH 7.50 (2H, m), 7.36 (m, 3H), 6.84 (s)], two olefinic [δH 7.43 (d, J = 15.8 Hz), 6.25 (d, J = 15.8 Hz)], two methyl [δH 2.75 (s, 3H), 2.35 (s, 3H)], seven sugar [δH 5.12 (d, J = 9.2 Hz), 5.59 (t, J = 9.2 Hz) 4.01 (m), 3.45 (m), 3.67 (m), 3.91(m), 3.71 (m)], and a methoxy [δH 3.88 (s, 3H)] signal. The 13C NMR and DEPT-135 spectra indicated the presence of 29 carbon signals, containing 3 ketone (δC 203.7, 182.0, 167.4), 12 aromatic (δC 164.0, 162.3, 159.5, 144.9, 135.7, 131.6, 130.1, 129.3, 117.6, 113.1, 112.4, 111.6), 2 olefinic (δC 146.3, 118.5), 2 methyl (δC 25.0, 23.7), 6 sugar (δC 83.3, 78.0, 74.3, 72.6, 71.9, 63.5), and 1 methoxy (δC 57.2) proton (, Figures S2?4). The COSY correlation of δH 5.12 (d, J = 9.2 Hz)/5.59 (t, J = 9.2 Hz), 5.59 (t, J = 9.2 Hz)/4.01 (m), 4.01 (m)/3.45 (m), and 3.45 (m)/3.67 (m) also identified the coupling interaction of sugar protons. In the heteronuclear multiple-bond correlation (HMBC) spectra, the dioxygenated carbons (C-9) and ketone carbon (C-10) were established for the side chain of C-2 (δc 164.0) based on the correlation of H-3 (δH 6.65)/C-9 (δc 98.6) and H-11 (δH 2.35)/[C-9 (δc 98.6), C-10 (δc 203.7)]. The methoxy group (δH 3.88) was confirmed by correlation with C-7 (δH 162.3), and the correlation of the 2' sugar proton (δH 5.59) with C-1" (δc 167.4) confirmed that the ketone of the cinnamoyl group was connected to the oxygen atom in the 2' position of the sugar group (Figures S5?7). Additionally, these 1H-/13C-NMR results were compared with those of compounds 7–9. As shown in , the structure of compound 1 was elucidated and named 9-dihydroxyl-2'-O-(Z)-cinnamoyl-7-methoxy-aloesin.
Table 1. 1H and 13C-NMR data for compound 1.
To evaluate the inhibitory activity of the isolated compounds 1–9 toward tyrosinase in vitro, the amount of substrate hydrolyzed by the enzyme was detected in the presence or absence of inhibitors using a UV-Vis spectrophotometer. Compounds 1–9 were tested at 100 µM, and showed inhibitory effects ranging from 1.2 ± 2.5% to 95.2 ± 0.5% of the control value (). Among them, compound 9, which showed the greatest inhibitory effect on tyrosinase, was determined to have an IC50 value of 9.8 ± 0.9 µM, while 50% of the inhibitory effect of kojic acid, used as a positive control, was found to be 19.5 ± 1.5 µM ().
Figure 3. (A) Inhibitory activity of compound 9 on tyrosinase (IC50: 9.8 ± 0.9 μM; kojic acid =19.5 ± 1.5 μM). (B) Relationship of the hydrolytic activity of tyrosinase with enzyme concentration at a variety of inhibitor concentration. (C) Lineweaver–Burk plot (Competitive type) and (D) Dixon plot (Ki: 5.8 ± 0.9 μM) for the inhibition of compound 9.
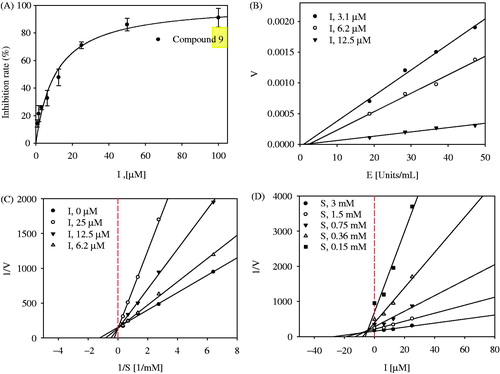
Table 2. Inhibition rate of isolated compounds 1–9 on tyrosinase and PMMoV.
