Abstract
Oxidative stress may be the major cause of induction of Shiga toxin-converting (Stx) prophages from chromosomes of Shiga toxin-producing Escherichia coli (STEC) in human intestine. Thus, we aimed to test a series of novel antioxidant compounds for their activities against prophage induction, thus, preventing pathogenicity of STEC. Forty-six compounds (derivatives of carbazole, indazole, triazole, quinolone, ninhydrine, and indenoindole) were tested. Fifteen of them gave promising results and were further characterized. Eleven compounds had acceptable profiles in cytotoxicity tests with human HEK-293 and HDFa cell lines. Three of them (selected for molecular studies) prevent the prophage induction at the level of expression of specific phage genes. In bacterial cells treated with hydrogen peroxide, expression of genes involved in the oxidative stress response was significantly less efficient in the presence of the tested compounds. Therefore, they apparently reduce the oxidative stress, which prevents induction of Stx prophage in E. coli.
Introduction
The significance of Shiga toxin-producing Escherichia coli (STEC) as a public health problem was recognized in 1982 in USA, during an investigation of the outbreak of hemorrhagic colitis associated with consumption of contaminated hamburgersCitation1. Since then, STEC strains have been implicated in many outbreaks of diarrhea worldwide. Quite recently (2011), the Shiga toxin-producing E. coli serotype O104:H4 was responsible for a serious epidemic outbreak in GermanyCitation2,Citation3. STEC pathogens can cause serious food poisoning with bloody diarrhea in humansCitation4. Their main virulence factors are Shiga toxins, encoded by stx genes located in genomes of bacteriophages, which occur in bacteria as prophagesCitation5. These bacteriophages are called Shiga toxin-converting or shortly Stx phages and belong to the lambdoid family of phagesCitation6. All phages within this group indicate high similarities in the lifecycle and genomic organization to bacteriophage λ, the most reviewed member of this familyCitation7. In the prophage state, most of phage genes, including stx genes, are not transcribed due to inhibition caused by the phage cI repressor. Consequently, Shiga toxins are not produced under such conditions. Expression of stx as well as other phage genes occurs effectively only after prophage induction. In most cases, this process requires activation of the RecA-dependent bacterial S.O.S. response, which is provoked by factors causing appearance of single-stranded DNA fragments. Activated RecA protein stimulates cleavage of both the SOS regulon repressor, the LexA protein, and the cI phage repressor. Prophage induction and subsequent phage lytic development lead to production of progeny phage particles and Shiga toxins, and then to their release from the lysed cellCitation8.
Induction of lambdoid prophages is easy and can be provoked by different inducers like treatment with targeted DNA antibiotics (e.g. mitomycin C), UV-treatmentCitation7, EDTACitation9, 60Co irradiationCitation10, high hydrostatic pressureCitation11, and many others. Such conditions do not naturally occur in the human gut, the place where the STEC infection initiates. Therefore, the oxidative stress conditions, which appear in human intestine as a response to STEC infection, has attracted attention of researchers. Indeed, it has been shown that hydrogen peroxide (which is excreted by neutrophils during infection) increases efficiency of Stx prophage induction and stimulates production of Shiga toxinsCitation12–14. In bacteria, H2O2 acts as a DNA-damaging agent that interacts with free cellular iron and form hydroxyl radicals OH• in Fenton reaction. DNA subjected to attack by OH• generates a wide range of base and sugar modifications and strand breaks, which in turn activate the RecA protein and SOS responseCitation15. According to hypothesis proposed by two different research groupsCitation16,Citation17, STEC strains may obtain advantage due to H2O2-mediated production of Shiga toxins. As demonstrated, these toxins are produced in a small fraction of STEC cells (usually less than 1%) which are sacrificed for the good of the whole bacterial population. Released toxins kill H2O2-producing protozoan predator, so allowing the rest of bacteria to survive its attack. It is suspected that STEC response to attack of neutrophils in human intestine might be analogousCitation16,Citation17.
The OxyR and SoxR regulators mainly control E. coli defense against oxidative stress. Interestingly, H2O2 activate genes of both regulons: OxyR (about 30 genes) and SoxRS (about 17 genes)Citation18. It was demonstrated that nitric oxide and its derivatives (NOs) as well as S-nitrosothiols (SNOs) may be involved in signaling in bacteria and may change properties of several bacterial proteins including OxyR, SoxR, NorR and Fur in Escherichia coliCitation19–22, as well as ResDE in Bacillus subtilisCitation23. As reported previously, certain SNOs are able to provoke a nitrosative stress in E. coli bacteria, evidenced by lowering of intracellular thiol and the transcriptional activation of OxyR by S-nitrosylationCitation24. OxyR is a thiol-containing transcriptional activator of antioxidant genes that are involved in H2O2 detoxification. The activation of E. coli antioxidant enzymes by OxyR in response to H2O2 has been widely described. The genes controlled by OxyR include catalase/hydroperoxidase (katG), glutathione reductase (gorA), and alkyl hydroperoxide reductase (ahpCF). Their expression is essential to ensure resistance to H2O2Citation24–26. As demonstrated previously, S-nitrosylation, S-hydroxylation, and S-glutathionylation of wild-type OxyR activate this protein and appear to serve as the functional switches in the activation of OxyR-regulated genesCitation24,Citation27. On the other hand, nitric oxide (NO) donors are able to activate SoxRS regulon and induce the expression of enzymes involved in the oxidative stress response of E. coli bacteria, such as the superoxide dismutase (sodA)Citation28,Citation29.
It was previously proposed that OxyR may be responsible for the lambdoid prophage maintenance under conditions of the oxidative stressCitation8. Other studies indicated that under such conditions, activated OxyR is able to compete effectively with the cI protein (the main repressor of phage lytic development) for the binding to the oR3 operator of the pM-pR promoter regionCitation30. These results allowed to suggest that OxyR might enhance repression of pR (the major promoter for expression of genes of phage lytic lifecycle) and enhance activation, and at the same time also downregulate repression, of the transcription of the cI gene from the pM promoter. This would lead to higher (relative to normal growth conditions) activity of pM, increased production of cI repressor and lower efficiency of prophage induction under the oxidative stress.
In the light of the importance of the oxidative stress in induction of Stx prophages and Shiga toxin production (summarized above), we analyzed in this study impact of the different antioxidant compounds, on viability of STEC and development of the Stx phage Φ24B. Since virulence of STEC depends on the prophage induction, and in the light of the fact that most of antibiotics should not be used for treatment of STEC-infected patients, development of novel drugs that would impair survival of STEC (without causing prophage induction) or prophage induction/development or both would be highly desirable. Although development of novel compounds which might be potentially used to combat E. coli infections, exemplified by those based on furanyl-derived sulfonamides, has been reportedCitation31, and sophisticated techniques become available to either engineer E. coli surfaceCitation32 or to use nanoparticles as carriers of compoundsCitation33, the anti-bacterial activity of these molecules were moderateCitation31. Therefore, we have tested a series of novel compounds that are derivatives of carbazole, indazole, triazole, quinolone, ninhydrin, and indenoindole.
