Abstract
Oregonin is an open-chain diarylheptanoid isolated from Alnus incana bark that possesses remarkable antioxidant and anti-inflammatory properties, inhibits adipogenesis, and can be used in the prevention of obesity and related metabolic disorders. Here, we aimed to investigate the effects of oregonin on the epigenetic regulation in cells as well as its ability to modulate DNA methylating enzymes expression and mitochondrial DNA (mtDNA) copies. Our results show that oregonin altered the expression of DNA methyltransferases and mtDNA copy numbers in dependency on concentration and specificity of cells genotype. A close correlation between mtDNA copy numbers and mRNA expression of the mtDnmt1 and Dnmt3b was established. Moreover, molecular modeling suggested that oregonin fits the catalytic site of DNMT1 and partially overlaps with binding of the cofactor. These findings further extend the knowledge on oregonin, and elucidate for the first time its potential to affect the key players of the DNA methylation process, namely DNMTs transcripts and mtDNA.
1. Introduction
Oregonin 1,7-bis-(3,4-dihydroxyphenyl)-3- hydroxyheptane-5-O-b-D-xylopyranoside () is an open-chain diarylheptanoid glycoside that contains 3-carbonyl and 5-xylosyloxy groups, and is the predominant compound in the hydrophilic extracts from the Alnus incana barkCitation1,Citation2. This natural compound possesses noticeable anti-oxidative and anti-inflammatory properties, inhibits the adipogenesis and can be used in the prevention of obesity and related metabolic disordersCitation3–8.
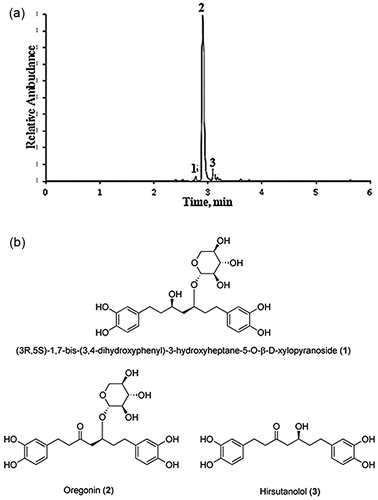
Figure 1. (a) Base peak ion chromatogram of purified (90% oregonin) alder bark extracts obtained by negative ion UPLC-ESI-MS. Oregonin (2), (3R,5S)-1,7-bis-(3,4-dihydroxyphenyl)-3-hydroxyheptane-5-O-β-d-xylopyranoside (1) and hirsutanolol (3). (b) Chemical structure of compounds 1–3.
The effects of oregonin, isolated from the bark of grey alders growing in Latvia, have been tested in vitro on blood samples from volunteers with clinically confirmed metabolic syndromes. The total and low-density cholesterol as well as the level of triacylglycerols is reduced by 30%, 20% and 30%, respectively, after incubation with alder bark ethyl acetate extract containing 60% of oregonin or with purified oregonin for 30 minCitation1. The effect has also been confirmed in white rats in vivoCitation1,Citation9. Pure oregonin from A. incana increases glutathione peroxidase and superoxide dismutase activity, and decreases the activity of lipase, catalase and malondialdehyde level (indicator of lipid peroxidation) in blood in vivoCitation1,Citation9. Also, it reduces lipid accumulation, inflammation and ROS production in primary human macrophages, indicating its anti-inflammatory activityCitation10.
Oregonin shares chemical features and substructure with the well-known dietary antioxidant curcumin, which is widely used as a bioactive constituent of food supplements. Some studiesCitation9,Citation11 have shown that oregonin expressed higher antioxidant activity and some others biological activities, particularly antibacterial and antifungal, than curcumin. While curcumin potential role in the epigenetic changes is describedCitation12,Citation13, to the best of our knowledge no reports on the effects of oregonin from A. incana on the epigenetic regulation in cells are available to date.
Epigenetic changes cover altered DNA methylation, histone modification, non-coding RNAs and chromatin remodelingCitation14,Citation15. DNA methylation is regulated by enzymes belonging to the DNA methyltransferases (DNMTs) family, which transfer methyl groups from S-adenosyl-l-methionine (SAM) to the 5-position of the cytosine pyrimidine ringCitation16. Hyper-methylation of DNA often occurs in gene-specific promoters mostly in gene-rich genomic regions (CpG-islands) and can lead to the inhibition of gene expression (gene silencing), while hypo-methylation is observed in repetitive elements all over the genomeCitation14,Citation17. Increasing evidences are accumulating on the importance of mitochondria in the epigenetic regulation of cells. It is known that mitochondria are able to influence cytosine methylation levels in the nucleus by modulating the flux of one-carbon units for the generation of SAM, the methyl donor in DNA methylationCitation18. As reported by Smiraglia et al.Citation19, many genes undergo changes in their methylation status in response to the depletion and repletion of mtDNA. Mitochondrial dysfunction could cause alterations in redox (reduced oxygen) reactions and SAM-CH3 production in the cell, leading to perturbed methylation of nuclear encoded genomic DNACitation18.
A strong correlation between epigenetic alterations, including mitochondrial factors, and human diseases, especially cancer, has been observedCitation19–21. Experimental evidences have pointed that polyphenols have great potential as epigenetic modulators, influencing various epigenetic factors such as histone acetyltransferasesCitation22, DNTMs and miRNAsCitation23–26. These findingsCitation13,Citation27–29 have increased the enthusiasm in developing therapeutic strategies for many diseases through targeting the various epigenetic factors by polyphenols. Accordingly, the identification of new natural and nontoxic polyphenols, able to affect reversibly the epigenetic deregulation, is nowadays a well-established and promising scientific topic.
