Abstract
Amidinobenzimidazole derivatives connected to 1-aryl-substituted 1,2,3-triazole through phenoxymethylene linkers 7a–7e, 8a–8e, and 9a–9e were designed and synthesised with the aim of evaluating their anti-bacterial and anti-trypanosomal activities and DNA/RNA binding affinity. Results from anti-bacterial evaluations of antibiotic-resistant pathogenic bacteria revealed that both o-chlorophenyl-1,2,3-triazole and N-isopropylamidine moieties in 8c led to strong inhibitory activity against resistant Gram-positive bacteria, particularly the MRSA strain. Furthermore, the non-substituted amidine and phenyl ring in 7a induced a marked anti-bacterial effect, with potency against ESBL-producing Gram-negative E. coli better than those of the antibiotics ceftazidime and ciprofloxacin. UV–Vis and CD spectroscopy, as well as thermal denaturation assays, indicated that compounds 7a and 8c showed also binding affinities towards ctDNA. Anti-trypanosomal evaluations showed that the p-methoxyphenyl-1,2,3-triazole moiety in 7b and 9b enhanced inhibitory activity against T. brucei, with 8b being more potent than nifurtimox, and having minimal toxicity towards mammalian cells.
Graphical Abstract
Introduction
The benzimidazole derivatives, which contain fused heterocyclic nuclei within their structures, are structural isosteres of purine bases. This allows them to interact with biopolymers and they, therefore, have diverse biological and clinical applicationsCitation1–5. Much research effort has been aimed at targeting DNA with benzimidazole ligands, with the goal of designing agents that have therapeutic applicationsCitation5–9. Although RNA is a well-established target of current antibiotics, designing new compounds that selectively recognise RNA has also been a difficult task, particularly when focused on the treatment of a variety of infectionsCitation10–12. The challenge is to produce drug-like molecules with high affinity for DNA/RNA, while maintaining sufficient sequence selectivity. While there are many areas of therapy that might benefit from DNA-directed intervention, there is currently an urgent need for new antimicrobials with novel modes of action.
Antibiotic resistance is a global public threat because of its effect on health care with prolonged hospitalisations and increased mortality. The increasing prevalence of hospital and community-acquired infections caused by multidrug-resistant (MDR) bacterial pathogens is limiting the options for effective antibiotic therapyCitation13,Citation14. Drug-resistant Gram-positive bacterial pathogens, including methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant enterococci (VRE), have become a serious clinical problem that impinges on the treatment of various nosocomial and community-acquired infectionsCitation15,Citation16. In addition, an increased incidence of MDR Gram-negative bacteria, such as Pseudomonas aeruginosa, Escherichia coli, and Klebsiella pneumoniae, coupled with the lack of novel antibiotics, represents one of the biggest threats to the control of respiratory and other infectionsCitation17. In order to overcome these emerging bacterial resistance problems, novel anti-bacterial drugs need to be developed. Accordingly, in recent years, numerous efforts have focused on discovering novel benzimidazole-based anti-bacterial agentsCitation18–24. The importance of a protonable chemical moiety within anti-bacterial drugs has been investigated in different studiesCitation25,Citation26. These have revealed the significant uptake of amidine-containing DNA ligands into bacteria, and also into the nuclei of eukaryotic cellsCitation27. In addition, the structural features of 1,2,3-triazole also enable it to mimic different functional groups, justifying its wide use as a bioisostere for the design of antimicrobial drug analogsCitation28,Citation29. For example 1,4-disubstituted 1,2,3-triazoles are good Z-amide isosteres, because the C-4 atom can act as an electrophilic site; the CH bond (in the 5-position) acts as a hydrogen bond donor, and the lone pair of N-3 electrons acts as a hydrogen bond acceptorCitation30.
A wide range of pharmacological activities has been attributed to the unusual chemical features of azole rings, such as benzimidazole and 1,2,3-triazole. These are able to interact in a non-covalent way with a range of targets, due to the presence of an electron-rich aromatic system and heteroatomsCitation31,Citation32, and act as promising moieties for the design of novel scaffolds with anti-bacterial activity. Thus, among the series of [1,2,4-triazolyl]phenyl-substituted 4,6-difluorobenzimidazoles ICitation33, analogues with electronegative substituents emerged as promising antimicrobials, while 2-thiobenzimidazole with [(1,2,4-triazolyl)ethylthio]phenyl moiety IICitation34 exhibited anti-bacterial properties that were selective for Helicobacter pylori (). Benzimidazole–1,2,3-triazole conjugates III with aromatic (p-chlorophenyl and p-fluorophenyl) 4-substituted triazoles exhibited selective anti-Moraxella catarrhalis activityCitation35. Furthermore, triazole-bearing monobenzimidazoles IV and V inhibited growth of Gram-positive bacteria, including two MRSA strains, and displayed E. coli DNA topoisomerase I inhibitionCitation36.
Figure 1. Representatives of benzimidazoles containing triazole moiety I–V as potential antibacterial agents.
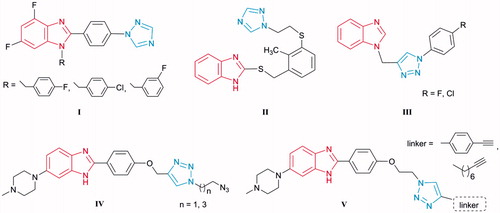
The increasingly important role of benzimidazole and triazole derivatives has been also demonstrated by their in vivo evaluations against Gram-positiveCitation37–42 and Gram-negative bacteriaCitation43. Bis-benzimidazole compound (ridinilazole, SMT-19969)Citation44 recently entered phase III human clinical trials for the treatment of Clostridium difficile.
Besides anti-bacterial activity, benzimidazole containing compounds have shown good anti-protozoal potencyCitation45–49. Human African trypanosomiasis (HAT), also known as sleeping sickness, is a fatal parasitic disease caused by two subspecies of Trypanosoma brucei. It has been estimated that over 50 million people are at risk of infection with HAT in more than 30 African countries, and there remains a clear need to develop new, safer, and more affordable agents to combat this fatal infectionCitation50. The efficacy of diarylamidines, such as pentamidineCitation51, berenylCitation52 and its orally active prodrug pafuramidineCitation53 (), in the treatment of protozoal diseases, especially trypanosomiasis, has been known for many years.
