Abstract
Small molecule inhibitors have a powerful blocking action on viral polymerases. The bioavailability of the inhibitor, nevertheless, often raise a significant selectivity constraint and may substantially limit the efficacy of therapy. Phosphonoacetic acid has long been known to possess a restricted potential to block DNA biosynthesis. In order to achieve a better affinity, this compound has been linked with natural nucleotide at different positions. The structural context of the resulted conjugates has been found to be crucial for the acquisition by DNA polymerases. We show that nucleobase-conjugated phosphonoacetic acid is being accepted, but this alters the processivity of DNA polymerases. The data presented here not only provide a mechanistic rationale for a switch in the mode of DNA synthesis, but also highlight the nucleobase-targeted nucleotide functionalization as a route for enhancing the specificity of small molecule inhibitors.
Graphical Abstract
Introduction
Small molecule compounds, represented by analogues of pyrophosphate, a product of the DNA polymerisation reaction – phosphonoformic (PFA, foscarnet) and phosphonoacetic (PAA) acids – comprise one of the three types of principal inhibitors of viral polymerasesCitation1. PFA is the most effective small molecule inhibitor of HIV reverse transcriptase identified so far. Sensitivity of various viral polymerases towards PFA is 100- to 1000-fold higher compared to that of mammalian replicative polymerases. However, the latter compound is cytotoxic and is used for the treatment of HIV and HIV-associated viral infections in critical cases onlyCitation2. Despite the similar structure, PAA is far less effective against viral polymerases but does not exhibit observable cytotoxicityCitation2.
A relaxed specificity of an innate polymerase is vital to the survival of the virusCitation3. The ability to control virus proliferation by using compounds that are specific to a replication complex is a tempting and intensively explored approach. A combination of several compounds, reciprocally inhibiting the polymerase, constitutes the essence of the highly active antiretroviral therapy (HAART) – a revolutionary approach ensuring the control over HIVCitation4. This treatment relies on the established allosteric and competitive inhibition modes and resulted inhibition of the polymerase leads to an effective restrain of HIV in vivoCitation5,Citation6. Nucleoside- and nucleotide-based inhibitors of the reverse transcriptase (NRTIs) act either as terminators of viral DNA synthesis or inherent mutagens. Their application often suffers from an insufficient rejection by cellular replicative polymerases, including DNA polymerase alpha, beta and gammaCitation7, thus leading to the high cytotoxicity. Such drawback is addressed by combination of different inhibitors, including non-nucleoside reverse transcriptase inhibitors (NNRTIs) and NRTI, into one moleculeCitation8–10. However, further improvement of polymerase inhibition is restricted by a limited set of compounds and the target enzyme repertoire. In consequence, such compounds have not been translated into any therapeutic agents yet.
Natural nucleoside triphosphates had long been considered as being the only acceptable substrates for polymerases. A paradigm shift occurred when nucleoside monophosphates conjugated to amino acids via 5′-alpha-phosphate had been synthesised and investigated. Negatively charged 5′ structures were demonstrated to be permissive for DNA biosynthesis by HIV-1 reverse transcriptaseCitation11–13 and other polymerasesCitation14,Citation15. Such compounds were nevertheless largely discriminated by cellular replicative polymerases, requiring the integrity of the 5′-triphosphate moiety.
Altogether, the currently available data on NRTI allow identification of several positions in the nucleotides that are permissive for modification without a noticeable impact on the specificity towards the viral DNA polymerases. While the C5 position of pyrimidine nucleobase has long been used for the introduction of various groups for DNA labellingCitation16, 5′-monophosphate as a platform for modification by negatively charged compounds has been recognised only recentlyCitation11.
Here, we focussed on novel analogues of nucleoside triphosphates designed so as to be catalytically acceptable by the target polymerases for incorporation into DNA. Three different PAA-conjugated compounds were prepared, and they varied significantly in inhibition capacity towards different DNA polymerases, including reverse transcriptases. Nucleobase-conjugated phosphonoacetic acid restricted the processivity of DNA polymerases. Ternary complexes of DNA polymerase with DNA were found to adopt discernible conformations in the presence of different nucleotides, and provided the mechanistic rationale for a switch in the mode of DNA synthesis.