Materials and methods
Chemistry
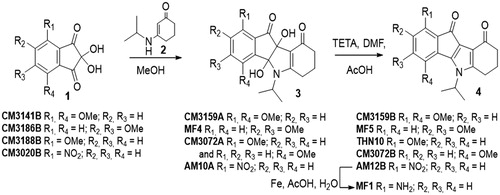
Chemical structures of all compounds tested in this report are presented in the supporting information (Supplementary Figures 1–4) with the corresponding references. Seven indenoindole derivatives are described in this paper (CM3159A, MF4, THN10, CM3072B, CM3159B, MF5, MF1). Melting points were determined on an Electrothermal 9200 capillary apparatus. The IR spectra were recorded on a PerkinElmer Spectrum Two IR spectrometer. The 1H and 13C NMR spectra were recorded on a Bruker DRX 400 spectrometer. Chemical shifts are expressed in ppm (δ) downfield from internal tetramethylsilane, and coupling constants J are reported in hertz (Hz). The following abbreviations are used: s, singlet; bs, broad singlet; d, doublet; t, triplet; dd, doubled doublet; q, quartet; sept, septuplet; m, multiplet; Cquat, quaternary carbons. The mass spectra were performed by direct ionization (EI or CI) on a ThermoFinnigan MAT 95 XL apparatus. Chromatographic separations were performed on silica gel columns by column chromatography (Kieselgel 300–400 mesh). All reactions were monitored by TLC on GF254 plates that were visualized under a UV lamp (254 nm). Evaporation of solvent was performed in vacuum with rotating evaporator. The purity of the final compounds (greater than 95%) was determined by uHPLC/MS on an Agilent 1290 system using a Agilent 1290 Infinity ZORBAX Eclipse Plus C18 column (2.1 × 50 mm, 1.8 μm particle size) with a gradient mobile phase of H2O/CH3CN (90:10, v/v) with 0.1% of formic acid to H2O/CH3CN (10:90, v/v) with 0.1% of formic acid at a flow rate of 0.5 ml/min, with UV monitoring at the wavelength of 254 nm. A run time of 10 min was applied. LC/HRMS data of all new indenoindole derivatives are presented in the supporting information.
General procedure for the synthesis of compound 3
A solution containing equimolar amounts of enaminone 2 and ninhydrin 1 dissolved in MeOH was stirred at room temperature for 19 h. Generally, a precipitate of compound 3 was formed. The solvent was evaporated, and the product triturated, filtrated and washed with EtOAc. A second quantity could be obtained from the filtrate by purification by silica gel column chromatography with CH2Cl2/acetone (96:4, v/v) as the eluent.
Mixture of regioisomers 4 b,9 b-Dihydroxy-5-isopropyl-1-methoxy-4 b,5,6,7,8,9 b-hexahydroindeno[1,2-b]indole-9,10-dione (major, MAJ) and 4 b,9 b-Dihydroxy-5-isopropyl-4-methoxy-4 b,5,6,7,8,9 b-hexahydroindeno[1,2-b]indole-9,10-dione (minor, min) CM3072A: beige solid. Yield 77%. IR (υ cm−1): 3368, 1714, 1595, 1532, 1491 cm−1. 1H NMR (300 MHz, DMSO-d6) δ: 7.73 (t, 1H, J = 8.1 Hz, H-2 MAJ), 7.54 (t, 1H, J = 7.9 Hz, H-3 min), 7.44 (d, 1H, J = 7.5 Hz, H-4 MAJ), 7.38 (d, 1H, J = 7.5 Hz, H-3 min), 7.26 (d, 1H, J = 7.3 Hz, H-2 min), 7.12 (d, 1H, J = 8.3 Hz, H-4 MAJ), 6.66 (s, 1H, OH MAJ), 6.32 (s, 1H, OH min), 5.73 (s, 1H, OH min), 5.54 (s, 1H, OH MAJ), 4.85 (sept, 1H, J = 7.1 Hz, CH(CH3)2 min), 4.54 (sept, 1H, J = 7.1 Hz, CH(CH3)2 MAJ), 3.95 (s, 3H, OCH3 min), 3.88 (s, 3H, OCH3 MAJ), 2.62–2.82 (m, 4H, H-6 MAJ + min), 2.06–2.13 (m, 4H, H-8 MAJ + min), 1.79–1.91 (m, 4H, H-7 MAJ + min), 1.45 (d, 3H, J = 6.8 Hz, CH3 MAJ), 1.39 (d, 3H, J = 7.2 Hz, CH3 min), 1.33 (d, 3H, J = 7.0 Hz, CH3 min), 1.25 (d, 3H, J = 7.1 Hz, CH3 MAJ).
4 b,9 b-Dihydroxy-5-isopropyl-1,4-dimethoxy-4 b,5,6,7,8,9 b-hexahydroindeno[1,2-b]indole-9,10-dione CM3159A: beige solid. Yield 66%. Mp 216 °C. IR (υ cm−1): 3347, 1717, 1497, 1270, 999. 1H NMR (400 MHz, DMSO-d6) δ: 7.30 (d, 1H, J = 8.9 Hz, H-3), 7.05 (d, 1H, J = 8.9 Hz, H-2), 6.27 (s, 1H, OH), 5.65 (s, 1H, OH), 4.82 (sept, 1H, J = 7.1 Hz, CH(CH3)2), 3.88 (s, 3H, OCH3), 3.80 (s, 3H, OCH3), 2.70–2.79 (m, 1H, CH2), 2.40–2.50 (m, 1H, CH2), 2.06 (t, 2H, J = 6.3 Hz, CH2), 1.75–1.84 (m, 2H, CH2), 1.36 (d, 3H, J = 7.1 Hz, CH3), 1.31 (d, 3H, J = 7.1 Hz, CH3). 13 C NMR + DEPT (100 MHz, DMSO-d6) δ: 194.7 (C = O), 188.7 (C = O), 165.5 (Cquat), 150.6 (Cquat), 149.5 (Cquat), 136.1 (Cquat), 124.3 (Cquat), 118.4 (CH), 112.9 (CH), 106.2 (Cquat), 95.8 (Cquat), 82.9 (Cquat), 55.7 (2 OCH3), 45.1 (CH), 36.7 (CH2), 24.5 (CH2), 22.4 (CH3), 22.2 (CH3), 21.8 (CH2). HRMS calcd for C20H24NO6 [M + H]+ 374.1598 found, 374.1599.
4 b,9 b-Dihydroxy-5-isopropyl-2,3-dimethoxy-4 b,5,6,7,8,9 b-hexahydroindeno[1,2-b]indole-9,10-dione MF4: white solid. Yield 87%. Mp 206 °C. IR (υ cm−1): 3071, 1727, 1712, 1488, 1284, 1174, 954. 1H NMR (400 MHz, DMSO-d6) δ: 7.39 (s, 1H, H-1), 7.09 (s, 1H, H-4), 6.60 (s, 1H, OH), 5.50 (s, 1H, OH), 4.64 (sept, 1H, J = 7.1 Hz, CH(CH3)2), 3.92 (s, 3H, OCH3), 3.83 (s, 3H, OCH3), 2.44–2.70 (m, 2H, CH2), 2.05 (t, 2H, J = 6.3 Hz, CH2), 1.72–1.85 (m, 2H, CH2), 1.43 (d, 3H, J = 7.1 Hz, CH3), 1.24 (d, 3H, J = 7.1 Hz, CH3). 13 C NMR + DEPT (75 MHz, DMSO-d6) δ: 196.2 (C = O), 188.8 (C = O), 164.2 (Cquat), 155.5 (Cquat), 151.0 (Cquat), 142.9 (Cquat), 127.9 (Cquat), 105.7 (CH), 105.5 (Cquat), 103.6 (CH), 95.4 (Cquat), 82.8 (Cquat), 56.3 (OCH3), 55.7 (OCH3), 44.7 (CH), 36.7 (CH2), 24.3 (CH2), 22.6 (CH3), 22.1 (CH3), 22.0 (CH2). HRMS calcd for C20H24NO6 [M + H]+ 374.1598, found 374.1603.