Using a combination of biological and computational modeling tools, the present study aims to investigate whether the isolated and purified polyphenol oregonin from A. incana bark can affect the key players of the DNA methylation process, namely DNA methylating enzymes and mitochondrial DNA (mtDNA) in mouse embryonic fibroblasts (MEFs).
2. Materials and methods
2.1. Oregonin extraction
Bark of A. incana (Grey alder) was collected in November 2015 from trees growing in Latvia. Diarylheptanoids were obtained using accelerated solvent extraction with ethyl acetate after removal of lipophilic extractives (EtOAc). The bark was extracted with pressurised ethyl acetate in an ASE 350 apparatus (Dionex, Sunnyvale, CA) by applying 3 × 5 min static cycles at 90 °C and 10.3 MPa. The solvent was evaporated in vacuo, the extract was freeze-dried and stored at −20 °C.
Analysis of A. incana extract was performed on ACQUITY UPLC H-Class (Waters, Singapore) with ACQUITY UPLC PDA Detector coupled with SYNAPT G2-Si High Definition Mass Spectrometry (Waters, Milford, Massachusetts, MA, USA). Column was Waters ACQUITY UPLC BEH C18, 2.1 × 50 mm, 1.7 µm, flow 0.5 ml/min, Mobile phase A: waters +0.1% formic acid, B: acetonitrile, gradient from 10% A to 100% B, injection 2 µL. Mass range (50–1200 Da), ESI negative mode, cone voltage 40 V, scan time 0.1 s, desolvation gas nitrogen (600 L/h), source temperature at 120 °C.
Oregonin was purified from alder bark EtOAc extract by SP1™ Purification System using Biotage column KP-C18-HS (12 × 150 mm, 35–70 μm) with solvent systems: solvent A = ethanol/water/acetic acid (200: 790: 10, v/v) and solvent B = ethanol. The gradient was from 0 to 25% solvent B. Identification and purity state of oregonin was determined using UPLC-PDA-MS/MS, NMR methods. The proof quality is assessed by comparison to the oregonin reference standard [≥95% (LC/MS-ELSD)] purchased from Sigma-Aldrich (Saint Louis, MA, USA). For the investigation of antioxidant activity, the methods of the deactivation of 2,2-diphenyl-1-picrylhydrazyl (DPPH•) and 2,2′-azinobis-(3-ethylbenzothiazoline-6-sulfonic acid (ABTS•+) radicals were applied.
2.2. Animals
The animals used for obtaining the mouse embryonic cells were reared and handled in vivarium of the IBIR-BAS in accordance with Bulgarian Veterinary Law (25/01/2011) regarding the life conditions and welfare of experimental animals, which is adapted to the European Union regulation 86/609 and Directive 2010/63/EU. The experimental protocols were approved by the National Ethics commission for animals (“Permission for using animals in the experiments”, N84/04.10.2013, expiry date 04.10.2018). The individual responsible for the animal welfare, which works directly with the animals used in the study, is licensed by the Bulgarian veterinary services for animal experimentation. Animals were anesthetised before sacrificing them. All efforts were made to minimise suffering.
2.3. Cell cultures
MEFs were prepared from 13- to 14-day-old embryos of ICR mice. After dissection of the red organs, the tissues were minced and digested with 0.25% trypsin/EDTA (Sigma-Aldrich, Saint Louis, MA, USA). Dissociated cells were grown in monolayers at 37 °C in a humidified atmosphere of 5% CO2 and 95% air using Dulbecco’s Modified Eagle Medium (DMEM) with 1.0 g/L glucose (PAN-Biotech, Aidenbach, Germany) as the culture medium. DMEM was supplemented with 10% fetal bovine serum (Sigma-Aldrich , Saint Louis, MA, USA) and penicillin, streptomycin, amphotericin B mix (PAN-Biotech, Aidenbach, Germany). Mouse embryo fibroblast cell line, NIH/3T3, was obtained from the American-Type Culture Collection (Manassas, VA, USA). Cells were cultured under the conditions used for MEFs.
2.4. Cell viability assay
Cell viability was estimated by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (Sigma-Aldrich, Saint Louis, MA, USA), which is based on the cleavage of a tetrazolium salt by mitochondrial dehydrogenases in viable cells. NIH/3T3 cells and MEFs were seeded in sterile flat bottom 96-well plates at a density of approximately 2 × 104 cells/well and allowed to attach overnight at 37 °C. The cells were then exposed to 50 and 100 μM oregonin dissolved in culture medium for 24 and 48 h. Untreated cells and blank wells (without cells) received 100 μl of the culture medium. After incubation with oregonin, the culture medium was removed and 100 μl of MTT at the concentration of 500 μg/ml in culture medium was added to the wells. Cells were incubated in a humidified CO2 incubator for another 3 h. Then, the MTT solution was removed and 100 μl of dimethyl sulfoxide (DMSO, Merck, Darmstadt, Germany) was added to dissolve formazan crystals. Absorbance at 544 nm was monitored with a plate reader (FLUOstar Optima, BMG Labtech, Offenburg, Germany) to obtain the number of viable cells relative to the control population. Data are expressed as mean values ± SD and obtained from three different experiments against each cell type.
2.5. Quantitative real-time PCR (qRT-PCR) assay
Fibroblasts were plated in sterile six-well plates at a density 2 × 105 cells/well and were left to adhere overnight. Cells were treated with 50 and 100 μM oregonin in culture medium for 24 h. The samples were collected and subjected by TriReagent (Sigma-Aldrich, Saint Louis, MA, USA) in accordance with producers’ instruction for total RNA and genomic DNA extractions.