However, current drugs have problems, such as toxicity, poor efficacy, and increasing resistance by the parasites. Although the precise anti-protozoan mechanisms of action of aromatic diamidines have not been fully elucidated, there is considerable evidence that direct interaction with the pathogen genome is important for activity. Recently, a diamidine containing a 1,2,3-triazole ring as central core was synthesised, which displayed better anti-trypanosomal efficacy than melarsoprol, curing all infected miceCitation54. It was found that incorporation of different hydrophobic aromatic head groups linked to the rest of the molecule by an amidine moiety improved both anti-bacterial activity and affinity to DNACitation27.
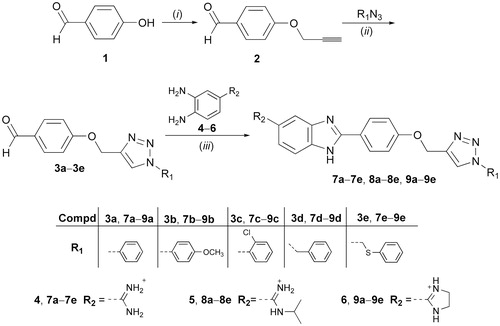
In view of the wide applications of the benzimidazole and 1,2,3-triazole moieties in drug development, and encouraged by the activity profile of both scaffoldsCitation35,Citation55, we synthesised molecules that contained both units attached through a phenoxymethylene linker as the central core, thereby expanding the electronic environment of chemical space ().
Figure 3. Design and synthesis of amidinobenzimidazoles connected to 1-aryl-substituted 1,2,3-triazole via phenoxymethylene unit.
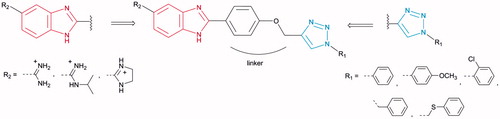
Targeted compounds were designed to contain a non-substituted amidine, N-isopropylamidine, and imidazoline moiety at the C-5 position of the benzimidazole core, as the hydrophilic end, and an aromatic unit at the N-1 position of the 1,2,3-triazole ring, as the hydrophobic end. It was anticipated that selected 5-amidinobenzimidazoles connected to 1-aryl-substituted 1,2,3-triazole would exhibit enhanced affinity for DNA/RNA compared to other aromatic amidines that we have recently studiedCitation56,Citation57. Therefore, interactions between 5-amidinobenzimidazoles 7a–7e, 8a–8e, and 9a–9e and DNA/RNA were assessed and their activities against Gram-positive, Gram-negative, and antibiotic-resistant, as well as their trypanocidal properties, were evaluated.
Materials and methods
General
All chemicals and solvents were purchased from Aldrich and Acros. Pre-coated Merck silica gel 60F-254 plates and Fluka (0.063–0.2 mm) silica gel using an appropriate solvent system were employed for thin layer chromatography (TLC) and column chromatography, respectively. Melting points were determined using Kofler micro hot-stage (Reichert, Vienna, Austria). 1H and 13 C NMR spectra were recorded on a Varian Gemini 300 and 600 spectrometers. All NMR spectra were recorded in DMSO-d6 at 298 K. Mass spectra were recorded on an Agilent 6410 instrument with electrospray interface and triple quadrupole analyser. Microwave-assisted syntheses were carried out in a microwave oven (Milestone start S) at 80 °C and pressure of 1 bar. The ultrasound-assisted reactions were performed in a bath cleaner (Bandelin, Sonorex digital 10 P, Berlin, Germany) with frequency of 35 kHz and power of 1000 W.
Experimental procedures for the synthesis of compounds
Amidino-substituted o-phenylenediamines (4–6)Citation58, 4-(prop-2-ynyloxy)benzaldehyde (2)Citation59, 4–(1,2,3-triazol-4-yl)methoxy)benzaldehyde (3b)Citation60, 4–(1,2,3-triazol-4-yl)methoxy)benzaldehydes (3a, 3d), and 5-amidinobenzimidazoles (7a, 7d, 8a, 8d, 9a, and 9d)Citation55 were prepared according to described procedures.
General procedure for the synthesis of compounds 3a–e
The reaction mixture of compound 2, Cu(0) (0.8 eq), 1 M CuSO4 (0.3 eq) and the corresponding azide (1.2 eq) was dissolved in 1 ml DMF and a mixture of t-BuOH: H2O = 1: 1 (3 ml). Method A: The reaction mixture was stirred under microwave irradiation (300 W) at 80 °C during 1.5 h. Method B: The reaction mixture was placed in an ultrasonic bath cleaner (1000 W, 35 kHz) at 80 °C for 1.5 h. The solvent was removed under reduced pressure and purified by column chromatography with CH2Cl2.
4-((1-(4-Methoxyphenyl)-1H-1,2,3-triazol-4-yl)methyleneoxy)benzaldehyde (3b)
Compound 3b was prepared using the above mentioned procedure from 2 (200 mg, 1.15 mmol) and 1-azido-4-methoxybenzene (2.76 ml, 1.38 mmol) to obtain 3b as white crystals (Method A: 149.3 mg, 42%; Method B: 271.2 mg, 76%; m.p. 127–130 °C) (m.p. lit.Citation60 = 126–127 °C). 1H NMR (300 MHz, DMSO) δ 9.89 (1H, s, CHO), 8.87 (1H, s, H5′), 7.90 (2H, d, J = 8.8 Hz, Ph), 7.81 (2H, d, J = 9.1 Hz, Ph), 7.28 (2H, d, J = 8.7 Hz, Ph), 7.14 (2H, d, J = 9.1 Hz, Ph), 5.36 (2H, s, OCH2), 3.83 (3H, s, OCH3). 13 C NMR (75 MHz, DMSO) δ 191.55, 162.99, 159.49, 143.08, 131.96, 130.03, 130.00, 123.25, 122.03, 115.35, 115.03, 61.47, 55.68.