4 b,9 b-Dihydroxy-5-isopropyl-1-nitro-4 b,5,6,7,8,9 b-hexahydroindeno[1,2-b]indole-9,10-dione AM10A: yellow solid. Yield 92%. Mp 227 °C. IR (υ cm−1): 3553, 1746, 1589, 1536, 1491. 1H NMR (500 MHz, DMSO-d6) δ: 8.28 (d, 1H, J = 7.6 Hz, H-4), 8.04 (d, 1H, J = 7.6 Hz, H-2), 7.99 (t, 1H, J = 7.6 Hz, H-3), 7.04 (s, 1H, OH), 5.87 (s, 1H, OH), 4.61 (sept, 1H, J = 7.2 Hz, CH(CH3)2), 2.41–2.77 (m, 2H, CH2), 2.10 (t, 2H, J = 6.0 Hz, CH2), 1.84 (m, 2H, CH2), 1.45 (d, 3H, J = 6.9 Hz, CH3), 1.27 (d, 3H, J = 6.9 Hz, CH3). 13 C NMR + DEPT (125 MHz, DMSO-d6) δ: 192.8 (C = O), 188.7 (C = O), 164.9 (Cquat), 148.8 (Cquat), 145.1 (Cquat), 136.1 (CH), 128.9 (CH), 126.5 (Cquat), 124.8 (CH), 104.5 (Cquat), 94.6 (Cquat), 83.3 (Cquat), 44.9 (CH(CH3)2), 36.7 (CH2), 24.2 (CH2), 22.5 (CH3), 22.4 (CH3), 21.9 (CH2). HRMS calcd for C18H19N2O6 [M + H]+, 359.1238 found: 359.1240.
General procedure for the synthesis of compound 4
A solution containing 5.92 mmol of 3 and 11.5 mmol (2 equiv) of TETA dissolved in 18 ml of DMF and 3.5 ml of AcOH was stirred at room temperature for 22 h. The solution was then poured into 50 ml of ice and water and stirred for 30 min. The resulting precipitate was filtered, washed with water and dried to give a first quantity of 4. EtOAc was added to the filtrate, and the organic product extracted. The organic phase was washed with water, dried over anhydrous Na2SO4 and evaporated in vacuum to give a second quantity of 4 which was purified by silica gel column chromatography with CH2Cl2/acetone (95:5, v/v) as the eluent.
5-Isopropyl-1-methoxy-5,6,7,8-tetrahydroindeno[1,2-b]indole-9,10-dione THN10: brown solid. Yield 50%. Mp 186 °C. IR (υ cm−1): 1694, 1661, 1590, 1261, 1023, 803. 1H NMR (400 MHz, DMSO-d6) δ: 7.38 (dd, 1H, J1 = 8.6 Hz, J2 = 7.5 Hz, H-3), 7.04 (d, 1H, J = 7.3 Hz, H-4), 6.98 (d, 1H, J = 8.8 Hz, H-2), 4.77 (sept, 1H, J = 6.9 Hz, CH(CH3)2), 3.86 (s, 3H, OCH3), 2.93 (m, 2H, CH2), 2.37 (m, 2H, CH2), 2.08 (m, 2H, CH2), 1.57 (d, 6H, J = 7.1 Hz, CH3). 13 C NMR + DEPT (100 MHz, DMSO-d6) δ: 191.2 (C = O), 182.2 (C = O), 156.8 (2 Cquat), 150.1 (2 Cquat), 136.8 (2 Cquat), 135.1 (CH), 121.2 (Cquat), 114.7 (CH), 112.8 (CH), 55.6 (OCH3), 49.5 (CH(CH3)2), 37.7 (CH2), 22.8 (2 CH2), 21.3 (2 CH3). HRMS calcd for C19H20NO3 [M + H]+ 310.1438, found: 310.1438.
5-Isopropyl-4-methoxy-5,6,7,8-tetrahydroindeno[1,2-b]indole-9,10-dione CM3072B: red solid. Yield 24%. Mp 224 °C. IR (υ cm−1): 3436, 1704, 1660, 1600, 1488, 1270. 1H NMR (400 MHz, DMSO-d6) δ: 7.19–7.28 (m, 2H, H-1 and H-2), 7.02 (d, 1H, J = 8.8 Hz, H-3), 5.67 (m, 1H, CH(CH3)2), 2.94 (s, 3H, OCH3), 3.00 (m, 2H, CH2), 2.38 (t, 2H, J = 6.3 Hz, CH2), 2.07 (t, 2H, J = 6.0 Hz, CH2), 1.56 (d, 6H, J = 7.0 Hz, CH3). 13 C NMR + DEPT (100 MHz, DMSO-d6) δ: 191.4 (C = O), 183.1 (C = O), 150.2 (Cquat), 149.8 (2 Cquat), 139.6 (2 Cquat), 130.6 (CH), 121.2 (2 Cquat), 119.2 (CH), 116.3 (CH), 56.3 (OCH3), 51.0 (CH(CH3)2), 37.7 (CH2), 24.5 (CH2), 23.4 (CH2), 21.6 (2 CH3). HRMS calcd for C19H20NO3 [M + H]+ 310.1438, found: 310.1435.
1,4-Dimethoxy-5-isopropyl-5,6,7,8-tetrahydroindeno[1,2-b]indole-9,10-dione CM3159B: orange-red solid. Yield 75%. Mp 247 °C. IR (υ cm−1): 1693, 1661, 1494, 1262. 1H NMR (400 MHz, DMSO-d6) δ: 7.19 (d, 1H, J = 9.3 Hz, H-3), 6.99 (d, 1H, J = 9.3 Hz, H-2), 5.71 (m, 1H, CH(CH3)2), 3.90 (s, 3H, OCH3), 3.81 (s, 3H, OCH3), 2.99 (m, 2H, CH2), 2.36 (m, 2H, CH2), 2.06 (m, 2H, CH2), 1.54 (d, 6H, J = 7.1 Hz, CH3). 13 C NMR + DEPT (75 MHz, DMSO-d6) δ: 191.2 (C = O), 181.7 (C = O), 151.9 (2 Cquat), 149.9 (Cquat), 144.3 (2 Cquat), 122.8 (Cquat), 122.2 (Cquat), 121.5 (CH), 117.1 (CH), 56.0 (2 OCH3), 50.7 (CH(CH3)2), 37.7 (2 CH2), 23.4 (CH2), 21.6 (CH3). HRMS calcd for C20H21NNaO4 [M + Na]+ 362.1363, found: 362.1364.
2,3-Dimethoxy-5-isopropyl-5,6,7,8-tetrahydroindeno[1,2-b]indole-9,10-dione MF5: red solid. Yield 74%. Mp 245 °C. IR (υ cm−1): 1696, 1659, 1268. 1H NMR (400 MHz, DMSO-d6) δ: 7.04 (s, 1H, H-1), 6.97 (s, 1H, H-4), 4.77 (sept, 1H, J = 6.9 Hz, CH(CH3)2), 3.93 (s, 3H, OCH3), 3.84 (s, 3H, OCH3), 2.94 (m, 2H, CH2), 2.38 (t, 2H, J = 6.0 Hz, CH2), 2.10 (t, 2H, J = 5.9 Hz, CH2), 1.61 (d, 6H, J = 6.5 Hz, CH3). 13 C NMR + DEPT (100 MHz, DMSO-d6) δ: 190.6 (C = O), 182.6 (C = O), 151.4 (Cquat), 150.6 (Cquat), 148.8 (Cquat), 148.1 (Cquat), 130.8 (Cquat), 128.6 (Cquat), 118.6 (Cquat), 116.9 (Cquat), 109.2 (CH), 106.0 (CH), 56.3 (OCH3), 55.9 (OCH3), 48.9 (CH(CH3)2), 37.3 (CH2), 22.6 (CH2), 22.5 (CH2), 21.2 (2 CH3). HRMS calcd for C20H21NNaO4 [M + Na]+ 362.1363, found: 362.1366.