2.5.1. Dnmts mRNA expression
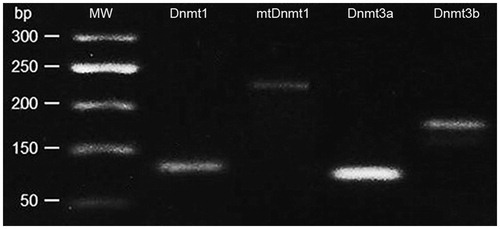
The extracted RNA was used for the synthesis of the first cDNA strands using cDNA synthesis kit (Thermo Fisher Scientific, Waltham, CA, USA). qRT-PCR was performed on Dnmt1; mtDnmt1, Dnmt3a and Dnmt3b genes using SYBR Green PCR Master Mix (Life Technologies, Waltham, CA, USA). The primers are presented in . β-actin was selected as a housekeeping gene.
Table 1. Primer sequences for mouse Dnmt genes and β-actin.
All primers were obtained from Sigma-Aldrich, Saint Louis, MA, USA. PCR analysis was performed using a two-step cycling protocol, which was optimised under the following conditions: cycles consisted of a 10-min initial denaturation phase at 95 °C, a 15-s denaturation phase at 95 °C and a 20-s annealing phase based on the Tm of the primers (55–60 °C) with the last two steps repeated for a total of 40 cycles. A melting curve was made after cycling by changing the temperature from 60 to 90 °C and read and recorded every 1 °C. The process was implemented on Mastercycler® ep realplex (Eppendorf, Hamburg, Germany). The presence of all Dnmts forms in the PCR product was confirmed by gel electrophoresis ().
2.5.2. mtDNA copy number evaluation
The purity and concentration of extracted DNA was measured using Nano Drop 1000 (Thermo Fisher Scientific, Waltham, MA USA). A ratiometric assayCitation30 of the levels of a single mitochondrial gene cytochrome B (Cytb) against a single copy nuclear gene (β-actin) was used to estimate the average copy value of mtDNA/per cell. This technique allows the calculation of average mtDNA copy number/cell without the necessity to estimate cell number. Primers and probes were obtained from Sigma-Aldrich, Saint Louis, MA, USA on the base of sequences mouse genes used by Aiken et al.Citation31 (). Probes were labelled fluorescently with carboxyfluorescein (FAM) (5′ end) and carboxytetramethylrhodamine (TAMRA) (3′ end). TaqMan Master Mix (Applied Biosystems, Foster City, CA, USA) was used for the reaction. The PCR conditions were following: initial steps at 50 °C for 2 min and 95 °C for 15 min, followed by 40 cycles of 15 s at 95 °C and 1 min at 65 °C. Standard curves were created separately for every sample from a dilution series to ensure the efficiency of reaction. The cycle threshold (CT) value for β-actin was subtracted from that for Cytb to give the value ΔCt. Average mtDNA copy numberCitation30 per nuclear genome (two actin gene copies) is calculated as 2 × 2−(ΔCt).
Table 2. Primers and probes for mtDNA copy analysis.
2.6. Statistical development of the results
All analyses were performed with Statistica software (StatSoft10, Tulsa, OK, USA). Student’s t-test was used for comparison between groups. Pearson’s correlation test was applied to measure the relationship between investigated parameters. Values of p < .05 were considered significant. Data are presented as a mean ± SD, and triplicate repetition of the experiments was done.
PCR data were analysed by DataAssist Sft. (Ver. 3.01, Applied Biosystems, Foster City, CA, USA) using ΔCt methodCitation32. The results were expressed as the mean of target to control gene expression ratio [fold changes (FC)]. For the evaluation of the oregonin effect, ΔΔCt methodCitation32 was applied for the comparison between control and experimental groups. Genes with fold change cutoff of ≥ 2 and p values ≤ 0.05 compared with the control were considered to be upregulated, while with FC ≤ 0.5 and p ≤ 0.05 were considered to be downregulated. FC values between 0.5 and 2 indicated no significant change in gene expression.
2.7. Molecular modelling
The possible binding mode of oregonin to DNMT1 was predicted by molecular docking. The crystallographic structure of human DNMT1 in complex with S-adenosyl-l-homocysteine (SAH) coded by PDB 4WXXCitation33 was retrieved from the Protein Data Bank (www.rcsb.org/pdb) and used as rigid receptor. As in our previous workCitation34, molecular docking simulations were performed by FRED program from OpenEye (version 3.0.1)Citation35,Citation36. The receptor was prepared with the make_receptor utility by centring the binding site on the SAH ligand. The 3D structures of curcumin (reference control) and oregonin were downloaded from PubChemCitation37. Ligands conformational analysis was carried out with OMEGA from OpenEye (version 2.5.1.4)Citation38,Citation39 by storing up to 600 conformers including the initial structure, sampling hydrogen positions for –OH groups and using all other parameters at their default values.
3. Results
3.1. Oregonin
The lyophilic dried oregonin isolated from A. incana bark EtOAc extract and used in all investigations contained 90 ± 0.8% of oregonin (2) (C24H30O10, UV λmax 281 nm. ES-: m/z 477 [M-H]−, M = 478.50 g/mol, 5 ± 0.1% of 1,7-bis-(3,4-dihydroxyphenyl)-3-hydroxyheptane-5-O-β-d-xylopyranoside (1) (C24H32O10, UV λmax 282 nm. ES-: m/z 479 [M-H]−, M = 480.52 g/mol) and 4 ± 0.1% of hirsutanolol (3) (C19H22O6, UV λmax 281 nm. ES-: m/z 345 [M-H]−, M = 346.38 g/mol) ().