4-((1–(2-Chlorophenyl)-1H-1,2,3-triazol-4-yl)methoxy)benzaldehyde (3c)
Compound 3c was prepared using the above mentioned procedure from 2 (200 mg, 1.15 mmol) and 1-azido-2-chlorobenzene (2.76 ml, 1.38 mmol) to obtain 3c as white solid (Method A: 194.2 mg, 53%; Method B: 231.67 mg, 64%; m.p. = 127–129 °C). 1H NMR (600 MHz, DMSO) δ 9.90 (1H, s, CHO), 8.76 (1H, s, H5′), 7.91 (2H, d, J = 8.7 Hz, Ph), 7.79 (1H, dd, J = 8.0, 1.2 Hz, Ph), 7.73 (1H, dd, J = 7.8, 1.6 Hz, Ph), 7.65 (1H, td, J = 7.8, 1.6 Hz, Ph), 7.60 (1H, td, J = 7.6, 1.3 Hz, ph), 7.30 (2H, d, J = 8.7 Hz, Ph), 5.40 (s, 2H, OCH2).13C NMR (151 MHz, DMSO) δ 191.84, 163.15, 142.48, 134.54, 132.14, 132.10, 130.84, 130.19, 128.81, 128.74, 128.63, 127.35, 115.54, 61.40.
4-((1-((Phenylthio)methyl)-1H-1,2,3-triazol-4-yl)methoxy)benzaldehyde (3e)
Compound 3e was prepared using the above mentioned procedure from 2 (200 mg, 1.15 mmol), azidomethyl phenyl sulfide (0.19 ml, 1.38 mmol), Cu (0) (59.8 mg, 0.94 mmol), 1 M CuSO4 (0.24 ml, 0.05 mmol) in DMF (1 ml), t-BuOH: H2O = 1: 1 (4 ml) to obtain 3e as yellow oil (Method A: 214.3 mg, 57%; Method B: 273.6 mg, 73%). 1H NMR (300 MHz, DMSO) δ 9.88 (1H, s, CHO), 8.21 (1H, s, 1H, H5′), 7.87 (2H, d, J = 8.8 Hz, Ph), 7.43–7.26 (5H, m, Ph), 7.21 (2H, d, J = 8.7 Hz, Ph), 5.96 (2H, s, 2H, CH2), 5.26 (2H, s, CH2). 13 C NMR (75 MHz, DMSO) δ 191.58, 162.94, 142.57, 132.36, 131.92, 130.68, 129.98, 129.43, 127.90, 124.72, 115.36, 61.33, 51.87.
General procedure for the synthesis of compounds 7a–7e, 8a–8e, and 9a–9e
The reaction mixture of 4-triazolylbenzaldehyde derivatives (3a–3e), o-phenylenediamine (4, 5, or 6) and 40% NaHSO3 was dissolved in 15 ml EtOH and stirred under reflux for 6–8 h. After completion of the reaction NaHSO3 (aq) was filtered and the reaction mixture was evaporated to dryness. Water was added (5 ml) and the mixture was stirred overnight and filtered. The crude residue was dissolved in HCl saturated MeOH (8–10 ml) and stirred overnight. Addition of ether resulted in precipitation of products 7a–7e, 8a–8e, and 9a–9e. Solid was collected by filtration, washed with anhydrous ether, and dried under vacuum.
2–(4-((1–(4-Methoxyphenyl)-1H-1,2,3-triazol-4-yl)methoxy)phenyl)-1H-benz[d]imidazole-5-carboximidamide dihydrochloride (7b)
Compound 7b was prepared using the above described method from 3b (200 mg, 0.65 mmol) and 4 (87.39 mg, 0.58 mmol) to obtain 7b as white powder (122.7 mg, 53%, m.p. = 195–197 °C). 1H NMR (600 MHz, DMSO) δ 9.35 (2H, s, NH), 8.94 (2H, s, NH), 8.90 (1H, s, H5′), 8.26 (2H d, J = 8.2 Hz, Ph), 8.15 (1H, s, H4), 7.84–7.80 (3H, m, Ph; H7), 7.71 (1H, d, J = 8.0 Hz, H6), 7.34 (2H, d, J = 8.7 Hz, Ph), 7.15 (2H, d, J = 9.0 Hz, Ph), 5.37 (2H, s, OCH2), 3.84 (3H, s, OCH3). 13 C NMR (75 MHz, DMSO) δ 165.85, 160.58, 159.42, 153.78, 143.21, 140.84, 134.73, 129.97, 129.13, 123.14, 122.69, 121.93, 120.61, 115.55, 114.97, 61.34, 55.63. MS (ESI, m/z) 440.1 [M + H]+. Anal. calcd. for C24H21N7O2 × 2 HCl ×2.5 H2O (Mr = 557.44): C 51.71, H 5.06, N 17.59; found: C 51.60, H 4.72, N 17.34%.
2–(4-((1–(2-Chlorophenyl)-1H-1,2,3-triazol-4-yl)methoxy)phenyl)-1H-benz[d]imidazole-5-carboximidamide dihydrochloride (7c)
Compound 7c was prepared using the above described method from 3c (200 mg, 0.64 mmol) and 4 (96.51 mg, 0.64 mmol) to obtain white powder 7c (210.3 mg, 58%, m.p. = 176–177 °C). 1H NMR (300 MHz, DMSO) δ 9.36 (2H, s, NH), 8.96 (2H, s, NH), 8.77 (1H, s, 1H, H5′), 8.28 (2H, d, J = 8.8 Hz, Ph), 8.16 (1H, s, H4), 7.88–7.55 (6H, m, Ph; H5; H6), 7.36 (2H, d, J = 8.9 Hz, Ph), 5.40 (2H, s, OCH2). 13 C NMR (151 MHz, DMSO) δ 165.62, 161.33, 152.72, 142.25, 134.41, 131.84, 131.26, 130.60, 129.84, 128.51, 127.23, 123.76, 123.22, 122.24, 115.90, 115.70, 114.82, 61.23. MS (ESI, m/z) 444.0 [M + H]+. Anal. calcd. for C23H18ClN7O × 2 HCl ×3 H2O (Mr = 570.86): C 48.39, H 4.59, N 17.17; found: C 48.11, H 4.47, N 17.38%.