5-Isopropyl-1-nitro-5,6,7,8-tetrahydroindeno[1,2-b]indole-9,10-dione AM12B: red solid. Yield 80%. Mp 244 °C. IR (υ cm−1): 1707, 1666, 1605, 1536. 1H NMR (400 MHz, DMSO-d6) δ: 7.63 (d, 1H, J = 7.5 Hz, H-2), 7.59 (t, 1H, J = 7.5 Hz, H-3), 7.49 (d, 1H, J = 7.0 Hz, H-4), 4.80 (sept, 1H, J = 7.0 Hz, CH(CH3)2), 2.95 (m, 2H, CH2), 2.37 (t, 2H, J = 6.0 Hz, CH2), 2.06 (m, 2H, CH2), 1.56 (d, 6H, J = 7.0 Hz, CH3). 13 C NMR + DEPT (100 MHz, DMSO-d6) δ: 190.8 (C = O), 177.8 (C = O), 155.7 (2 Cquat), 149.7 (Cquat), 145.5 (Cquat), 136.0 (2 Cquat), 134.9 (CH), 127.4 (Cquat), 121.6 (CH), 121.0 (CH), 49.5 (CH(CH3)2), 37.3 (CH2), 22.8 (CH2), 22.5 (CH2), 21.2 (2 CH3). HRMS calcd for C18H16N2NaO4 [M + Na]+, 347.1002 found: 347.0989.
1-Amino-5-isopropyl-5,6,7,8-tetrahydroindeno[1,2-b]indole-9,10-dione MF1: To a stirred solution of nitro derivative AM12B (485 mg, 1.50 mmol) in 4.9 ml of glacial acetic and 3.4 ml of water at 90 °C was added iron (686 mg, 12.2 mol) in portions. The resulting suspension was stirred at the same temperature for 45 min. Then, the reaction mixture was cooled to room temperature and basified with saturated NaHCO3 aqueous solution. The organic phase was extracted with dichloromethane washed with brine, dried with Na2SO4, filtered and evaporated to dryness. The residue was purified by silica gel column chromatography with CH2Cl2/acetone (1:1, v/v) as the eluent to afford 254 mg of MF1: yellow solid. Yield 58%. Mp 238 °C. IR (υ cm−1): 3448, 3351, 1673, 1625, 1463. 1H NMR (400 MHz, DMSO-d6) δ: 7.07 (t, 1H, J = 7.7 Hz, H-3), 6.64 (d, 1H, J = 7.1 Hz, H-2), 6.57 (d, 1H, J = 8.6 Hz, H-4), 6.34 (bs, 2H, NH2), 4.72 (sept, 1H, J = 7.0 Hz, CH(CH3)2), 2.93 (t, 2H, J = 6.0 Hz, CH2), 2.38 (t, 2H, J = 6.0 Hz, CH2), 2.07–2.13 (m, 2H, CH2), 1.57 (d, 6H, J = 6.8 Hz, CH3). 13 C NMR + DEPT (100 MHz, DMSO-d6) δ: 191.0 (C = O), 186.4 (C = O), 149.1 (Cquat), 148.3 (Cquat), 146.8 (Cquat), 134.8 (Cquat), 133.3 (CH + Cquat), 118.8 (CH), 116.3 (Cquat), 114.0 (Cquat), 109.0 (CH), 48.7 (CH(CH3)2), 37.5 (CH2), 22.7 (2 CH2), 20.9 (2 CH3). HRMS calcd for C18H18N2NaO2 [M + Na]+ 317.1260, found: 317.1259.
Prophage induction experiment
E. coli MG1655 strainCitation34 lysogenic for phage Φ24B Δstx2::catCitation35 was cultured in LB medium at 37 °C with one of the analyzed compounds (Supplementary Figures S1–S4), in the presence or absence of the prophage inductor: 0.2 µg/ml; 0.5 µg/ml mitomycin C or 1 mM H2O2. In control experiments, DMSO (a solvent used for preparation of stock solutions) was added instead of the tested compound. Each analyzed compound was added at the beginning of the culture (time 0), at three different concentrations (50, 100 and 200 μM), and selected prophage inductor was added after 3 h. Measurements of the bacterial cultures optical density at 600 nm (A600) were performed at indicated times (0, 3, 5, 7, and 9 h). Six hours after an induction (at the ninth hour of the experiment), 50 μl samples were withdrawn, mixed with 10 µl of chloroform and centrifuged (4,500 rpm, 5 min). Serial dilutions were prepared in TM buffer (10 mM Tris–HCl, 10 mM MgSO4; pH 7.2). Phage titer (number of phages per ml) was determined by spotting 2.5 μl of each dilution of the phage lysate on a freshly prepared LB agar (1.5%) with 2.5 μg/ml chloramphenicol, to obtain visible plaques formed on bacterial lawn (according to a procedure described previouslyCitation36), with a poured mixture of 1 ml indicator E. coli MG1655 strain culture and 2 ml of 0.7% nutrient agar (prewarmed to 45 °C), supplemented with MgSO4 and CaCl2 (to a final concentration of 10 mM each). Plates were incubated at 37 °C overnight. Each experiment was repeated three times. The relative phage titer (PFU/ml) representing ratios of phage titers in induced and non-induced cultures was calculated and presented as percentage of relative phage titer obtained in control experiments, with DMSO added instead of the analyzed compound.
MTT cell viability assay: 3 × 103 HEK-293 (Human Embryonic Kidney) or HDFa (Human Dermal Fibroblast, adult) cells were passaged in each well of 96-well plate, and allowed to attached overnight. Cells were then treated with DMSO (control cells) or 50, 100 or 200 µM of tested compound at 37 °C. After 48 h incubation, 25 µl of MTT solution (4 mg/ml) was added to each well. Following 3 h incubation at 37 °C, formazan crystals, formed in living cells, were dissolved in 100 µl of DMSO. Absorbance was measured at 570 and 620 nm (reference wavelength) in a VictorCitation3 microplate reader.
Bacterial RNA preparation: For the extraction of total RNA from E. coli strain MG1655 lysogenic for Φ24B bacteriophage, the procedure described in previous subsection was employed with slight modification. Three selected compounds: CM092, CM032D, and CM3186 were added to the bacterial culture at time 0 to a final concentration 200 µM. The control experiment was treated with DMSO solution, which allows estimation of impact of tested compounds on prophage induction. After 3 hours of bacterial cultivation at 37 °C, phage lytic development was provoked by addition of 1 mM H2O2. At indicated time after prophage induction, 1 × 109 bacterial cells were treated with 10 mM NaN3 (Sigma-Aldrich), harvested and frozen in a liquid nitrogen. The RNA samples were prepared using the High Pure RNA Isolation Kit (Roche Applied Science) and TURBO DNA-freeTM Kit (Life Technologies) according to the manufacturer’s guidelines. The amount and quality of RNA were evaluated by using NanoDrop spectrophotometer (Eppendorf). In addition, the band patterns of total RNA was visualized by agarose gel electrophoresis. Moreover, the absence of DNA was verified by PCR amplification, and by quantitative real-time reverse transcription-PCR (qRT-PCR).
cDNA synthesis via reverse transcription: Each cDNA was prepared using Transcriptor Reverse Transcriptase and random hexamer primers (Roche Applied Science) following the protocols supplied from the provider. 1.25 µg of the total RNA sample was used as a template. The resulting cDNA mixture was diluted 10-fold and analyzed in qRT-PCR.