The oregonin powder from A. incana showed high antioxidant activity. The following IC50 concentrations (required to scavenge 50% of free radicals) were reported: in ABTS•+ 3.14 ± 0.13 mg·mL−1 and in DPPH• 4.52 ± 0.16 mg·mL−1 respectively compared to synthetic antioxidant trolox (a water-soluble vitamin E analogue), whose IC50 values in tests were 4.01 ± 0.12 (ABTS) mg·mL−1 and 4.72 ± 0.12 (DPPH) mg·mL−1.
3.2. Effect of oregonin on viability of cells in vitro
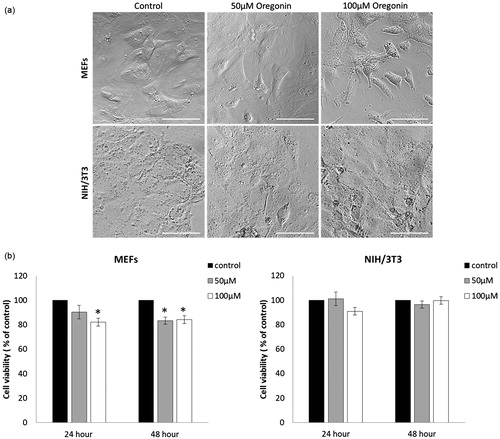
Untreated control MEFs exhibited normal fibroblast morphology at 48 h (). Upon incubation of MEFs with increasing concentrations of oregonin for 48 h, apoptotic morphological changes in a few cells were observed. The shapes of the oregonin treated NIH/3T3 were similar to those of the untreated cells. The potency of oregonin to reduce cell viability of MEFs and NIH/3T3 is shown in . After 24 h treatment with 100 µM oregonin, MEFs viability was decreased by 18% (t-test p < .05). After 48 h of oregonin treatment in concentrations 50 and 100 µM, MEFs viability declined significantly by 16.7% and 15.8%, respectively. Both concentration of oregonin showed no growth inhibiting effect in NIH/3T3 cells after treatment of 24 and 48 h.
3.3. Effect of oregonin on the Dnmts expression and mtDNA copy number
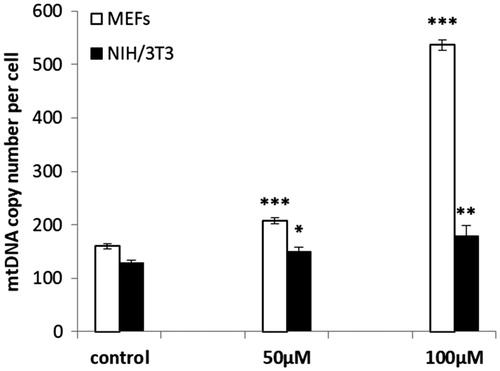
A significant increase of mtDNA copy numbers in oregonin treated MEF and NIH/3T3 cells was found (. The highest effect was observed in MEFs incubated with 100 μM oregonin (increase with 3.37 fold). The number of the mtDNA copy in NIH/3T3 cell at 50 μM and 100 μM increased only with 0.17 and 0.39 fold, respectively.
Figure 4. mtDNA copy numbers in MEF and NIH/3T3 cells treated with oregonin. The data are presented as a mean ± SD of three independent experiments. *p < .05; **p < .01; ***p < .001 compared to control.
Purified oregonin in dose 50 μM down-regulated mRNA transcripts of Dnmt1 in both NIH/3T3 and MEFs and had no effect on the Dnmt3a at 24 h of incubation ().
Figure 5. DNA methyltransferases mRNA expression in MEF (a) and NIH3T3 (b) cells incubated with oregonin in doses 50 and 100 μM for 24 h. The data are presented as a mean ± SD of three independent experiments. Fold change cut-off of ≥2 and p values ≤ 0.05 compared with the control were considered to be upregulated, while with FC ≤ 0.5 and p ≤ .05 were considered to be downregulated. *p < .05.
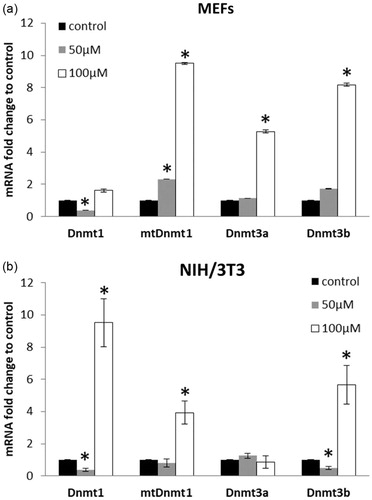
The mtDnmt1 transcripts in NIH/3T3 cells were unchanged (FC = 0.8) while in MEFs their level increased (FC = 2.31, p < .05). The Dnmt3b transcripts were unaffected in MEFs, but downregulated in NIH/3T3 cells. Oregonin in dose 100 μM up-regulated mtDnmt1 and Dnmt3b in both cell types (FC > 2.0, p < .05). MEFs treated with this concentration of oregonin showed increased expression of Dnmt3a, but had no effect on Dnmt1 mRNA transcripts. At the same experimental conditions (100 μM) in NIH3T3 cells, a drastic increase of Dnmt1 (FC = 9.51, p < .05) was observed, whereas no alterations were found on Dnmt3a mRNA transcripts.
The correlative analysis showed the different relationships between expressed Dnmts in MEF and NIH/3T3 cells. A high positive correlation was determined between mtDnmt1 and Dnmt3b as well as between Dnmt3b and Dnmt3a in MEFs (). In NIH/3T3 cells, we observed a similar relationship between mtDnmt1 and Dnmt3b. Additionally, the changes in the expression of total Dnmt1 closely correlated with changes in the expression of mtDnmt1 and Dnmt3b.
Table 3. Correlative relationship between expression of Dnmts in NIH/3T3 and MEF cells.