Materials and methods
Chemistry
All commercial chemicals and solvents were purchased from Sigma-Aldrich or Merck and used without further purification. Thin-layer chromatography (TLC) was carried out on aluminium sheets coated with silica gel 60 F254 (Merck). Reverse-phase chromatography was carried out on Grace flash cartridges C-18, ion-exchange chromatography on DEAE-Sephadex A25 columns. NMR spectra were recorded in D2O on Bruker Ascend 400. 1H NMR – 400 MHz, 31 P NMR – 162 MHz. Chemical shifts were reported in ppm relative to solvent resonance signal as an internal standard. HPLC-MS analyses were performed using a high performance liquid chromatography system, equipped with a photo diode array detector (SPD-M20A) and a mass spectrometer (LCMS-2020, Shimadzu) equipped with an ESI source. The chromatographic separation was conducted using a Hydrosphere C18 column, 4 × 150 mm (YMC) at 40 °C and a mobile phase that consisted of 10 mM ammonium acetate water solution (solvent A), and acetonitrile (solvent B). Mass spectrometry data was acquired in both positive and negative ionisation mode and analysed using the LabSolutions LCMS software. The nucleotides linked with phosphonoacetic acid were synthesised according to the published procedures with modificationsCitation17,Citation18.
General procedure for the synthesis of dTMP-PAA (PA5T)
To a solution of thymidine 5′-monophosphate morpholidate (203 mg, 0.5 mmol) in 2 ml of dimethylformamide (DMF), 700 µL (3 mmol) of tributylamine and 140 mg (1 mmol) of phosphonoacetic acid were added and the reaction mixture was stirred at 50 °C temperature for 24 h. After the reaction was completed (TLC), DMF was evaporated under reduced pressure. The residue was dissolved in water and purified twice by reverse-phase chromatography (C18 cartridge, methanol/water mixture, 10:0 → 10:2). The solvents were removed under reduced pressure to afford colourless oil reaction product. Yield 0.035 mmol (7%). Rf = 0.22 (1,4-dioxane/water/2-propanol/ammonia water, 4:3:2:1). UV λmax: 267 nm; MS (ESI+): m/z 445.00 [M + H]+, 443.00 [M-H]–. 1H NMR (D2O, ppm): δ = 2.13 (d, J = 0.9 Hz, 3H), 2.21 (m, 1H), 2.42 (m, 1H), 2.62 (d, J = 5.9 Hz, 2H), 3.90 (m, 2H), 4.15 (m, 1H), 4.46 (m, 1H), 6.36 (t, J = 6.3 Hz, 1H), 7.21 (d, J = 0.9 Hz, 1H). 31 P NMR (D2O, ppm): δ = –11.53 (d, J = 27.5 Hz, Pα), 9.38 (d, J = 27.5 Hz, Pβ).
General procedure for the synthesis of dTTP-PAA (PA5TT)
To a solution of thymidine 5′-triphosphate tributylamonium salt (0.48 mmol) in 5 ml of DMF, 67 mg (0.48 mmol) of phosphonoacetic acid, 98 mg (0.48 mmol) of dicyclohexylcarbodiimide and 58 mg (0.48 mmol) of dimetylaminopyridine were added and the reaction mixture was stirred at room temperature for 24 h. After the reaction was completed (TLC), the precipitate was filtered, filtrate was evaporated under reduced pressure. The residue was quenched by addition of water and purified by ion-exchange chromatography on DEAE-Sephadex A25 column (20 ml) with a linear gradient (0.05–0.4 M) of LiCl as the mobile phase. The product was eluted with 0.3–0.35 M LiCl, the solution was concentrated under reduced pressure to several millilitres and poured into 40 ml acetone/methanol mixture (4/1). The formed precipitate was collected by centrifugation (4000 rpm, 10 min) and washed twice with a mixture of acetone/methanol, 4/1. The nucleotide was dissolved in 2 ml of water and evaporated under reduced pressure. Slightly acidic solution of nucleotide was neutralised with 1 M sodium hydroxide solution to pH 7.0. Yield 0.060 mmol (12.5%). Rf = 0.19 (1,4-dioxane/water/2-propanol/ammonia water, 4:5:2:1). UV λmax: 267 nm; MS (ESI+: m/z 604.90 [M + H]+, 602.90 [M − H]–. 1H NMR (D2O, ppm): δ = 2.05 (m, 1H), 2.15 (d, J = 0.9 Hz, 3H), 2.48 (m, 1H), 2.62 (d, J = 5.9 Hz, 2H), 3.91 (m, 2H), 4.14 (m, 1H), 4.43 (m, 1H), 6.36 (t, J = 6.3 Hz, 1H), 7.21 (d, J = 0.9 Hz, 1H). 31 P NMR (D2O, ppm): δ = –22.75–22.15 (2 t, Pβ, γ), –11.31 (d, J = 16.7 Hz, Pα), 10.95 (d, J = 23.4 Hz, Pδ).