Real-time PCR assay: qRT-PCR was performed with the LightCycler 480 Real-Time PCR System (Roche Applied Science) according to the procedure described previouslyCitation14,Citation37,Citation38. Reactions were prepared in Roche 96-well plates containing: 10 µl 2 × SYBR Green I Master Mix (Roche Applied Science), 6.25 µl/ml cDNA, and 200 nM specific primers (). Real-time PCR amplification were carried out for 55 cycles and cycling conditions were as follows: an initial incubation at 95 °C for 5 min, followed by denaturation at 95 °C for 10 s, annealing at 60 °C for 15 s, and extension at 72 °C for 5 s. No-template controls and a melting curve analysis were performed for each reaction. The 16S rRNA gene was used as reference gene that showed no changes in expression levels in the presence of H2O2 and tested compounds. Each experiment was repeated three times.
Table 1. Primers used in the real-time PCR assay.
Real-time PCR analysis: For precise relative quantification of changes in the gene expression, the E-Method with efficiency correction was chosen. This method has been used and described in detail by us previouslyCitation37–39. The sample before addition of the induction agent (3 h after inoculation) was a calibrator. The final results were calculated using the following formula: Normalized relative ratio = Et CT(t) calibrator − CT(t) sample/Er CT(r) calibrator − CT(r) sample, where E is efficiency, t is target and r is reference. The raw run data for E. coli strain MG1655 and Φ24B genes were transferred to the LC480Conversion software and the efficiency and PCR efficiency was calculated for each gene by LinRegPCR program.
Results
We have analyzed effects of 46 compounds (Supplementary Figures S1–S4) on growth of E. coli MG1655 bacteria lysogenic with phage Φ24B (a Shiga-toxin converting phage). Density of bacterial culture was measured at indicated time after prophage induction with 0.2 µg/ml mitomycin C in the presence and absence of analyzed compounds. The results are presented in .
Table 2. Differences in OD values resulting from comparison of bacterial growth (E. coli strain MG1655 lysogenic for Φ24B) in control culture (DMSO) and cultures carried out with the analyzed compounds. Each experiment was conducted during 9 h, in the presence or absence of mitomycin C (MITC).
Based on the analysis of growth inhibition of bacterial cultures, 15 compounds were selected to further studies. Most of them were able to increase OD value of the mitomycin C-treated bacterial culture, above 100% in at least one analyzed concentration.
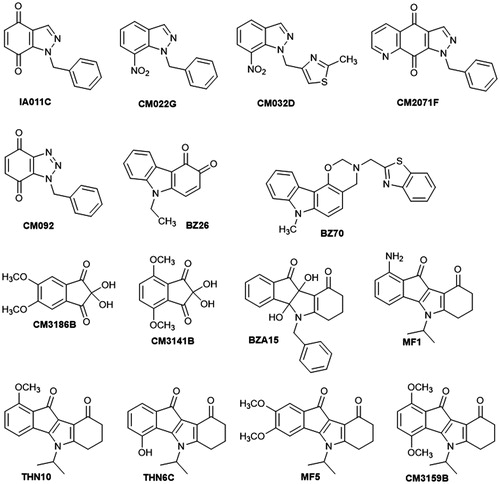
The 15 compounds selected for further biological exploration are presented in . Indazole-4,7-dione IA011C was synthesized in two steps from CM022G: reduction of the nitro group by hydrogenation in acidic medium followed by oxidation by PIFACitation40. Nitroindazoles CM022GCitation40 and CM032DCitation41 were prepared by N-alkylation of the commercially available 7-nitro-1H-indazole with respectively benzyl bromide or 4-(chloromethyl)-2-methylthiazole as alkylating agent. Pyrazolo[4,5-g]quinoline-4,9-dione CM2071F was obtained by a Diels-Alder reaction between N,N-dimethylhydrazone and indazole-4,7-dione IA011CCitation42. Benzotriazole-4,7-dione CM092 was obtained by a 1,3-dipolar cycloaddition of benzyl azideCitation43 on commercial para-benzoquinoneCitation40. Carbazole-3,4-quinone BZ26 was obtained in two steps from the commercially available 9-ethylcarbazol-3-amine. The formation of the corresponding diazonium salt at 0 °C followed by heating the latter in highly acidic aqueous solution gave 9-ethylcarbazol-3-ol, which was finally oxidized with Frémy’s saltCitation44.
Oxazinocarbazole BZ70 was prepared by a Mannich type condensation of benzo[d]thiazol-2-ylmethanamine and formaldehyde with 9-methylcarbazol-4-ol. This latter was obtained by a chemoselective N-alkylation of the commercially available carbazol-4-ol which was achieved by generating the N,O-dianion with NaH in a DMF/THF mixture under argon atmosphere at room temperature and subsequent treatment with methyl iodideCitation45. Dimethoxynin-hydrins (2,2-dihydroxydimethoxyindane-2,3-diones) CM3186B and CM3141B were prepared in a single step from the corresponding commercial substituted indan-1-ones by microwave-assisted selenium oxidationCitation46. Dihydroxyindeno[1,2-b]indole-9,10-dione BZA15Citation47 was prepared by condensation of ninhydrin and 3-(benzylamino)cyclohex-2-en-1-one. 5-Isopropylindeno[1,2-b]indole-9,10-dione derivatives MF1Citation48, THN10, THN6CCitation49, MF5, and CM3159B, were synthesized in two steps according to the method previously describedCitation47: first, condensation between 3-(isopropylamino)cyclohex-2-en-1-one 2Citation47 and the corresponding substituted ninhydrin, then deoxygenation using tetraethylthionylamide (TETA) ().
Use of dimethoxyninhydrins CM3141B and CM3186B afforded the dihydroxydimethoxyindeno[1,2-b]indole-9,10-diones CM3159A and MF4, respectively, which were easily deoxygenated to give the expected dimethoxyindenoindoles CM3159B and MF5.
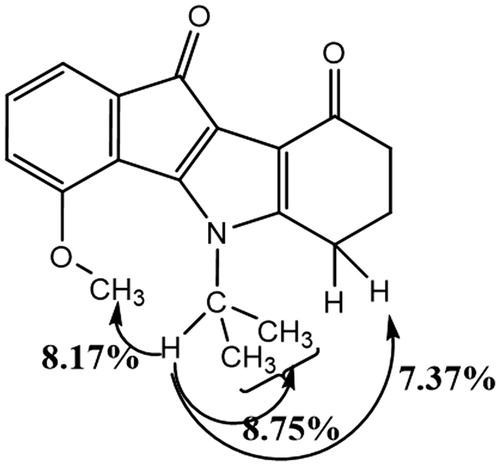
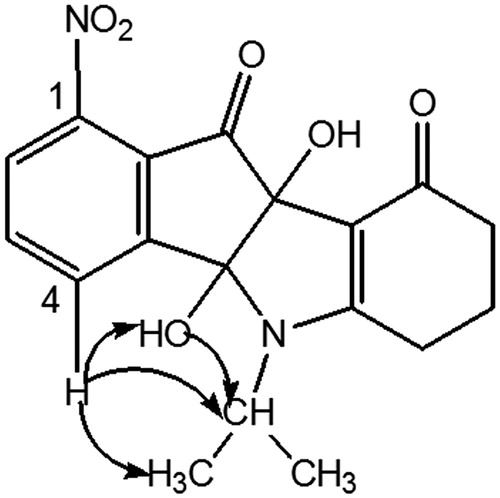
Condensation of enaminone 2 with 4-methoxyninhydrin CM3188BCitation40 led to a mixture of the two regiosiomers 1- and 4-methoxy (ratio 69/31) which could not be separated under classical conditions. After deoxygenation of the mixture, the two regioisomers 1- and 4-methoxyindenoindoles were easily separated by column chromatography and their assignment established by NMR experiments. NOE experiment for the regioisomer CM3072B () showed a correlation between the CH of the isopropyl group and the methoxy group.