The graph of enzyme activity versus enzyme concentration (18.8–47.3 U/mL) in the presence of different concentrations of compound 9 gave a series of straight lines with varying slopes with the similar point on the abscissa (). Lineweaver–Burk plots indicated that the plot of reverse velocity versus the concentration of substrate (ranging from 0.15 to 3.00 mM) in the presence of different concentrations of compound 9 (6.2 µM, 12.5 µM, and 25 µM) crossed the same point on the y-axis (). Moreover, all of the straight lines in a Dixon plot passed through one point in the negative x-axis and positive y-axis plane (). Overall, the enzyme kinetic studies revealed that compound 9 is a reversible-competitive inhibitor of tyrosinase with a Ki value of 5.8 ± 0.9 µM.
We also simulated molecular docking with the Autodock 4.2 program. The enzyme kinetics revealed that compound 9 inhibits the catalytic reaction by directly interacting with the active site of tyrosinase. Based on this, molecular docking was set up with a grid containing the active site for the molecular simulation. The lowest Autodock score, which was obtained with 50 docking runs, allowed us to determine the best position of the enzyme-ligand complex (Autodock score: −7.43 kcal/mol). As shown in , the chromone group of compound 9 is directed towards the two copper ions in the pocket of the active site of tyrosinase. At the same time, the p-coumaroyl group of compound 9 is situated on the left side of the active site. Compound 9 closely associates with 17 amino acids of tyrosinase through hydrogen bonding at distances of 3.09, 2.99, and 3.08 Å with His244, Thr261, and Val283, respectively (, ).
Figure 4. Docking pose of 9 at the lowest energy with enzyme indicated as ribbon (A). The green dotted line represents hydrogen-bond interactions between compound 9 and enzyme (B).
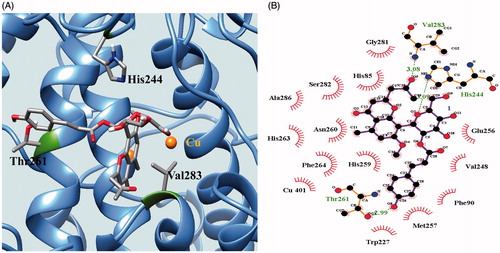
Table 3. Interaction and Autodock score on tyrosinase of 9.
To characterize the inhibitory activity of compounds 1–9 isolated from the CHCl3 and EtOAc extracts of A. vera toward PMMoV, an anti-viral assay was performed in vitro using the half-leaf method. Ribavirin (34.5 ± 3.5% at 1.5 mg/mL) and skim milk (10.5 ± 7.8% at 1.5 mg/mL) were used as positive controls. As shown in , compounds 1–9 exhibited inhibitory effects ranging from 7.5 ± 7.1% to 45.2 ± 4.1% of the control value at 1.5 mg/mL (). Compounds 3 and 7 exhibited the greatest inhibition (45.2 ± 4.1% and 37.5 ± 6.5% compared to the positive controls).
Conclusion
A previous study reported that methanol extracts of A. vera L. gel had IC50 values ranging from 6 to 10 mg/mL against mushroom tyrosinaseCitation18. We performed this study to isolate the constituents from this plant and assess their inhibitory activity toward tyrosinase. One new compound, 1, and eight known compounds, 2–9, were isolated from A. vera. Compound 1, which was identified using the spectroscopy data, was named 9-dihydroxyl-2'-O-(Z)-cinnamoyl-7-methoxy-aloesin. Among them, compound 9 was found to exhibit inhibitory activity toward tyrosinase with an IC50 value of 9.8 ± 0.9 µM, and behaved as a reversible-competitive inhibitor (Ki: 5.8 ± 0.9 µM). Moreover, a docking simulation suggested that this compound was best anchored as an L-shaped structure around the entrance and outside the active site of tyrosinase. Furthermore, the inhibitor (compound 9) interacted with three amino acids (His244, Thr261 and Val283) on the outside of tyrosinase via hydrogen bonding instead of interacting with residues in the active site. Moreover, compounds 3 and 7 were shown to suppress infection by PMMoV in plants using a half-leaf assay. Finally, this study confirms components of A. vera as potential tyrosinase and PMMoV inhibitors.