2–(4-((1-((Phenylthio)methyl)-1H-1,2,3-triazol-4-yl)methoxy)phenyl)-1H-benz[d]imidazole-5-carboximidamide dihydrochloride (7e)
Compound 7e was prepared using the above described method from 3e (200 mg, 0.64 mmol) and 4 (96.51 mg, 0.64 mmol) to obtain 7e as white powder (324.8 mg, 90%, m.p. = 173–176 °C). 1H NMR (300 MHz, DMSO) δ 9.46 (2H, s, NH), 9.08 (2H, s, NH), 8.34 (2H, d, J = 8.9 Hz, Ph), 8.25 (1H, s, H5′), 8.20 (1H, d, J = 1.1 Hz, H4), 7.89 (2H, t, J = 7.8 Hz, Ph), 7.80 (1H, dd, J = 8.6 Hz, H6), 7.45–7.29 (6H, m, H7; Ph), 5.99 (2H, s, CH2), 5.30 (2H, s, CH2).13C NMR (75 MHz, DMSO) δ 165.70, 161.12, 153.11, 142.62, 139.17, 132.38, 131.84, 130.59, 129.59, 129.38, 127.82, 124.66, 123.55, 122.98, 115.72, 115.30, 61.27, 51.80. MS (ESI, m/z) 456.1 [M + H]+. Anal. calcd. for C24H21N7OS ×2 HCl ×1.7 H2O (Mr = 559.09): C 51.56, H 4.76, N 17.54; found: C 51.80, H 4.68, N 17.22%.
N-isopropyl-2–(4-((1–(4-methoxyphenyl)-1H-1,2,3-triazol-4-yl)methoxy)phenyl)-1H-benz[d]imidazole-5-carboximidamide dihydrochloride (8b)
Compound 8b was prepared using the above described method from 3b (200 mg, 0.65 mmol) and 5 (115.2 mg, 0.65 mmol) to obtain 8b as brown powder (225.8 mg, 58%, m.p. = 188–191 °C). 1H NMR (300 MHz, DMSO) δ 9.66 (1H, d, J = 7.7 Hz, NH), 9.51 (1H, s, NH), 9.08 (1H, s, NH), 8.92 (1H, s, H5′), 8.39 (2H, d, J = 8.7 Hz, Ph), 8.08 (1H, s, H4), 7.88 (1H, d, J = 8.5 Hz, H7), 7.83 (2H, d, J = 9.0 Hz, Ph), 7.69 (1H, d, J = 7.6 Hz, H6), 7.39 (2H, d, J = 8.8 Hz, Ph), 7.15 (2H, d, J = 9.0 Hz, Ph), 5.39 (2H, s, CH2), 4.15–4.02 (1H, m, CH), 3.84 (3H, s, OCH3), 1.32 (6H, d, J = 6.3 Hz, CH3CHCH3).13C NMR (151 MHz, DMSO) δ 162.18, 161.13, 159.50, 152.96, 143.20, 130.02, 129.64, 124.28, 124.21, 123.74, 123.30, 122.03, 115.77, 115.36, 115.05, 61.43, 55.71, 45.25, 21.38. MS (ESI, m/z) 482.1 [M + H]+. Anal. calcd. for C27H27N7O2 × 2 HCl ×2.6 H2O (Mr = 601.32): C 53.93, H 5.73, N 16.30; found: C 53.62, H 5.71, N 16.39%.
N-isopropyl-2–(4-((1–(2-chlorophenyl)-1H-1,2,3-triazol-4-yl)methoxy)phenyl)-1H-benz[d]imidazole-5-carboximidamide dihydrochloride (8c)
Compound 8c was prepared using the above described method from 3c (200 mg, 0.64 mmol) and 5 (101.0 mg, 0.57 mmol) to obtain 8c as white powder (105.5 mg, 28%, m.p. = 210–213 °C). 1H NMR (300 MHz, DMSO) δ 9.58 (1H, d, J = 8.2 Hz, NH), 9.43 (1H, s, NH), 8.99 (1H, s, NH), 8.78 (1H, s, H5′), 8.30 (2H, d, J = 8.6 Hz, Ph), 8.04 (1H, s, H4), 7.87–7.57 (6H, m, H7; H6; Ph), 7.37 (2H, d, J = 8.4 Hz, Ph), 5.40 (2H, s, OCH2), 4.13–4.00 (1H, m, CH), 1.31 (6H, d, J = 6.1 Hz, CH3CHCH3). 13 C NMR (151 MHz, DMSO) δ 162.41, 160.38, 153.72, 142.46, 134.42, 131.84, 130.61, 128.93, 128.57, 128.55, 128.47, 127.09, 123.10, 127.09, 122.58, 120.24, 116.13, 115.53, 61.14, 45.06, 21.31. MS (ESI, m/z) 486.1 [M + H]+. Anal. calcd. for C26H24ClN7O × 2 HCl ×2.3 H2O (Mr = 600.33): C 52.02, H 5.14, N 16.33; found: C 52.22, H 5.03, N 16.59%.
N-isopropyl-2–(4-((1-(phenylthiomethyl)-1H-1,2,3-triazol-4-yl)methoxy)phenyl)-1H-benz[d]imidazole-5-carboximidamide dihydrochloride (8e)
Compound 8e was prepared using the above described method from 3e (200 mg, 0.61 mmol) and 5 (108.9 mg, 0.61 mmol) to obtain 8e as yellow powder (73.1 mg, 21%, m.p. = 152–154 °C). 1H NMR (600 MHz, DMSO) δ 9.66 (1H, d, J = 7.5 Hz, NH), 9.51 (1H, s, NH), 9.09 (1H, s, NH), 8.39 (2H, d, J = 7.9 Hz, Ph), 8.25 (1H, s, H5′), 8.08 (1H, s, H4), 7.87 (1H, d, J = 8.3 Hz, H7), 7.68 (1H, d, J = 8.5 Hz, H6), 7.40 (2H, d, J = 7.5 Hz, Ph), 7.36–7.15 (5H, m, Ph), 5.98 (2H, s, CH2), 5.29 (2H, s, CH2), 4.24–3.99 (1H, m, CH), 1.31 (6H, d, J = 6.4 Hz, CH3CHCH3).13C NMR (75 MHz, DMSO) δ 162.93, 160.52, 154.42, 145.37, 143.19, 132.82, 130.98, 129.76, 129.09, 128.18, 124.99, 123.19, 122.71, 122.05, 115.86, 61.55, 52.18, 45.44, 21.75. MS (ESI, m/z) 498.1 [M + H]+. Anal. calcd. for C27H27N7OS ×2 HCl ×0.3 H2O (Mr = 575.95): C 56.31, H 5.18, N 17.02; found: C 56.33, H 5.37, N 17.28%.