The interesting finding was the definition of the close relationship between mtDNA copy number and expression of mtDnmt1 and Dnmt3b in MEFs ().
Table 4. Correlation between mtDNA copy number and Dnmts expression in mouse fibroblasts.
3.4. Molecular docking of oregonin to DNMT1
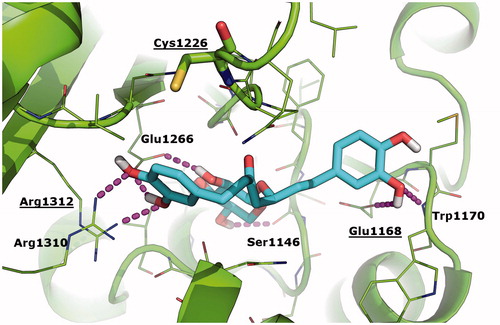
The work of Liu et al.Citation12 has clearly substantiated the possible covalent binding of curcumin to DNMT1 in correspondence of Cys1226 as a mechanism of hypomethylation. Based on the chemical similarity between curcumin and oregonin, and sustained by biological activity data described above, here we aimed to elucidate the possible binding of oregonin to DNMT1 and the respective pharmacophores. To this aim, we performed molecular docking simulations of curcumin (used as reference control) and oregonin towards the crystallographic structure of human DNMT1 in complex with SAHCitation33. Results of our study were nicely in agreement with the binding mode of curcumin predicted by Liu et al.Citation12 towards the DNMT1 homology model (data not shown), and suggested for the first time the geometric fitting of oregonin within the catalytic site of DNTM1 (. In particular, oregonin binds in proximity of the cofactor binding cleft of DNMT1 and partially overlaps with the crystallographic binding conformation of SAH. This interaction is reinforced by a number of H-bonds established with residues Ser1146, Glu1168, Trp1170, Glu1266, Arg1310, and Arg1312 (. The sugar moiety occupies a sub-pocket of DNMT1 and nicely overlaps with the homocysteine moiety of SAH, thus further suggesting that DNMT1 inhibition by oregonin may be due to the competition with the cofactor SAH, as already observedCitation34 for different type of epigenetics regulators. Similar to curcumin, oregonin does not interact directly with the key residue Cys1226, even though it is located in close proximity of oregonin in docking results (minimum distance between the thiol group of Cys1226 and oregonin is 3.7 Å). Moreover, oregonin establishes H-bonds with Glu1168 and Arg1312, which have been already highlighted as crucial anchor points for the binding of curcumin derivatives in the work of Liu et al.Citation12 Different from curcumin, oregonin does not possess chemically reactive groups that might interact covalently with Cys1226, thus the binding of oregonin to DNMT1 is expected to be non-covalent and reversible. Finally, the Chemgauss4 affinity score calculated by the docking program is slightly better for curcumin than oregonin (−11.25 vs. −10.99, respectively; the lower the better).
Figure 6. Binding mode of oregonin to DNMT1 crystallographic structure as predicted by molecular docking simulations. Oregonin is shown as cyan sticks; the crystallographic structure of DNMT1 (PDB: 4 WXX)Citation31 is shown as green cartoon; residues within 5 Å from Oregonin are showed as green lines; Cys1226 is shown as green sticks. H-bonds are highlighted as magenta dashed lines; DNMT1 residues H-bonded to oregonin are labelled. Residues that are contacted by curcumin and its analogues in Liu et al.’s studyCitation12 are labelled and underlined.
4. Discussion
4.1. Cell viability
The results of the cell viability test are in agreement with data obtained for the oregonin extracted from others genus of Alnus. The period of incubation 72 h and the concentrations of oregonin from Alnus japonica between 0 and 50 µM had no negative effects on the viability and morphology of endothelial cellsCitation40. Under the in vivo studies have shown the low toxicity of oregonin from A. glutinosaCitation41 in the experiments with brine shrimps and from A. japonicaCitation7 – with specific pathogen-free ICR mice. It was reportedCitation42 that diarylheptanoids from Alnus nepalensis show high biological activities without any side effect at high oral dose. Compared to other polyphenol with similar structure, particularly curcumin, oregonin is less toxic. In fact, curcumin in dose 50 μM in in vitro experimentsCitation43 reduced the cell viability more than 50%, whereas in our study the lowest percent of viable cells treated with oregonin in dose 50 μM was more than 80%. The results from our study clearly showed that oregonin from bark of A. incana in doses of 50 and 100 μM had no effect on the viability of NIH3T3 cells in the frame of 48 h and expressed middle toxically effect on MEFs (up to 18%).
4.2. mtDNA copy number and Dnmts
Changes in mtDNA copy number have been reported in a broad range of human diseasesCitation44, such as diabetes and its complications, obesity, cancer, HIV complications and ageing. Our interest in the investigation of this parameter was triggered by the evidence of important role of mitochondria in the DNA methylation process and remarkable antioxidant properties of oregonin. Results described in this work clearly show the ability of oregonin to increase the mtDNA content in the investigated cells although this effect is quantitatively dependent on the cell type, as NIH/3T3 cells showed more genomic stability than MEFs. The drastic increase of mtDNA content in MEFs treated with 100 μM was accompanied by a decrease of cell viability at 24 h. Most likely, there is an adaptive response to increase oxidative stress by an enhancement of mitochondrial biogenesis due to toxic dose of oregoninCitation44. On the other hand, these changes also correspond to the increase of Dnmts expression in MEFs. Our study highlighted for the first time the close relationship between changes in mtDNA content and expression of Dnmts mRNAs, which could be explained by considering that DNMTs enzymatic function is depended on the methyl donor SAM, whose proper production is guaranteed also by mitochondriaCitation18. The dependence of the DNA methylation process on the mitochondria content is confirmed by Smiraglia et al.Citation19, who analyse the methylation status of several genes in response to the depletion and repletion of mtDNA. However, the mechanism provoked by oregonin to increase the mtDNA replication is still unclear. The replication of the mitochondrial genome is regulated via the actions of a combination of gene expression coding in mtDNA and nuclear DNA such as d-loop region of mtDNA, TFAM, RNA polymerase (POLRMT), DNA polymerase POLG, transcription factors 1 and 2 (TFB1Mand TFB2M), nuclear respiratory factors 1 and 2 (NRF-1, NRF-2) and peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC1-α)Citation45–48. Oregonin could affect these genes via genetic or epigenetic pathways due to its ability to change the expression of Dnmts. Additionally, the anti-oxidative properties of oregonin result in an alteration of the mRNA expression of several antioxidant-related genes and oxidative stress responsive genesCitation10, which could explain the increase in mtDNA copy number through the communication between nuclear and mitochondrial genes.