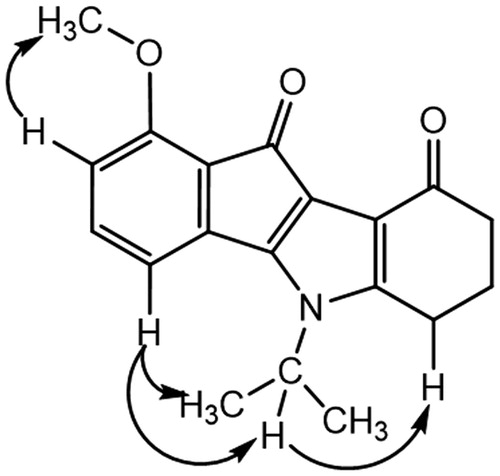
This correlation was not observed for the other regioisomer, compound THN10. NOESY experiment for this latter showed correlations () between the CH of the isopropyl group and an aromatic proton (H-4), confirming unambiguously the assignment.
Amino derivative MF1 was prepared from 4-nitroninhydrin CM3020BCitation40. The first coupling step led to the single 1-nitro regioisomer AM10A. Assignment was established by NMR experiments: 2 D NMR HSQC and 1H-13 C HMBC correlations, using the characteristic long-range 3J and 4J couplings and NOESY. All of the cross couplings observed are reported in the , and the decisive correlations for the structural assignment are summarized in .
Table 3. 2 D 1H–13 C HMBC correlations for AM10A (DMSO, 500.13 MHz).
The carbonyls C-9 and C-10 were firstly attributed due to the 3J and 4J couplings between the protons 6-H and 7-H with C-9 on the one hand, and OH-9 b and 2-H with C-10 on the other hand. Then assignment of the C-4 b and C-9 b was allowed. In fact, only the OH-9 b proton correlated with C-10 (3JH,C). Next, the cross couplings found in the NOESY between the protons 4-H with OH-4 b and the CH of isopropyl group allowed us to identify unambiguously the proposed regiochemistry (). This derivative AM10A was then dedihydroxylated with TETA to afford AM12B. Finally, the nitro group was reduced into amine MF1 using iron in acidic conditionsCitation50.
Detailed analysis of effects of the tested 15 compounds on bacterial growth revealed various activities of different substances. Induction of the lysogenic bacterial culture with mitomycin C caused prophage induction and subsequent lysis of the host cells, seen in as a decrease in the cell culture optical density.
Figure 6. Growth of E. coli MG1655 lysogenic with Φ24BΔstx2::cat at 37 °C in LB medium after induction with 0.5 µg/ml mitomycin C (added to the culture at time 3 h) in the absence or presence of tested compounds at indicated concentrations (added to the culture at time 0). Bacterial growth was monitored by measurement of A600 at indicated times. Presented results are mean values from three experiments with SD indicated as error bars.
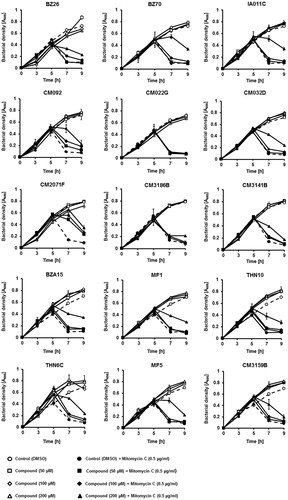
However, majority of the selected compounds (BZ26, BZ70, IA011C, CM092, CM032D, CM2071F, CM3141B, BZA15, MF1, THN10, THN6C, MF5, CM3159B) ameliorated this effect, preventing bacterial cell lysis after treatment with mitomycin C, in at least one used concentration (). These results suggested that such compounds might prevent either effective prophage induction or phage lytic development. Hence, we have measured titers of bacteriophages in cultures treated with mitomycin C with and without the presence of tested compounds. In addition, hydrogen peroxide was used as an inducing agent, as this substance was found previously as a prophage inducer occurring naturally in human gut. Contrary to mitomycin C, this inducer did not cause detectable changes in the density of bacterial culture (data not shown). In mitomycin C-treated lysogenic E. coli cell culture, among the 15 selected compounds, the presence of most of them in the culture caused a significant decrease in the titer of bacteriophage (). Importantly, in hydrogen peroxide-treated cells, the effects of these compounds on phage titer were dramatic ().
Figure 7. Relative phage titer in cultures of E. coli MG1655 lysogenic with Φ24BΔstx2::cat treated with 0.5 µg/ml mitomycin C (A) or 1 mM H2O2 (B) (inducers were added to the culture at time 3 h) in the absence (control experiments) or presence of tested compounds at indicted concentrations (added to the culture at time 0). Presented results are mean values from three experiments with SD indicated as error bars.
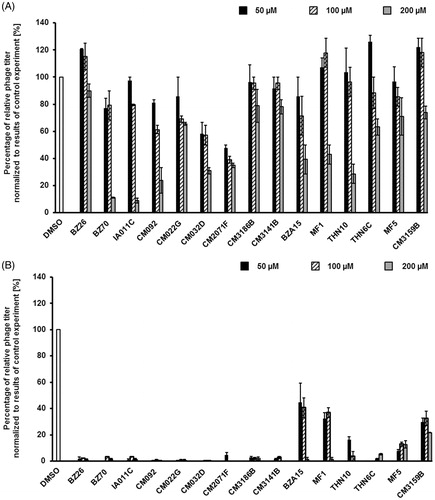
These results indicate that in the presence of compounds BZ26, BZ70, IA011C, CM092, CM022G, CM032D, CM2071F, CM3186B, CM3141B, THN6C, Φ24B prophage induction and/or phage lytic development is/are impaired. This suggest that at least some of them might be considered as potential anti-STEC drugs.
Since any potential therapeutic must be safe for humans, we have tested the selected 15 compounds for toxicity to human cells. Model HEK-293 and HDFa cells were treated with these compounds for 48 h in cultures, and their viability was assessed in the MTT test ().
Figure 8. Viability of human HEK-293 and HDFa cells in cultures treated with tested compounds at indicated concentrations for 48 h. Cell viability was tested in the MTT test. Presented results are mean values from three experiments with SD indicated as error bars. The significance of differences between control and cells treated with tested compounds was assessed by the ANOVA test. Differences were marked by asterisks (*) and considered significant when the p value was <0.05.
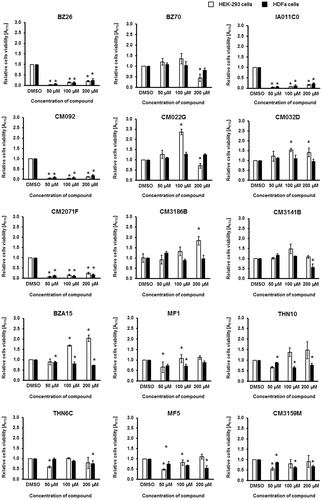
Compounds BZ26, IA011C, CM092, and CM2071F were evidently toxic to human cells which precludes their use as therapeutics (). Among the rest, there were substances neutral for human cells’ viability or those even stimulating their growth (compounds: BZ70, CM022G, CM032D, CM3186B, CM3141B, BZA15, MF1, THN10, THN6C, MF5, and CM3159B) ().
To learn about molecular mechanisms of impairment of prophage induction and/or phage lytic development, expression of selected phage and host genes was monitored quantitatively by using qRT-PCR. Three compounds belonging to different groups of human cell growth inhibitors (CM092), and neutral or weakly stimulating agents (CM032D, CM3186B), were chosen for further investigations ().