5–(4,5-Dihydro-1H-imidazol-2-yl)-2–(4-((1–(4-methoxyphenyl)-1H-1,2,3-triazol-4-yl)methoxy)phenyl)-1H-benz[d]imidazole dihydrochloride (9b)
Compound 9b was prepared using the above described method from 3b (200 mg, 0.65 mmol) and 6 (114.9 mg, 0.65 mmol) to obtain 9b as yellow powder (279.1 mg, 70%, m.p. = 197–199 °C). 1H NMR (300 MHz, DMSO) δ 10.68 (2H, s, NH), 8.91 (1H, s, H5′), 8.37 (1H, s, H4), 8.32 (2H, d, J = 8.3 Hz, Ph), 7.88 (1H, d, J = 4.3 Hz, H7), 7.82 (2H, d, J = 8.5 Hz, Ph), 7.35 (2H, d, J = 8.1 Hz, Ph), 7.28 (1H, d, J = 8.3 Hz, H6), 7.15 (2H, d, J = 8.1 Hz, Ph), 5.37 (2H, s, OCH2), 4.03 (4H, s, CH2CH2), 3.83 (3H, s, OCH3).13C NMR (151 MHz, DMSO) δ 165.49, 160.68, 159.51, 154.27, 143.30, 143.09, 136.43, 131.97 (C4), 130.01 (Ph-q), 129.27 (Ph), 123.21 (C6), 123.00 (C5′), 122.04 (Ph), 120.62 (C5), 116.05, 115.63, 115.37, 115.06, 61.38, 55.72, 44.44. MS (ESI, m/z) 466.1 [M + H]+. Anal. calcd. for C26H23N7O2 × 2 HCl ×0.9 H2O (Mr = 554.65): C 56.30, H 4.87, N 17.68; found: C 56.04, H 4.72, N 17.97%.
5–(4,5-Dihydro-1H-imidazol-2-yl)-2–(4-((1–(2-chlorophenyl)-1H-1,2,3-triazol-4-yl)methoxy)phenyl)-1H-benz[d]imidazole dihydrochloride (9c)
Compound 9c was prepared using the above described method from 3c (200 mg, 0.64 mmol) and 6 (112.9 mg, 0.64 mmol) to obtain 9c as red powder (115.3 mg, 30%, m.p. = 194–196 °C). 1H NMR (600 MHz, DMSO) δ 10.82 (2H, s, NH), 8.81 (1H, s, H5′), 8.45 (1H, s, H4), 8.40 (2H, d, J = 8.6 Hz, Ph), 7.98 (1H, d, J = 8.5 Hz, H7), 7.93 (1H, d, J = 8.3 Hz, H6), 7.81 (1H, dd, J = 8.0, 1.2 Hz, Ph), 7.75 (1H, dd, J = 7.8, 1.6 Hz, Ph), 7.67 (1H, td, J = 7.8, 1.6 Hz, Ph), 7.62 (1H, td, J = 7.7, 1.3 Hz, Ph), 7.40 (2H, d, J = 8.9 Hz, Ph), 5.43 (2H, s, OCH2), 4.04 (4H, s, CH2CH2).13C NMR (75 MHz, DMSO) 165.20, 161.07, 153.69, 142.40, 138.84, 136.92, 134.44, 131.90, 130.66, 130.15, 129.62, 128.63, 128.52, 127.21, 123.57, 119.46, 116.66, 116.38, 115.69, 61.24, 44.42. MS (ESI, m/z) 470.1 [M + H]+. Anal. calcd. for C25H20ClN7O × 2 HCl ×1.9 H2O (Mr = 577.08): C 52.03, H 4.51, N 16.99; found: C 52.31, H 4.66, N 16.63%.
5–(4,5-Dihydro-1H-imidazol-2-yl)-2–(4-((1-(phenylthiomethyl)-1H-1,2,3-triazol-4-yl)methoxy)phenyl)-1H-benz[d]imidazole dihydrochloride (9e)
Compound 9e was prepared using the above described method from 3e (200 mg, 0.61 mmol) and 6 (108.9 mg, 0.61 mmol) to obtain 9e as brown powder (149.4 mg, 41%, m.p. = 162–164 °C). 1H NMR (300 MHz, DMSO) δ 10.57 (2H, s, NH), 8.34–8.18 (4H, m, H5′; H4; Ph), 7.85 (2H, s, H7; H6), 7.44–7.21 (7H, m, Ph), 5.97 (2H, s, CH2), 5.26 (2H, s, CH2), 4.03 (4H, s, CH2CH2).13C NMR (151 MHz, DMSO) δ 165.13, 162.84, 160.80, 153.75, 142.60, 132.38, 131.79, 130.48, 129.86, 129.40, 129.32, 127.72, 124.63, 116.37, 115.52, 115.23, 61.17, 51.69, 44.33. MS (ESI, m/z) 482.0 [M + H]+. Anal. calcd. for C26H23N7OS ×2 HCl ×2.2 H2O (Mr = 594.14): C 52.56, H 4.99, N 16.50; found: C 52.64, H 5.12, N 16.29%.
Spectroscopic experiments
Polynucleotides
Polynucleotides were purchased as follows: polyG–polyC, polyA–polyU (Sigma-Aldrich, St. Louis, MO), calf thymus ctDNA (Aldrich). Polynucleotides were dissolved in PBS buffer, I = 0.05 mol dm−3, pH 7.0. The calf thymus ctDNA was additionally sonicated and filtered through a 0.45 mm filter. The polynucleotide concentration was spectroscopically determined as the concentration of nucleobasesCitation62.
UV–Vis spectroscopy
All UV–Vis absorbance measurements were conducted on a Perkin Elmer Lambda 25 spectrophotometer (Perkin Elmer, Waltham, MA). A quartz cell with a 1 cm path length was used for all absorbance studies. Compound stock solutions were 1 mM. The DNA/RNA at increasing ratios was then titrated into the compound buffer solution (1.48 × 10−5 mol dm−3) and the corresponding absorption spectra were recorded under the same conditions. All data were graphed and analysed using Origin software version 9.0 (OriginLab Corporation, Northampton, MA). The stability constants (Ks) and [bound compound]/[polynucleotide phosphate] ratios (n) were calculated according to the Scatchard EquationCitation63,Citation64. Values for Ks and n given in all have satisfying correlation coefficients (0.99).
Table 1. Hypochromic effects (H/%)Table Footnotea, binding constants (log Ks)Table Footnoteb and ratios nTable Footnotec ([compound]/[polynucleotide phosphate]) calculated from the UV–Vis titrations of compounds with ds-DNA/RNA (PBS, I = 0.015 M, and pH = 7).