Changes in the mtDNA content and expression of Dnmts mRNA transcripts may be also related to mtDNA methylation. There are very few studies that describe the role of DNMTs in the regulation of the mtDNA methylation, and their results are often questionable by the lack of clear understanding of the regulation mechanism. However, the localisation of DNMTs to mitochondria in different cells has been reported. The mtDNMT1 was defined in the mitochondria of MEF and human colon carcinoma cellsCitation49, the DNMT3a localises to mitochondria in mouse brain and spinal cordCitation50 and to mitochondria in muscle tissue of humanCitation51. Our results also show that in MEF cells there is the closest correlation between changes of the mtDNA content and expression of mtDnmt1 mRNA transcripts.
The novelty of our study is the analysis of the oregonin effect on the expression of the mRNA transcripts of mitochondrial isomeric form of DNA methyltransferase1-mtDnmt1. Both oregonin concentrations up-regulated the level of the mtDnmt1 transcripts in MEFs. In NIH/3T3 cells, the increased level of mitochondrial isoform mRNA was provoked only by 100 μM of oregonin. Recently accumulated dataCitation21,Citation52 reveal that external factors can alter mtDNA methylation followed by changing the mitochondrial genes expression. This process is mediated by mtDnmt1. In accordance with data of Shock et al.Citation49, hyper expression of mtDnmt1 in mouse fibroblasts related to the alteration in the expression of Nd1 and Nd6 genes, coding in mitochondria. Additionally, authors underlined that mtDnmt1 is sensitive to regulation by activators that respond to oxidative stress. The well-known anti-oxidative properties of oregoninCitation1,Citation10 may further explain the overexpression of the mtDnmt1 transcripts, as observed in our work.
Here, we showed that polyphenol oregonin affected the expression of DNMTs transcripts in both NIH/3T3 cells and MEFs but in some different ways. Its effect is depended on concentration and specific of cells genotype. The drastic increase of the DNA methyltransferase mRNA expression (FC between 4.9 and 9.5) in both types of cells treated with 100 μM of oregonin allows proposing that the molecule exerts some epigenetic toxicity. The increased level of DNMTs could lead to DNA hypermethylation, which occurrence at promoter CpG islands is a major epigenetic mechanism in silencing the genes expressionCitation53,Citation54. That could have both side effects in dependence of silenced genes. The silence of the viable important genes could disturb the cells and provoke the diseases development. However, suppression of “negative” genes could have therapeutic effects.
The main dataCitation13,Citation56,Citation57 about the influence of polyphenols, including curcumin, on the DNA methylation stressed a hypomethylation effect due to downregulation of the Dnmt1 mRNA transcripts, which directly correlated with decrease of the DNMT1 protein expression. Here, we observed similar effects for Dnmt1 mRNA in two different cell lines treated with oregonin at 50 μM. In silico studyCitation12 of the interaction between curcumin and DNMT1 suggested that curcumin covalently blocks the catalytic thiolate of DNMT1 to exert its inhibitory effect on DNA methylation. In line with Zheng et al.Citation55 results, reported the suppression of DNA methyltransferase 3 b by curcumin, we also defined the decrease of Dnmt3b mRNA transcripts (FC = 0.5, p < .05) in NH/3T3 cells treated with 50 μM of oregonin. In analogy to Liu et al.Citation12, here we used molecular modeling to predict the possible binding of oregonin to DNMT1. Although oregonin does not possess chemically reactive groups as curcumin, docking results clearly showed that the two compounds might bind within the catalytic site of DNMT1 in a similar conformation, and partially overlapping with the binding of the cofactor SAH/SAM. Based on the chemical similarity between curcumin and oregonin, here we pinpointed the pharmacophores that may be relevant for binding to DNMT1 and inhibiting its catalytic function to provide hypomethylation effects.
5. Conclusion
The new properties of the purified oregonin, extracted from the bark of A. incana, were confirmed in this study. The oregonin altered the epigenetic marks related to the DNA methylation, especially, expression of DNMTs mRNAs and mtDNA content in MEFs in dependence on used concentration and cell genotype. Molecular docking predicted the ability of oregonin to bind within the catalytic active site of DNMT1, which suggest that the compound may inhibit DNMT1 enzymatic activity.
However, the development of oregonin as a substance with potential to regulate the epigenetic changes, needs further investigations, particularly to elucidate the pathways of other epigenetic mechanisms (histone acetylation, chromatin remodelling) and genes involved in these processes. Additionally, research is needed to determine the appropriate dose for treatment without cytotoxic effect.