General procedure for the synthesis of aadUTP-PAA (PAT)
To an activated NHS ester of phosphonoacetic acid (0.24 mmol), 40 µL (0.16 mmol) of tributylamine and a concentrated (up to 190 mM) solution of aadUTP (0.08 mmol) in water were added and the reaction mixture was stirred at room temperature for 24 h. After the reaction was completed (TLC), the reaction mixture was quenched by addition of water and purified by ion-exchange chromatography on DEAE-Sephadex A25 column (10 ml) with a linear gradient (0.05–0.4 M) of LiCl as the mobile phase. The product was eluted with 0.3 M LiCl, the solution was concentrated under reduced pressure to 1–2 ml and poured into a mixture of acetone/methanol (4/1, 20 ml). The formed precipitate was collected by centrifugation (4000 rpm, 10 min) and washed twice with a mixture of acetone/methanol, 4/1. The nucleotide was dissolved in 1 ml of water and evaporated under reduced pressure. Slightly acidic solution of nucleotide was neutralised with 1 M sodium hydroxide solution to pH 7.0. Yield 0.030 mmol (37.5%). Rf = 0.26 (1,4-dioxane/water/2-propanol/ammonia water, 4:5:2:1). UV λmax: 240 and 286 nm; MS (ESI+: m/z 645.99 [M + H]+, 643.99 [M-H]–. 1H NMR (D2O, ppm): δ = 2.05 (m, 1H), 2.39 (m, 1H), 2.57 (d, J = 5.9 Hz, 2H), 3.99 (m, 2H), 4.26 (m, 3H), 4.46 (m, 1H), 5.88 (m, 1H), 6.36 (t, J = 6.3 Hz, 1H), 6.54 (d, J = 6.5 Hz, 1H), 8.05 (s, 1H). 31 P NMR (D2O, ppm): δ = –22.42 (t, J = 18.6 Hz, Pβ), –10.83 (d, J = 18.5 Hz, Pα), –5.19 (d, J = 18.8 Hz, Pγ), 11.88 (s, Pδ).
Polymerases
Natural nucleoside triphosphates, Klenow exo- and Taq (under the brand name DreamTaq) polymerases were purchased from Thermo Fisher Scientific. The recombinant HIV-1Citation19 and M.MuLVCitation20 reverse transcriptases were purified from E.coli as N-terminal His-tagged proteins using standard protocols and HisPur Cobalt Resin (Thermo Scientific) according to the manufacturer’s recommendations.