Thermal melting (Tm)
Tm experiments were conducted with a Perkin Elmer Lambda 25 spectrophotometer in 1 cm quartz cuvettes. The absorbance of the DNA/RNA-compound complex was monitored at 260 nm as a function of temperature. The absorbance of the ligands was subtracted from every curve, and the absorbance scale was normalised. The ΔTm values were calculated by subtracting Tm of the free nucleic acid from Tm of the complex. Every reported ΔTm value was the average of at least two measurements. The error of ΔTm is ±0.5 °C. All data were graphed and analysed using Origin software version 9.0.
Circular dichroism (CD)
The CD spectra of DNA/RNA (concentration in cuvette 2 × 10−5 M) were recorded with a JASCO J-800 spectrometer (JASCO UK Ltd., Dunmow, United Kingdom) at different ratios r = 0.1, 0.3, 0.5, and 0.7 (r = [compound]/[polynucleotide]) at 25 °C in aqueous buffer solution (pH = 7, PBS, and I = 0.05 mol dm−3). Titrations were carried out by addition of aliquots of 1 mM stock solutions of the relevant compound (at increasing ratios) to the buffered polynucleotide (DNA/RNA) solution in a 1 cm quartz cuvette and scanned over a wavelength range 220–450 nm. All data were graphed and analysed using Origin software version 9.0.
Biological evaluations
Anti-bacterial screening
The compounds were evaluated for their in vitro anti-bacterial activity against Gram-positive bacteria: S. aureus (ATCC 25923), MRSA, methicillin-sensitive S. aureus (MSSA), E. faecalis, vancomycin-resistant E. faecium (VREF), and Gram-negative bacteria: E. coli (ATCC 25925), P. aeruginosa (ATCC 27853), A. baumannii (ATCC 19606) and ESBL-producing K. pneumoniae (ATCC 27736). Standard broth microdilution method as recommended in guidelines of Clinical and Laboratory Standards InstituteCitation61,Citation65,Citation66 was applied and the minimum inhibitory concentration (MIC) of compounds was tested. In short, testing was performed in U-bottomed 96-well sterile plastic microdilution trays (Falcon 3077, Becton Dickinson Labware, Franklin Lakes, NJ) in cation (Ca2+ and Mg2+) adjusted Mueller–Hinton broth medium (Becton Dickinson and Co., Cockeysville, MD). All testings were performed in triplicate.
Anti-trypanosomal screening and cytotoxicity assays
Bloodstream form T. brucei (strain 221) were cultured in modified Iscove’s medium, as outlinedCitation67 and trypanocidal assays were performed using 96-well microtitre plates. The compound concentrations that inhibited growth by 50% (IC50) and 90% (IC90) were determined. Parasites were sub-cultured at 2.5 × 104 ml−1, compounds were added at range of concentrations, and the plates incubated at 37 °C. Resazurin was added after 48 h, the plates incubated for a further 16 h, and then read in a Spectramax plate reader (Molecular Devices Corporation, San Jose, CA). The data were analysed using GraphPad Prism (GraphPad, La Jolla, CA). Each drug concentration was tested in triplicate.
Cytotoxicity against mammalian cells was also assessed using microtitre plates. Briefly, L6 cells (a rat myoblast line) were seeded at 1 × 104 ml−1 in 200 µl of growth medium containing different compound concentrations. The plates were then incubated for 6 d at 37 °C and 20 µl resazurin added to each well. After a further 8 h incubation, the fluorescence was determined using a Spectramax plate reader, as outlined above.
Results and discussion
Chemistry
1,2,3-Triazole-linked 5-amidinobenzimidazoles 7a–7e, 8a–8e, and 9a–9e are synthesised as outlined in Scheme 1. 4-Hydroxybenzaldehyde was propargylated to give 4-(prop-2-ynyloxy)benzaldehyde (2), which subsequently via the regioselective Cu(I) catalysed cycloaddition with aromatic azides resulted in 4–(1,2,3-triazol-1-yl)benzaldehyde derivatives (3a–3e) comprising an N-1-aryl-substituted 1,2,3-triazole subunit. An efficient and environmentally benign synthetic protocolCitation68, applying microwave and ultrasound irradiation, was used in the synthesis of 3a–3e. The efficiency of both ultrasound and microwave conditions were compared and indicated that ultrasound-assisted syntheses of 3a–3e resulted in higher yields than those of microwave-assisted reactions. Amidino-substituted 1,2-phenylenediamines (4–6) that were used for the synthesis of the target 5-amidinobenzimidazoles 7a–7e, 8a–8e, and 9a–9e were synthesised from the corresponding nitrile by the Pinner methodCitation58. 4-Amidino 1,2-phenylenediamines (4–6) reacted with the bisulfite adduct of the 4–(1,2,3-triazol-1-yl)benzaldehyde derivatives (3a−e) to produce amidine (7a–7e), N-isopropylamidine (8a–8e), and imidazoline-substituted (9a–9e) benzimidazole derivativesCitation69.
Spectroscopic characterisation of compounds
5-Amidinobenzimidazole derivatives 7a–7e, 8a–8e, and 9a–9e were synthesised and characterised by UV–Vis spectroscopy. UV–Vis spectra displayed two absorption maxima at around 260 and 315 nm (Table S1, Supporting Information). Absorbancies of solutions were proportional to their concentrations up to 1 × 10−4 moldm−3, indicating that there is no significant intermolecular stacking that could give rise to hypochromic effects. Furthermore, the UV–Vis spectra of 7a–7e, 8a–8e, and 9a–9e revealed negligible temperature dependent changes (25–90 °C) and excellent reproducibility upon cooling to 25 °C. The results showed that all evaluated compounds were stable and suitable for further spectroscopic and biological investigations.
Interactions with double-stranded polynucleotides
Spectrophotometric titrations of compounds with ds-DNA/RNA
UV–Vis absorption spectroscopy is simple, widely used and one of the most effective methods for detecting the interaction of small molecules with DNA. In general, these interactions and the subsequent formation of a new complex leads to changes in UV–Vis spectraCitation70. Therefore, UV–Vis spectroscopy was applied to investigate the interaction of compounds 7a–7e, 8a–8e, and 9a–9e with ds-DNA/RNA. UV–Vis titration with ctDNA showed a hypochromic effect indicating the disappearance of free molecule and the formation of a new compound-DNA species (Figure S1, Supporting Information). The hypochromic effect (12–44%) was accompanied by a small bathochromic shift (Δλ = 3–9 nm) that was found to originate from the stabilisation of DNA secondary structure due to the interaction with small moleculesCitation71.