IENZ_1235568_Supplementary_Material.pdf
Download PDF (471.2 KB)Disclosure statement
This study was supported partly by a basic research project (PJ010878) of National Institute of Horticultural and Herbal Science, RDA.
References
- Mandrioli R, Mercolini L, Ferranti A, et al. Determination of aloe emodin in Aloe vera extracts and commercial formulations by HPLC with tandem UV absorption and fluorescence detection. Food Chem 2011;126:387–93.
- Benítez S, Achaerandio I, Pujolà M, Sepulcre F. Aloe vera as an alternative to traditional edible coatings used in fresh-cut fruits: A case of study with kiwifruit slices. LWT-Food Sci Technol 2015;61:184–93.
- Zhang X-F, Wang H-M, Song Y-I, et al. Isolation, structure elucidation, antioxidative and immunomodulatory properties of two novel dihydrocoumarins from Aloe vera. Bioorg Med Chem Lett 2006;16:949–53.
- Ray A, Ghosh S, Ray A, Aswatha SM. An analysis of the influence of growth periods on potential functional and biochemical properties and thermal analysis of freeze-dried Aloe vera L. gel. Ind Crop Prod 2015;76:298–305.
- Sun YN, Kim JH, Li W, et al. Soluble epoxide hydrolase inhibitory activity of anthraquinone components from Aloe. Bioorg Med Chem 2015;23:6659–65.
- Lucini L, Pellizzoni M, Pellegrino R, et al. Phytochemical constituents and in vitro radical scavenging activity of different Aloe species. Food Chem 2015;170:501–7.
- Zhong J-S, Huang Y-Y, Zhang T-H, et al. Natural phosphodiesterase-4 inhibitors from the leaf skin of Aloe barbadensis Miller. Fitoterapia 2015;100:68–74.
- Hu Y-H, Liu X, Jia Y-L, et al. Inhibitory kinetics of chlorocinnamic acids on mushroom tyrosinase. J Biosci Bioeng 2014;177:142–6.
- Si Y-X, Ji SY, Wang W, et al. Effects of boldine on tyrosinase: inhibition kinetics and computational simulation. Process Biochem 2013;48:152–61.
- Ha YM, Park YJ, Lee JY, et al. Design, synthesis and biological evaluation of 2-(substituted phenyl)thiazolidine-4-carboxylic acid derivatives as novel tyrosinase inhibitors. Biochimie 2012;94:533–40.
- Chen X-X, Zhang J, Chai W-M, et al. Reversible and competitive inhibitory kinetics of amoxicillin on mushroom tyrosinase. Int J Biol Macromol 2013;62:726–33.
- Wang W, Curtis-Long MJ, Lee BW, et al. Inhibition of tyrosinase activity by polyphenol compounds from Flemingia philippinensis roots. Bioorg Med Chem 2014;24:1115–20.
- Lee GY, Kim JH, Choi S-K, Kim YH. Constituents of the seeds of Cassia tora with inhibitory activity on soluble expoxide hydrolease. Bioorg Med Chem Lett 2015;25:5097–101.
- Kim JH, Ryu YB, Lee WS, Kim YH. Neuraminidase inhibitory activities of quaternary isoquinoline alkaloids from Corydalis turtschaninovii rhizome. Bioorg Med Chem 2014;22:6047–52.
- Speranza G, Manitto P, Cassara P, Monti D. Feralolide, a dihydroisocoumarin from cape Aloe. Phytochemistry 1993;22:175–8.
- Okamura N, Hine N, Harada S, et al. Three chromone components from Aloe vera leaves. Phytochemistry 1996;43:495–8.
- Schmidt J, Blitzke T. Structural investigation of 5-methylchromone glycosides from Aloe species by liquid chromatography/electrospray tandem mass spectrometry. Eur J Mass Spectrom 2001;7:481–90.
- Gupta SD, Masakapalli SK. Mushroom tyrosinase inhibition activity of Aloe vera L. gel from different germplasms. Chin J Nat Med 2013;11:616–20.