Product composition analysis
For the acceptance of nucleotides for DNA biosynthesis, reactions were performed as describedCitation21 with further modifications. In short, a duplex DNA comprising 5′ 33 P labelled primer oligodeoxyribonucleotide 5′ TAATACGACTCACTATAGGGAGA and template oligodeoxyribonucleotide 5′ CCGGAATTAAAATCTCCCTATAGTGAGTCGTATTA was annealed by heating for 5 min at 95 °C and gradually (> 2 h) cooled down to room temperature. A DNA duplex (5 nM) was subjected to primer extension reactions in the presence of 25–50 nM of HIV-1, M.MuLV, Klenow exo- or Taq polymerases, in a reaction mix comprising 20 mM sodium glutamate, pH 8.2, 10 mM DTT, 0.5% Triton X-100, 20 mM sodium chloride and 1 mM magnesium chloride. The reactions were performed in the presence of 0.1–10 µM of dTTP, dATP, or nucleotide-PAA conjugates at 37 °C for 5–60 min. The reactions were terminated by adding an equal volume of STOP solution, containing 95% deionised formamide and 25 mM EDTA. The products were resolved on a 15% 19:1 denaturing (8 M urea) polyacrylamide gel. The gel was dried on Whatman paper and autoradiographed using a FUJIFILM phosphorimager screen. For equilibrium analysis, the termination step of the primer extension reaction was omitted and the samples were immediately loaded on a native 8% 37.5:1 polyacrylamide gel. Subsequent processing of the gel was performed as described above.
Results and discussion
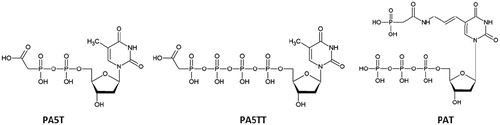
The activity of viral polymerases may be affected both by the conformational change or the change in kineticsCitation22–24. In this study, we focussed on phosphonoacetic acid-nucleotide conjugates designed so as to achieve a high affinity for the target polymerases. Three different PAA conjugates, such as dTMP conjugated with the phosphono moiety of PAA via 5′-phosphate (referred as PA5T), dTTP conjugated with the phosphono moiety of PAA via 5’-gamma-phosphate (PA5TT), and 5-aminoallyl-dUTP conjugated with the carboxy group of PAA via 5-amino group (PAT), were prepared and analysed (). In the case of PA5T, the gamma-phosphate of a natural dTTP was exchanged for the acetic group, thus preserving catalytically important linkage between alpha and beta phosphates. After the addition of phosphonoacetate, PA5TT retained the structure of a natural dTTP. For PAT, the modification was introduced at a nucleobase of the natural nucleotide.
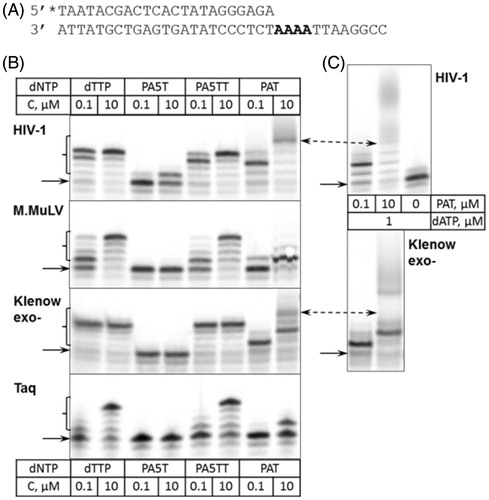
The ability of selected DNA polymerases to accept the aforementioned conjugates was analysed at a single-nucleotide resolution. Radioactively labelled DNA primer was hybridised to the template strand to form a fully complementary duplex with a 3′ single-stranded tail, thus directing primer extension by dTTP up to four consecutive events in a row (). The reaction of a DNA biosynthesis in the presence of HIV-1 and M.MuLV reverse transcriptases, as well as Klenow exo- and Taq DNA polymerases was analysed. For a better resolution of products, the reaction time of 5 min was chosen (). It became immediately evident that all aforementioned polymerases differed in their ability to accept the selected conjugates as a substrate. In the case of all polymerases tested, only the incorporation of PA5TT was comparable to that of dTTP. In contrast, PA5T was accepted exclusively by HIV-1 reverse transcriptase (), and only a negligible amount of this compound was incorporated by M.MuLV and Klenow exo- after a prolonged incubation (data not shown). Whereas Taq polymerase showed no ability to incorporate PA5T into DNA (data not shown). Performance of PAT was found to be ambiguous. The nucleotide was accepted by HIV-1 reverse transcriptase as well as the Klenow exo- DNA polymerase, although at a slightly lower rate. In the case of both M.MuLV reverse transcriptase and Taq DNA polymerase, however, only a negligible amount of PAT was incorporated. To test whether incorporation of PAA disturbed a structure of the resulted DNA fragment enough to compromise a further elongation of the DNA primer, a single-nucleotide resolution assay was modified by including an additional step. For enzymes that utilised PAT (HIV-1 and Klenow exo-), dATP was added after the completion of PAT incorporation, and the reaction was continued for additional five minutes. The elongation proceeded only when the primer contained four residues of PAT (, lanes for 10 µM PAT). Whereas the reaction products containing three or less residues of PAT incorporated remained un-extended ().