To assess the sequence selectivity of the compounds, the experiment was repeated with ds-RNA polynucleotides (polyA-polyU and polyC-polyG). The addition of polyA-polyU in most cases led to hypochromic (19–58%) and small bathochromic (2–11 nm) changes in the visible absorption spectra as a result of complex formation. Absorption spectra obtained by adding aliquots of polyC-polyG to the compound solutions were recorded until saturation was achieved. In general, it was observed that addition of polyC-polyG resulted in a pronounced decrease of UV–Vis absorption maxima at 300–320 nm (27–50%), followed by small bathochromic shifts (Δλ = 2–7 nm). No further studies were conducted with compounds whose UV–Vis spectra showed minimal changes (ΔA < 0.08 at r = 1–0.1) during titration with DNA/RNA polynucleotides. It can be inferred that these compounds interact with polynucleotides only through a very weak electrostatic and external mode (). During titration with polyA-polyU, a clear isosbestic point was observed in UV–Vis spectra of 7a and 9c, pointing to the formation of one dominant type of complex.
The binding constants logKs and the density of the binding sites n were calculated using Scatchard plot analysis. In addition, the binding constants Ks for compounds 8b and 8e were calculated only for titration data taken at the r ≥ 0.1, because below that ratio changes in absorption maxima were too small for accurate calculation (ΔA ≤ 0.04) (. The binding constants Ks and ratios n obtained by processing UV–Vis titration data using the Scatchard equation are summarised in .
Thermal denaturation experiments
Thermal melting enables the rapid qualitative evaluation of the relative binding affinities of the compounds towards selected polynucleotides ()Citation72,Citation73. The melting temperature (Tm) is defined as the differences between the melting temperatures of free polynucleotides and their complexes with small non-covalently bound molecules. The correlation between binding constant and the increase of Tm was found to be quite complex, because the number of binding sites, positive charge of compounds, potential cooperativity, and the affinity for the unfolded polynucleotide have also to be taken into accountCitation74.
Table 2. ΔTm values (°C) of studied ds-polynucleotides upon addition of compounds 7a, 7c–7e, 8a–8e, and 9b–9e at different ratio rTable Footnoteb (PBS and pH = 7)a.
Denaturation experiments were carried out at different amounts of the compounds (r = 0.1, 0.3, 0.5, and 0.7 eq; r = [compound]/[polynucleotide]) with ctDNA and polyA-polyU. The results of the denaturation experiments are listed in .
Generally, results correlated with those of UV–Vis experiments. Strong non-linear dependence of ΔTm values on the ratio r was revealed, suggesting saturation of binding sites at r = 0.5–0.7 (for 7c–7d, 8a–8e, and 9d), r = 0.3–0.5 (for 7a, 9c, and 9e), in good accordance with the calculated values presented in . Results showed that compounds 8a–8e stabilised ctDNA slightly better than compounds 7a–7e and 9a–9e. Biphasic curves for interactions of compounds 7e, 8b, and 9b–9e with polyA-polyU at higher ratios r indicated additional binding modes. The above-mentioned compounds have monophasic curves at r ≤ 0.3, which together with results on UV–Vis titration confirmed intercalation as the dominant binding mode, while above that, ratio biphasic curves indicated agglomeration of compounds along the polynucleotide chains.
Circular dichroism (CD) experiments
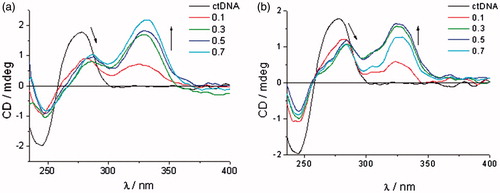
CD spectroscopy has been extensively employed for the investigation of small molecule–polynucleotide (DNA/RNA) interactionsCitation75,Citation76. Binding of achiral small molecules within the chiral DNA/RNA helix results in an induced CD spectrum (ICD)Citation77,Citation78. The appearance of ICD bands upon titration (r = 0.1–0.7) at λ > 300 nm was used to estimate the orientation of the chromophore in the ctDNA/RNA binding site and for the determination of binding mode. ctDNA features approximately 40% GC and 60% AT base pairs and adopts a B-helix with a narrow, deep, well-accessible minor groove and a rather broad, and shallow major groove. The two RNA polymers, polyA-polyU and polyG-polyC, form a typical A-helix with a broad minor and narrow major groove. The main difference among ds-RNAs is the presence of the amino group at N-2 in guanine which protrudes into the grooves and thereby may influence the affinity and binding mode of compounds being studied. The addition of the compounds being investigated resulted in a decrease of the ds-DNA/RNA CD bands (λ = 220–400 nm, S2). Observed changes in the intensity of CD bands for ds-DNA/RNA indicated the partial disruption of the polynucleotide helical chirality upon binding of a small molecule. The addition of the compounds 7a, 7c–7e, 8a–8e, and 9c–9d in solution with ctDNA generated strong, positive ICD signals in the range of 300–350 nm ( and S2, Supporting Information). This may arise due to groove binding being the dominant binding mode for this class of compoundsCitation79,Citation80.
ICD spectra of polyA-polyU and polyC-polyG with the addition of evaluated compounds, except for 7a, 8a–8e, and 9d, showed a decrease of CD band in the range of 220–300 nm, followed by appearance of new negative signal at >300 nm ( and S2, Supporting Information). This indicates that intercalation is the dominant binding mode. Compounds 7b–9b showed higher affinity for dsRNA than ctDNA. While 8b and 9b bound to polyA-polyU, 7b showed higher affinity for polyC-polyG ( and S2, Supporting Information).
Figure 5. ICD spectra of RNA polynucleotides with 5-amidinobenzimidazoles with p-methoxyphenyl-1,2,3-triazole unit: compound 7b (a), compound 8b (b), and compound 9b (c).
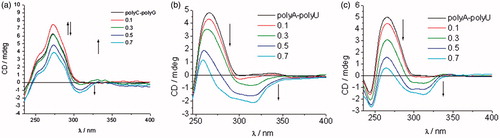
ICD spectra of 8b and 9b, in the presence of polyA-polyU, showed an intense increase of signal above r = 0.3, while ICD spectra of 7b with polyC-polyG showed a weaker intercalation signal above r = 0.5. This is in agreement with the results obtained by UV–Vis and thermal melting methods. Minimal changes of the intensity of the CD bands of polyC-polyG upon titration with compounds 7a, 8a–8e, and 9d, suggest a non-specific binding mode. Most probably compounds bind on the outside of the polyC-polyG polynucleotide. The intensity of negative ICD bands in polyA-polyU ICD spectra was also observed to be more intense than those in polyC-polyG spectra obtained with the same compound.