Acknowledgements
The research was implemented under frame of collaboration in COST Action FA1403 and bilateral project between Bulgarian Academy of Sciences and Latvian Academy of Sciences. We gratitude doctor M. Kicheva from “Progene-Ltd” company for technical support of the gene expression and mtDNA copy research.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Telysheva G, Dizhbite T, Bikovens O, et al. Structure and antioxidant activity of diarylheptanoids extracted from bark of grey alder (Alnus incana) and potential of biorefinery-based bark processing of European trees. Holzforschung 2011;65:623–9.
- Vidaković V, Novaković M, Popović Z, et al. Significance of diarylheptanoids for chemotaxonomical distinguishing between Alnus glutinosa and Alnus incana. Holzforschung 2017;72:9–16.
- Lee CJ, Lee SS, Chen SC, et al. Oregonin inhibits lipopolysaccharide-induced iNOS gene transcription and upregulates HO-1 expression in macrophages and microglia. Br J Pharmacol 2005;146:378–88.
- Martineau L, Hervé J, Muhamad A, et al. Anti-adipogenic activities of Alnus incana and Populus balsamifera bark extracts, part I: sites and mechanisms of action. Planta Medica 2010;76:1439–46.
- Martineau L, Hervé J, Muhamad A, et al. Anti-adipogenic activities of Alnus incana and Populus balsamifera bark extracts, part II: bioassay-guided identification of actives salicortin and oregonin. Planta Medica 2010;76:1519–24.
- Choi SE, Kim KH, Kwon JH, et al. Cytotoxic activities of diarylheptanoids from Alnus japonica. Arch Pharm Res 2008;31:1287–9.
- Ra JC, Kim YH, Sohn DH, Antioxidative and hepatoprotective compositions containing diarylheptanoids from Alnus japonica, U.S. Patent no: 2011/0144039 A1; 2011.
- Dinić J, Ranelović T, Stanković T, et al. Chemo-protective and regenerative effects of diarylheptanoids from the bark of black alder (Alnus glutinosa) in human normal keratinocytes. Fitoterapia 2015;105:169–76.
- Ponomarenko J, Trouillas P, Martin N, et al. Elucidation of antioxidant properties of wood bark derived saturated diarylheptanoids: a comprehensive (DFT-supported) understanding. Phytochemistry 2014;103:178–87.
- Lundquist A, Magnusson LU, Ullstrom C, et al. Oregonin reduces lipid accumulation and proinflammatory responses in primary human macrophages. Biochem Biophys Res Commun 2015;458:693–9.
- Anand P, Thomas SG, Kunnumakkara AB, et al. Biological activities of curcumin and its analogues (Congeners) made by man and mother nature. Biochem Pharmacol 2008;76:1590–611.
- Liu Z, Xie Z, Jones W, et al. Curcumin is a potent DNA hypomethylation agent. Bioorg Med Chem Lett 2009;19:706–9.
- Boyanapalli SSS, Tony Kong AN. “Curcumin, the King of Spices”: epigenetic regulatory mechanisms in the prevention of cancer, neurological, and inflammatory diseases. Curr Pharmacol Rep 2015;1:129–39.
- Meng H, Cao Y, Qin J, et al. DNA methylation, its mediators and genome integrity. Int J Biol Sci 2015;11:604–17.
- Tajbakhsh J. DNA methylation topology: potential of a chromatin landmark for epigenetic drug toxicology. Epigenomics 2011;3:761–70.
- Aggarwal R, Jha M, Shrivastava A, Jha AK. Natural compounds: role in reversal of epigenetic changes. Biochemistry (Mosc) 2015;80:972–89.
- Chappell G, Pogribny IP, Guyton KZ, Rusyn I. Epigenetic alterations induced by genotoxic occupational and environmental human chemical carcinogens: a systematic literature review. Mutat Res Rev Mutat Res 2016;768:27–45.
- Minocherhomji S, Tollefsbol TO, Singh KK. Mitochondrial regulation of epigenetics and its role in human diseases. Epigenetics 2012;7:326–34.
- Smiraglia DJ, Kulawiec M, Bistulfi GL, et al. A novel role for mitochondria in regulating epigenetic modification in the nucleus. Cancer Biol Ther 2008;7:1182–90.
- Chen T, Hevi S, Gay F, et al. Complete inactivation of DNMT1 leads to mitotic catastrophe in human cancer cells. Nat Genet 2007;39:391–6.
- Ferreira A, Serafim TL, Sardao VA, Cunha Oliveira T. Role of mtDNA-related mitoepigenetic phenomena in cancer. Eur J Clin Invest 2015;45:44–9.
- Choi KC, Jung MG, Lee YH, et al. Epigallocatechin-3-gallate, a histone acetyltransferase inhibitor, inhibits EBV-induced B lymphocyte transformation via suppression of RelA acetylation. Cancer Res 2009;69:583–92.
- Stefanska B, Salame P, Bednarek A, Fabianowska-Majewska K. Comparative effects of retinoic acid, vitamin D and resveratrol alone and in combination with adenosine analogues on methylation and expression of phosphatase and tensin homologue tumor suppressor gene in breast cancer cells. Br J Nutr 2012;107:781–90.
- Day JK, Bauer AM, DesBordes C, et al. Genistein alters methylation patterns in mice. J Nutr 2002;132:2419S–23S.
- Fang MZ, Chen D, Sun Y, et al. Reversal of hypermethylation and reactivation of p16INK4a, RARbeta, and MGMT genes by genistein and other isoflavones from soy. Clin Cancer Res 2005;11:7033–41.
- Fernández-Bedmar Z, Anter J, Alonso-Moraga A, et al. Hepatocarcinogenic potential of hesperidin, a natural polyphenol of Citrus juices. Mol Carcinog 2017;56:1653–62.
- Szyf M. DNA methylation and demethylation as targets for anticancer therapy. Biochemistry (Mosc) 2005;70:533–49.