Figure 2. Incorporation of nucleotide-PAA conjugates into DNA. (A) DNA substrate used for a study. Asterisk at 5′ terminus of the primer denotes 33 P-labelled strand. Nucleotides utilised in primer extension are presented in bold. (B) Extension of the DNA substrate in the presence of various nucleotides. The concentrations are given in the table, above and below the corresponding lanes. In total, 5 nM of duplex DNA was incubated with 50 nM of polymerase (indicated on the left) in the presence of dTTP or conjugates at 37 °C for 5 min. Arrow indicate the position of the unreacted primer strands, brackets denote the products of primer extension. (C) Extension of PAT-containing DNA by dATP. Upon the completion of PAT incorporation reaction, dATP was added up to 1 µM and the reaction was continued at the same temperature for additional 5 min. PAT concentration is indicated in the table between the panels, the polymerases used in reactions are indicated on the right. Double-sided arrows mark a product of the DNA extension in the presence of PAT and further prolonged in the presence of dATP.
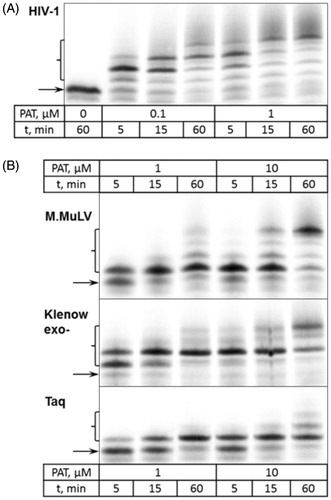
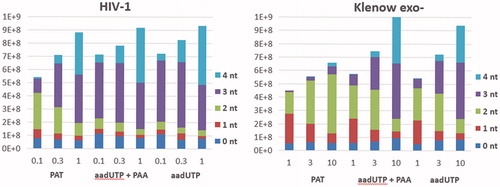
The observed differences in the ability to incorporate PAT among selected DNA polymerases were investigated in further detail. The time course experiments of primer extension were performed under varying concentrations of PAT. Based on our results, HIV-1 reverse transcriptase had the highest preference for PAT (). The other three enzymes tested exhibited lower ability to incorporate the aforementioned compound (). The intermediate products of primer extension involving one or two PAT moieties were well-preserved during the course of DNA biosynthesis. While the progressive formation of reaction intermediates suggested a distributive mode of DNA synthesis, the enzymes employed in this study showed much higher processivity with dTTP as a substrate for primer extension ().
Figure 3. DNA extension in the presence of PAT. (A) The incorporation of PAT by HIV-1 reverse transcriptase. PAT concentrations and duration of reaction are listed in the table below the gel picture. (B) The incorporation of PAT by polymerases denoted on the left. PAT concentrations (10-fold higher than those in A) and duration of reaction are listed in the table below the gel picture. Arrows indicates the position of the unreacted primer strand of DNA duplex, brackets – the primer extension products.