Figure 9. Expression of bacterial genes coding for proteins involved in the oxidative stress response and of selected bacteriophage genes in E. coli MG1655 lysogenic with Φ24BΔstx2::cat either non-treated (A) or after induction with 1 mM H2O2 (B) in the absence (control experiments) or presence of tested compounds added to final concentration of 0.2 mM. Levels of mRNAs were determined by RT-qPCR. Presented results are mean values from three experiments with SD indicated as error bars.
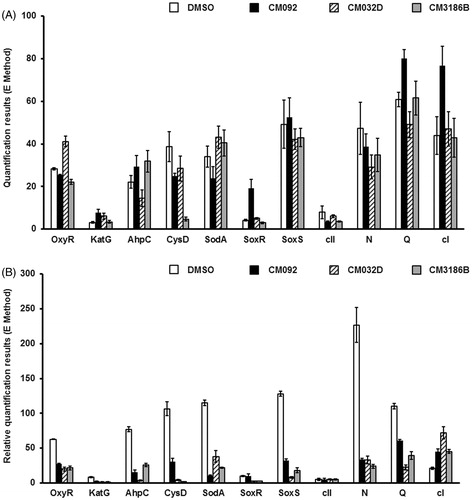
In the absence of the prophage inducing agent, the influence of compounds CM092, CM032D, and CM3186B on expression of bacterial genes involved in the oxidative stress response, as well as of bacteriophage genes crucial for either lytic (N, Q) or lysogenic (cI, cII) development was negligible (). However, addition of hydrogen peroxide caused activation of the host genes coding for the oxidative stress proteins, and phage genes coding for antitermination proteins (as expected), while having little effects on phage lysogenic proteins (also as expected) (). Importantly, simultaneous presence of hydrogen peroxide and any of the tested compounds caused a significant decrease of the efficiency of expression of the oxidative stress genes as well as phage genes (N, Q) supporting the lytic development (). In contrast to these genes, expression of the gene coding for the cI repressor was enhanced under these conditions. These results suggest that compounds CM092, CM032D, and CM3186B impair expression of bacterial genes involved in the oxidative stress response, or ameliorate the effects of oxidative stress. This leads to prevention of the prophage induction by hydrogen peroxide.
Discussion and conclusions
Infections by STEC strains are particularly dangerous due to both production of strong toxins (Shiga toxins) by these pathogens and unavailability of the use of antibiotic therapy because these antibacterial compounds may induce Stx prophage leading to enhanced toxin synthesis and releaseCitation2,Citation3. Stx prophage induction is required for effective expression of stx genes and production of Shiga toxinsCitation2,Citation17. Therefore, compounds which interfere with prophage induction might be potential anti-STEC agents. Since it was previously demonstrated that oxidative stress conditions are likely responsible for Stx prophage induction in human intestine infected with STEC (summarized inCitation8), we asked whether compounds revealing antioxidant features might be effective in inhibition of Stx prophage induction, thus, diminishing pathogenicity of STEC strains.
We have tested 46 compounds, derivatives of carbazole, indazole, triazole, quinolone, ninhydrine, and indenoindole, and after preliminary experiments, 15 of them were chosen for more detailed analysis. These chemicals prevented or alleviated induction of Stx prophages by hydrogen peroxide, thus, revealing an anti-STEC-pathogenicity potential. Importantly, out of 15 tested compounds, eleven had acceptable profiles in the cytotoxicity MTT test, i.e. they did not cause a decrease in viability of two human cell lines (HEK-293 and HDFa). These might be considered for further studies as potential anti-STEC drugs.
Studies on the mechanism of action of the selected compounds, CM092, CM032D, and CM3186B, indicated that in E. coli lysogenic with an Stx phage they had generally no significant effects on expression of tested host and phage genes under standard growth conditions. However, after addition of hydrogen peroxide to the bacterial culture, all three tested compounds caused drastic decrease in efficiency of expression of phage genes involved in the lytic development and an increase in expression of the cI gene, coding for the phage repressor, responsible for prophage maintenance. These results demonstrate that CM092, CM032D, and CM3186B prevent the prophage induction at the level of expression of specific phage genes. This may arise from interference with the oxidative stress, as in cells treated with hydrogen peroxide, expression of genes involved in the oxidative stress response was significantly less efficient in the presence of the tested compounds. Therefore, the tested compounds apparently reduce the oxidative stress in the presence of hydrogen peroxide, which prevents induction of Stx prophage in E. coli.
In conclusion, several compounds tested in this study, particularly BZ70, CM022G, CM032D, CM3186B, CM3141B, BZA15, MF1, THN10, THN6C, MF5, CM3159B, might be considered as potential anti-STEC drugs, and qualified for further studies. At least some of them, CM092, CM032D, and CM3186B, act by reduction of the oxidative stress, changes in gene expression and prevention of Stx prophage induction.
IENZ_1444610_Supplementary_Material.pdf
Download PDF (1.7 MB)Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Riley LW, Remis RS, Helgerson SD, et al. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N Engl J Med 1983;308:681–5.
- Muniesa M, Hammerl JA, Hertwig S, et al. Shiga toxin-producing Escherichia coli O104:H4: a new challenge for microbiology. Appl Environ Microbiol 2012;78:4065–73.
- Bloch SK, Felczykowska A, Nejman-Faleńczyk B. Escherichia coli O104:H4 outbreak-have we learnt a lesson from it? Acta Biochim Pol 2012;59:483–8.
- Nataro JP, Kaper JB. Diarrheagenic Escherichia coli. Clin Microbiol Rev 1998;11:142–201.
- Mizutani S, Nakazono N, Sugino Y. The so-called chromosomal verotoxin genes are actually carried by defective prophages. DNA Res 1999;6:141–3.
- Schmidt H. Shiga-toxin-converting bacteriophages. Res Microbiol 2001;152:687–95.
- Węgrzyn G, Licznerska K, Węgrzyn A. Phage λ–new insights into regulatory circuits. Adv Virus Res 2012;82:155–78.
- Licznerska K, Nejman-Faleńczyk B, Bloch S, et al. Oxidative stress in Shiga toxin production by enterohemorrhagic Escherichia coli. Oxid Med Cell Longev 2016;2016:3578368.
- Imamovic L, Muniesa M. Characterizing RecA-independent induction of Shiga toxin2-encoding phages by EDTA treatment. PLoS One 2012;7:e32393.
- Yamamoto T, Kojio S, Taneike I, et al. 60Co irradiation of Shiga toxin (Stx)-producing Escherichia coli induces Stx phage. FEMS Microbiol Lett 2003;222:115–21.
- Aertsen A, Faster D, Michiels CW. Induction of Shiga toxin-converting prophage in Escherichia coli by high hydrostatic pressure. Appl Environ Microbiol 2005;71:1155–62.
- Wagner PL, Acheson DWK, Waldor MK. Human neutrophils and their products induce Shiga toxin production by enterohemorrhagic Escherichia coli. Infect Immun 2001;69:1934–7.
- Łoś JM, Łoś M, Węgrzyn A, Węgrzyn G. Hydrogen peroxide-mediated induction of the Shiga toxin-converting lambdoid prophages ST2-8624 in Escherichia coli O157:H7. FEMS Immunol Med Microbiol 2010;58:322–9.
- Licznerska K, Dydecka A, Bloch S, et al. The role of the exo-xis region in oxidative stress-mediated induction of Shiga toxin-converting prophages. Oxid Med Cell Longev 2016;2016:8453135.