- Gupta SC, Kannappan R, Reuter S, et al. Chemosensitization of tumors by resveratrol. Ann N Y Acad Sci 2011;1215:150–60.
- Link A, Balaguer F, Goel A. Cancer chemoprevention by dietary polyphenols: promising role for epigenetics. Biochem Pharmacol 2010;80:1771–92.
- Nicklas JA, Brooks EM, Hunter TC, et al. Development of a quantitative PCR (TaqMan) assay for relative mitochondrial DNA copy number and the common mitochondrial DNA deletion in the rat. Environ Mol Mutagen 2004;44:313–20.
- Aiken CE, Cindrova-Davies T, Johnson MH. Variations in mouse mitochondrial DNA copy number from fertilization to birth are associated with oxidative stress. Reprod Biomed Online 2008;17:806–13.
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001;29:e45.
- Zhang ZM, Liu S, Lin K, et al. Crystal structure of human DNA methyltransferase 1. J Mol Biol 2015;427:2520–31.
- Navakauskienė R, Mori M, Christodoulou MS, et al. Histone demethylating agents as potential S-adenosyl-L-methionine-competitors. Med Chem Commun 2016;7:1245–55.
- FRED 3.0.1: OpenEye scientific software, Santa Fe, NM. Available from: http://www.eyesopen.com (accessed in February 2018.)
- McGann M. FRED pose prediction and virtual screening accuracy. J Chem Inf Model 2011;51:578–96.
- Kim S, Thiessen PA, Bolton EE, et al. PubChem substance and compound databases. Nucleic Acids Res 2015;44:D1202–13.
- OMEGA 2.5.1.4: OpenEye scientific software, Santa Fe, NM. Available from: http://www.eyesopen.com (accessed in February 2018.)
- Hawkins PC, Skillman AG, Warren GL, et al. Conformer generation with OMEGA: algorithm and validation using high quality structures from the Protein Databank and Cambridge Structural Database. J Chem Inf Model 2010;50:572–84.
- Han J, Lee WS, Kim J-R, et al. Effects of diarylheptanoids on the tumor necrosis factor-r-induced expression of adhesion molecules in human umbilical vein endothelial cells. J Agric Food Chem 2007;55:9457–64.
- Kumarasamy Y, Cox PJ, Jaspars M, et al. Bioactivity of hirsutanol, oregonin and genkwanin, isolated from the seeds of Alnus glutinosa (Betulaceae). Nat Prod Commun 2006;1:641. https://abdn.pure.elsevier.com
- Saxena A, Yadav D, Mohanty S, et al. Diarylheptanoids rich fraction of Alnus nepalensis attenuates malaria pathogenesis: in-vitro and in-vivo study. Phytother Res 2016;30:940–8.
- Oelkrug C, Lange C, Wenzel E, et al. Analysis of the tumoricidal and anti-cachectic potential of curcumin. Anticancer Res 2014;34:4781–8.
- Malik A, Czajka A. Is mitochondrial DNA content a potential biomarker of mitochondrial dysfunction? Mitochondrion 2013;13:481–92.
- Campbell CT, Kolesar JE, Kaufman BA. Mitochondrial transcription factor A regulates mitochondrial transcription initiation, DNA packaging, and genome copy number. Biochim Biophys Acta 2012;1819:921–9.
- Falkenberg M, Larsson NG, Gustafsson CM. DNA replication and transcription in mammalian mitochondria. Annu Rev Biochem 2007;76:679–99.
- Hock MB, Kralli A. Transcriptional control of mitochondrial biogenesis and function. Annu Rev Physiol 2009;71:177–203.
- Leigh-Brown S, Enriquez JA, Odom DT. Nuclear transcription factors in mammalian mitochondria. Genome Biol 2010;11:215.
- Shock LS, Thakkar PV, Peterson EJ, et al. DNA methyltransferase 1, cytosine methylation, and cytosine hydroxymethylation in mammalian mitochondria. Proc Natl Acad Sci USA 2011;108:3630–5.
- Chestnut BA, Chang Q, Lesuisse C, et al. Epigenetic regulation of motor neuron cell death through DNA methylation. J Neurosci 2011;31:16619–36.
- Wong M, Gertz B, Chestnut B, Martin L. Mitochondrial DNMT3A and DNA methylation in skeletal muscle and CNS of transgenic mouse models of ALS. Front Cell Neurosci 2013; 7:279.
- Iacobazzi V, Castegna A, Infantino V, Andria G. Mitochondrial DNA methylation as a next generation biomarker and diagnostic tool. Mol Genet Metab 2013;110:25–34.
- Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med 2003;349:2042–54.
- Fang MZ, Jin Z, Wang Y, et al. Promoter hypermethylation and inactivation of O(6)-methylguanine-DNA methyltransferase in esophageal squamous cell carcinomas and its reactivation in cell lines. Int J Oncol 2005; 26:615–22.
- Zheng J, Wu C, Lin Z, et al. Curcumin up-regulates phosphatase and tensin homologue deleted on chromosome 10 through microRNA-mediated control of DNA methylation – a novel mechanism suppressing liver fibrosis. FEBS J 2014; 281:88–103.
- Mirza S, Sharma G, Parshad R, et al. Expression of DNA methyltransferases in breast cancer patients and to analyze the effect of natural compounds on DNA methyltransferases and associated proteins. J Breast Cancer 2013;16:23–31. Mar.
- Sharma V, Jha AK, Kumar A, et al. Curcumin-mediated reversal of p15 gene promoter methylation: implication in anti-neoplastic action against acute lymphoid leukaemia cell line. Folia Biologica (Praha) 2015;61:81–9.