To decipher the reason(s) behind the reduced processivity of polymerases upon incorporation of PAT, the primer extension assays were performed with either (i) PAT alone, (ii) a mix of 5-aminoallyl-dUTP (precursor of PAT) and PAA, or (iii) 5-aminoallyl-dUTP alone. The performance of HIV-1 and Klenow exo- polymerases was evaluated by densitometric analysis of the formed products. Both the amount and a relative distribution of the reaction products within a given sample were calculated (). In the case of PAT and 5-aminoallyl-dUTP, the pronounced difference in the pattern of product formation during primer extension reactions was observed. As seen in , upon the incorporation of 5-aminoallyl-dUTP, the formation of higher molecular mass products was observed, starting from the lowest concentrations of the nucleotide and irrespectively of the presence of PAA in the reaction mixture (, series ii and iii). Such pattern is typical for the polymerases that incorporate nucleotides in a processive manner. Compared to 5-aminoallyl-dUTP, a lower yield, particularly at the lowest concentrations of the compound, was observed during the incorporation of PAT. Compared with 5-aminoallyl-dUTP (setup ii and iii), the primer extension reaction in the presence of PAT was highlighted by the prevalence of less-extended products, independent on PAT concentrations employed (setup i). The inhibition of the primer extension in the presence of free PAA was not observed (setup ii); moreover, a minor stimulation of the reaction took place in the case of Klenow exo-.
Figure 4. Primer extension in the presence of PAT and 5-aminoallyl-2′-deoxyUTP (aadUTP) with or without the equimolar amounts of PAA. Polymerases used in the primer extension reactions are indicated above the graph. PAT, aadUTP and PAA concentrations in µM are given below the graph. Reaction time was 5 min. For calculation of reaction outcome, a number of incorporated nucleotides is taken into account and is represented by the total height of the bar. The internal fractions of the bar represent a distribution of the respective products and are indicated in graph legend.
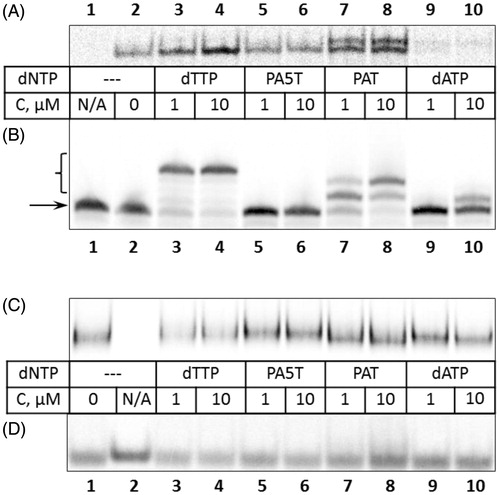
The binding of polymerase to a DNA substrate in a catalytically-competent complex mode was evaluated in order to investigate the overall conformation of the ternary complex. Two natural nucleotides (dTTP and dATP) along with PA5T and PAT were employed in this study, enabling the detection of both specific and non-specific complexes. The primer extension reaction was performed for five minutes, and the reaction products were resolved by native PAGE. In the absence of nucleotide triphosphates, the binding of Klenow exo- to the DNA substrate () was found to be somewhat weaker under the conditions employed (, lane 2). In contrast, the presence of nucleotide triphosphates either enhanced binding (, lanes 3, 4, 7, 8) or stimulated the formation of complexes of different mobility (, lanes 7–10 and Supplement, Part 1). Notably, the complexes of reduced mobility were observed upon incubation with PAT and dATP. The latter compound was incorporated into the primer rather inefficiently, as evidenced by denaturing gel electrophoresis (, lanes 9, 10). In comparison, an incorporation of PAT was more efficient (, lanes 7, 8). Our results indicate that, in the presence of DNA and PAT, the catalytically-competent ternary complex includes Klenow exo- polymerase with altered conformation. The analysis of ternary complexes formed by HIV-1 reverse transcriptase also indicated a conformational irregularity (, lanes 3–10). Here, a mobility of PAT-containing complex was similar to that of dTTP-containing one (, lanes 3, 4, 7, 8). In addition, the smears migrating just above free DNA substrate indicate that the ternary complexes assembled in the presence of PAT may be meta-stable (, lanes 7, 8 and Supplement, Part 2), disassembling shortly after the loading of samples on a gel.