- Aruoma OI, Halliwell B, Gajewski E, Dizdaroglu M. Damage to the bases in DNA induced by hydrogen peroxide and ferric ion chelates. J Biol Chem 1989; 264:20509–12.
- Mauro SA, Koudelka GB. Shiga toxin: expression, distribution, and its role in the environment. Toxins (Basel) 2011;3:608–25.
- Łoś JM, Łoś M, Węgrzyn G. Bacteriophages carrying Shiga toxin genes: genomic variations, detection and potential treatment of pathogenic bacteria. Future Microbiol 2011;6:909–24.
- Semchyshyn H, Bagnyukova T, Storey K, Lushchak V. Hydrogen peroxide increases the activities of soxRS regulon enzymes and the levels of oxidized proteins and lipids in Escherichia coli. Cell Biol Int 2005;9:898–902.
- González-Flecha B, Demple B. Role for the oxyS gene in regulation of intracellular hydrogen peroxide in Escherichia coli. J Bacteriol 1999;181:3833–6.
- Vasil'eva SV, Stupakova MV, Lobysheva II, et al. Activation of the Escherichia coli SoxRS-regulon by nitric oxide and its physiological donors. Biochemistry Mosc 2001;66:984–8.
- Marshall HE, Merchant K, Stamler JS. Nitrosation and oxidation in the regulation of gene expression. Faseb J 2000;14:1889–900.
- Mukhopadhyay P, Zheng M, Bedzyk LA, et al. Prominent roles of the NorR and Fur regulators in the Escherichia coli transcriptional response to reactive nitrogen species. Proc Natl Acad Sci USA 2004;101:745–50.
- Nakano MM. Induction of ResDE-dependent gene expression in Bacillus subtilis in response to nitric oxide and nitrosative stress. J Bacteriol 2002;184:1783–7.
- Hausladen A, Privalle CT, Keng T, et al. Nitrosative stress: activation of the transcription factor OxyR. Cell 1996;86:719–29.
- Storz G, Tartaglia LA, Ames BN. Transcriptional regulator of oxidative stress-inducible genes: direct activation by oxidation. Science 1990;248:189–94.
- Kullik I, Toledano MB, Tartaglia LA, Storz G. Mutational analysis of the redox-sensitive transcriptional regulator OxyR: regions important for oxidation and transcriptional activation. J Bacteriol 1995;177:1275–84.
- Kim SO, Merchant K, Nudelman R, et al. OxyR: a molecular code for redox-related signaling. Cell 2002;109:383–96.
- Vasil'eva SV, Stupakova MV, Lobysheva II, et al. Activation of the Escherichia coli SoxRS-regulon by nitric oxide and its physiological donors. Biochemistry (Mosc) 2001;66:984–8.
- D'Autreaux B, Touati D, Bersch B, et al. Direct inhibition by nitric oxide of the transcriptional ferric uptake regulation protein via nitrosylation of the iron. Proc Natl Acad Sci USA 2002;99:16619–24.
- Glinkowska M, Łoś JM, Szambowska A, et al. Influence of the Escherichia coli oxyR gene function on lambda prophage maintenance. Arch Microbiol 2010;192:673–83.
- Chohan ZH, Shaikh AU, Naseer MM, Supuran CT. In-vitro antibacterial, antifungal and cytotoxic properties of metal-based furanyl derived sulfonamides. J Enzyme Inhib Med Chem 2006;21:771–81.
- Del Prete S, Perfetto R, Rossi M, et al. A one-step procedure for immobilising the thermostable carbonic anhydrase (SspCA) on the surface membrane of Escherichia coli. J Enzyme Inhib Med Chem 2017;32:1120–8.
- Perfetto R, Del Prete S, Vullo D, et al. Production and covalent immobilisation of the recombinant bacterial carbonic anhydrase (SspCA) onto magnetic nanoparticles. J Enzyme Inhib Med Chem 2017;32:759–66.
- Jensen KF. The Escherichia coli K-12 “wild types” W3110 and MG1655 have an rph frameshift mutation that leads to pyrimidine starvation due to low pyrE expression levels. J Bacteriol 1993;175:3401–7.
- Allison HE. Immunity profiles of wild-type and recombinant Shiga-like toxin-encoding bacteriophages and characterization of novel double lysogens. Infect Immun 2003;71:3409–18.
- Łoś JM, Golec P, Węgrzyn G, et al. Simple method for plating Escherichia coli bacteriophages forming very small plaques or no plaques under standard conditions. Appl Environ Microbiol 2008;74:5113–20.
- Bloch S, Nejman-Faleńczyk B, Dydecka A, et al. Different expression patterns of genes from the exo-xis region of bacteriophage λ and Shiga toxin-converting bacteriophage Ф24B following infection or prophage induction in Escherichia coli. PLoS One 2014;9:e108233.
- Nejman-Faleńczyk B, Bloch S, Licznerska K, et al. A small, microRNA-size, ribonucleic acid regulating gene expression and development of Shiga toxin-converting bacteriophage Ф24B. Sci Rep 2015;5:10080.
- Nowicki D, Bloch S, Nejman-Faleńczyk B, et al. Defects in RNA polyadenylation impair both lysogenization by and lytic development of Shiga toxin-converting bacteriophages. J Gen Virol 2015;96:1957–68.
- Marminon C, Gentili J, Barret R, Nebois P. Synthesis of N-benzylated indole-, indazole- and benzotriazole-4,7-diones. Tetrahedron 2007;63:735–9.
- Zeinyeh W, Xia H, Lawton P, et al. Synthesis and modulation properties of imidazo[4,5-b]pyridin-7-one and indazole-4,7-dione derivatives towards the Cryptosporidium parvum CpABC3 transporter. Eur J Med Chem 2010;45:2480–8.
- Marminon C, Fenet B, Mignosi V, et al. Diels-Alder reactions between acrolein N,N-dimethylhydrazone and N-benzylated benzotriazole-, indazole- or indole-4,7-diones. Heterocycles 2009;78:2799–809.
- Saxon E, Luchansky SJ, Hang HC, et al. Investigating cellular metabolism of synthetic azidosugars with the Staudinger ligation. J Am Chem Soc 2002;124:14893–902.
- Compain-Batissou M, Latreche D, Gentili J, et al. Synthesis and Diels-Alder reactivity of ortho-carbazolequinones. Chem Pharm Bull (Tokyo) 2004;52:1114–16.
- Issa S, Walchshofer N, Kassab I, et al. Synthesis and antiproliferative activity of oxazinocarbazole and N,N-bis (carbazolylmethyl) amine derivatives. Eur J Med Chem 2010;45:2567–77.
- Marminon C, Nacereddine A, Bouaziz Z, et al. Microwave-assisted oxidation of indan-1-ones into ninhydrines. Tetrahedron Lett 2015;56:1840–2.
- Hemmerling HJ, Reiss G. Partially saturated indeno[1,2-b]indole derivatives via deoxygenation of heterocyclic α-hydroxy-N,O-hemiaminals. Synthesis 2009;2009:985–99.
- Perret F, Marminon C, Zeinyeh W, et al. Preparation and characterization of CK2 inhibitor-loaded cyclodextrin nanoparticles for drug delivery. Int J Pharm 2013;441:491–8.
- Jabor Gozzi G, Bouaziz Z, Winter E, et al. Converting potent indeno[1,2-b]indole inhibitors of protein kinase CK2 into selective inhibitors of the breast cancer resistance protein ABCG2. J Med Chem 2015;58:265–77.
- Alcalde E, Mesquida N, Frigola J, et al. Indene-based scaffolds. Design and synthesis of novel serotonin 5-HT6 receptor ligands. Org Biomol Chem 2008;6:3795–810.