Figure 5. Equilibrium (A, C and D, gel shift mode) and product composition analysis (B) in the presence of various nucleotides. The nucleotides and their concentrations are listed in the table between panels, N/A – no protein added. A. Binding of Klenow exo– to the DNA substrate under catalytic conditions as analysed by native electrophoresis. Only bound fraction of DNA is shown. (B) Analysis of the products of primer extension by Klenow exo–. The samples from panel A were resolved on a denaturing gel. Arrow indicates the position of the unreacted primer strand, bracket – the products of primer extension. C. Binding of HIV-1 reverse transcriptase to the DNA. The bound fraction of DNA is shown at 2-fold extended vertical axis. (D) Unbound fraction of DNA substrate in HIV-1 polymerase binding assays, presented in C. Full-length pictures of gels represented in A, C and D are included in the Supporting Information, Part 1 and Part 2.
Phosphonoacetic acid is generally recognised for a restricted capability to inhibit DNA biosynthesisCitation2, despite its highly analogous structure to pyrophosphate and foscarnet, a well-known small molecule inhibitor. To achieve a better affinity with selected polymerases and to prepare “a poison pill”, PAA was linked at different positions of natural nucleotide. Ideally, during DNA prolongation, a nucleotide-conjugated small-molecule inhibitor should be positioned at the active centre of polymerase. A vicinity to the catalytic moieties and a high local concentration of a small molecule inhibitor are expected to compensate for the low affinity of phosphonoacetic acid. However, the structure of 5′-phosphate region of the prepared conjugates was found to be crucial for the recognition of these substrates by all tested DNA polymerases. As such, at low levels of nucleotides, PA5T was incorporated by polymerases far less efficiently than PA5TT, which has an intact 5′-triphosphate structure. A discriminative incorporation of PA5T by HIV-1 reverse transcriptase hints of the potential of this compound for antiviral therapy.
The impaired performance of polymerases in the presence of PAT is rather surprising. The C5 position in pyrimidine has been found to be permissive for the incorporation of various functionalities into DNA decades agoCitation16. Moreover, PAA conjugated to the 5-aminoallyl group is not as extreme, both in terms of size and charge patterns, as many other nucleobase modifications efficiently accepted by polymerasesCitation16. Our results revealed that upon binding of PAT, the ternary complex of Klenow exo- polymerase as well as that of HIV-1 reverse transcriptase adopt an alternative conformation similar to that originated from the binding of an incorporation-deficient nucleotide. However, the exact mechanism of phosphonoacetic acid-induced alterations in the structure of the productive complex is yet to be elucidated. Nevertheless, our results suggested that in the presence of PAT, polymerases switched from a processive to a somewhat distributive mode of action.
Given the pressure that the host cell puts on the virus – to replicate efficiently or die – even minute alterations in the activity of viral replicases has fatal consequences for the virusCitation1. Previously neglected small molecules with low affinity for polymerases reappear as a valuable scaffold for the development of new generation inhibitors. Therefore, the purposeful conjugation of these compounds with nucleotides may provide a basis for further development of novel antiviral drugs.
IENZ_1417275_Supplementary_Material.pdf
Download PDF (146.6 KB)Acknowledgements
Authors acknowledge Dr. Stephen Le Grice for generous donation of plasmid for the expression of HIV-1 reverse transcriptase. We thank Dr. L. Kalinienė for assistance with the preparation of the manuscript.
Disclosure statement
The authors report no conflicts of interest.
Additional information
Funding
References
- Oberg B. Rational design of polymerase inhibitors as antiviral drugs. Antiviral Res 2006;71:90–5.
- Oberg B. Antiviral effects of phosphonoformate (PFA, foscarnet sodium). Pharmacol Ther 1989;40:213–85.
- Huang H, Chopra R, Verdine GL, Harrison SC. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science 1998;282:1669–75.
- Fauci AS, Folkers GK. Toward an AIDS-free generation. JAMA 2012;308:343–4.
- Menéndez-Arias L. Molecular basis of human immunodeficiency virus drug resistance: an update. Antiviral Res 2010;85:210–31.
- Menéndez-Arias L. Molecular basis of human immunodeficiency virus type 1 drug resistance: overview and recent developments. Antiviral Res 2013;98:93–120.
- Margolis AM, Heverling H, Pham PA, Stolbach A. A review of the toxicity of HIV medications. J Med Toxicol 2014;10:26–39.
- Velázquez S, Alvarez R, San-Félix A, et al. Synthesis and anti-HIV activity of [AZT]-[TSAO-T] and [AZT]-[HEPT] dimers as potential multifunctional inhibitors of HIV-1 reverse transcriptase. J Med Chem 1995;38:1641–9.
- Renoud-Grappin M, Fossey C, Fontaine G, et al. Imidazo[1,5-b]pyridazine-d4T conjugates: synthesis and anti-human immunodeficiency virus evaluation. Antivir Chem Chemother 1998;9:205–23.
- Pontikis R, Dollé V, Guillaumel J, et al. Synthesis and evaluation of “AZT-HEPT”, “AZT-pyridinone”, and “ddC-HEPT” conjugates as inhibitors of HIV reverse transcriptase. J Med Chem 2000;43:1927–39.
- Adelfinskaya O, Herdewijn P. Amino acid phosphoramidate nucleotides as alternative substrates for HIV-1 reverse transcriptase. Angew Chem Int Ed Engl 2007;46:4356–8.
- Giraut A, Song X-P, Froeyen M, et al. Iminodiacetic-phosphoramidates as metabolic prototypes for diversifying nucleic acid polymerization in vivo. Nucleic Acids Res 2010;38:2541–50.,
- De S, Groaz E, Margamuljana L, et al. Sulfonate derived phosphoramidates as active intermediates in the enzymatic primer-extension of DNA. Org Biomol Chem 2015;13:3950–62.
- Song Y, Zhan P, Li X, et al. Multivalent agents: a novel concept and preliminary practice in Anti-HIV drug discovery. Curr Med Chem 2013;20:815–32.
- Giraut A, Abu El-asrar R, Marlière P, et al. 2′-Deoxyribonucleoside phosphoramidate triphosphate analogues as alternative substrates for E. coli polymerase III. ChemBioChem 2012;13:2439–44.
- Hollenstein M. Nucleoside triphosphates-building blocks for the modification of nucleic acids. Molecules 2012;17:13569–91.
- Hutchinson DW, Cload PA, Haugh MC. Organophosphorus compounds as antiviral agents. Phosphorus Sulfur 1983;14:285–93.
- Hamilton CJ, Roberts SM. Synthesis of fluorinated phophonoacetate derivatives of carbocyclic nucleoside monophosphonates and activity as inhibitors of HIV reverse transcriptase. J Chem Soc Perkin Trans 1 1999; 1051–6.
- Le Grice SF, Naas T, Wohlgensinger B, Schatz O. Subunit-selective mutagenesis indicates minimal polymerase activity in heterodimer-associated p51 HIV-1 reverse transcriptase. EMBO J 1991;10:3905–11.
- Shinnick TM, Lerner RA, Sutcliffe JG. Nucleotide sequence of moloney murine leukaemia virus. Nature 1981;293:543–8.
- Tauraitė D, Ražanas R, Mikalkėnas A, et al. Synthesis of pyridone-based nucleoside analogues as substrates or inhibitors of DNA polymerases. Nucleos. Nucleot. Nucl 2016;35:163–77.
- Meyer PR, Matsuura SE, Zonarich D, et al. Relationship between 3′-azido-3′-deoxythymidine resistance and primer unblocking activity in foscarnet-resistant mutants of human immunodeficiency virus type 1 reverse transcriptase. J Virol 2003;77:6127–37.
- Marchand B, Tchesnokov EP, Götte M. The pyrophosphate analogue foscarnet traps the pre-translocational state of HIV-1 reverse transcriptase in a Brownian ratchet model of polymerase translocation. J Biol Chem 2007;282:3337–46.
- Zahn KE, Tchesnokov EP, Götte M, Doublié S. Phosphonoformic acid inhibits viral replication by trapping the closed form of the DNA polymerase. J Biol Chem 2011;286:25246–55.