Biological evaluations
In vitro anti-bacterial activity
The in vitro anti-bacterial activity of 5-amidinobenzimidazoles 7a–7e, 8a–8e, and 9a–9e was tested against Gram-positive bacteria including S. aureus (ATCC 25923), Enterococcus faecalis (ATCC 29212), and Gram-negative bacteria including E. coli (ATCC 25925), K. pneumoniae (ATCC 700803), P. aeruginosa (ATCC 27853), and Acinetobacter baumannii (ATCC 19606). The MICs were determined and compared with those of the antibiotics ceftazidime, ciprofloxacin, ampicillin, and gentamicin (Table S2, Supporting Information).
Generally, compounds showed better activities against Gram-positive than Gram-negative bacteria. Only 5-amidinobenzimidazoles 7a, 7d, and 7e proved to be active against three Gram-negative strains, particularly amidinobenzimidazole 7d, which has an N-1-benzyl substituent. The type of amidino moiety in 5-benzimidazole had impact on the anti-bacterial activities, with non-substituted amidinobenzimidazoles 7a–7e having the highest overall activities (Table S2, Supporting Information). Compounds that exhibited anti-bacterial activities with MIC <256 µg/ml were evaluated against antibiotic resistant Gram-positive clinical strains, such as MRSA, MSSA and VREF () and Gram-negative clinical strains including extended-spectrum β-lactamase (ESBL)-producing E. coli, K. pneumoniae, and P. aeruginosa ().
Table 3. Anti-bacterial activity of selected compounds against antibiotic-resistant Gram-positive clinical strains.
Table 4. Anti-bacterial activity of selected compounds against antibiotic-resistant Gram-negative clinical strains.
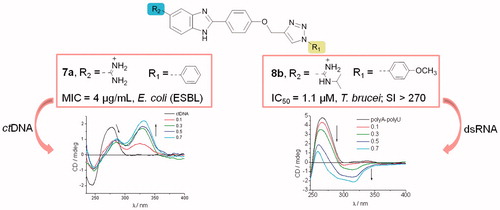
The evaluated compounds had a wide range of activity against MRSA, with the 5-N-isopropylamidinobenzimidazoles 8a–8e being the most active (MIC = 8–32 µg/mL) (). 8a–8e were also active against the MSSA strain, although to a lesser extent (MIC = 16–128 µg/ml). 8c also displayed modest activity against VRE-E. faecium. Among other compounds, benzimidazole imidazoline 9c had promising activity against the MRSA strain (MIC = 8 µg/ml). Against the antibiotic-resistant Gram-negative bacteria (), 5-amidinobenzimidazole 7a, with the N-1-phenyl-1,2,3-triazole, proved to be the most potent, with IC50 values of 4 µg/ml for E. coli, and 8 µg/ml for K. pneumoniae. However, this compound was only marginally effective against P. aeruginosa. Compounds 7d and 7e prove to be active against K. pneumoniae (MIC = 16 µg/ml). Introduction of a methylene (7d and 9d) and sulphide-bridge (7e and 9e) between 1,2,3-triazole and the phenyl ring reduced the activity against the antibiotic resistant E. coli and K. pneumoniae clinical strains. 7d, 7e, and 9e had slightly greater potency against P. aeruginosa compared with 7a. Overall, the results indicated that the o-chlorophenyl hydrophobic unit, with N-isopropylamidine, as the hydrophilic unit, in 8c contributed to anti-bacterial activity, particularly against the MRSA strain. Importantly, 7a was the most potent of the compounds against ESBL-producing E. coli, with higher activity than the reference antibiotics ceftazidime and ciprofloxacin.
One of our aims was to determine if there was a relationship between the affinity of compounds towards ds-DNA/RNA and their antimicrobial activity. UV–Vis and CD spectroscopy, as well as thermal denaturation assays, showed that compounds 7b, 9a, and 9b, which did not bind to ctDNA, had only marginal anti-microbial activities (MIC ≥128 µg/ml). Conversely, 5-amidinobenzimidazole 7a, which showed the highest affinity to ctDNA, exhibited high potency against ESBL-producing E. coli, which is in agreement with previous findingsCitation56,Citation81,Citation82.
Screening of the anti-trypanosomal activity
Results on the in vitro testing against the bloodstream form T. brucei of the 5-amidinobezimidazoles 7a–7e, 8a–8e, and 9a–9e with 1,4-disubstituted 1,2,3-triazole, and nifurtimox as reference drug, are summarised in . Similarly to anti-bacterial evaluations, we investigated how cationic moieties and aromatic substituents attached to the N-1 of the 1,2,3-triazole ring directly, or through the methylene and methylenesulphide spacer, influenced, anti-trypanosomal potencies.
Table 5. Anti-trypanosomal activitya of compounds 7a–7e, 8a–8e, and 9a–9e against Trypanosoma brucei strain.
Phenyl, p-methoxyphenyl, o-chlorophenyl, benzyl and (phenylthio)methyl substituents had a significant negative impact on IC50 values of anti-trypanosomal activity in the following order: p-OCH3 > Ph > PhSCH2 ≈ Bn > o-Cl. Except for 7c–9c, all compounds were active against T. brucei with IC50 values ranging from 1.1 to 13.5 µM. Interestingly, the o-chlorophenyl substituent in 7c–9c caused the loss of anti-trypanosomal activity (IC50 > 15 µM). The presence of the p-methoxyphenyl substituent in 7b–9b led to enhanced anti-trypanosomal potency, with the 5-N-isopropylamidinobenzimidazole analogue 8b being the most promising compound (IC50 = 1.1 µM, IC90 = 3.5 µM), which is 4-fold more potent than nifurtimox. UV–Vis titrations and thermal denaturation assays suggested that 7b–9b have low affinity to ctDNA () indicating that DNA is not the primary target for their anti-trypanosomal activity. Cytotoxicity assays against the rat myoblast cell line L6, revealed negligible activity, with three-figure selectivity index ().
Conclusions
The 1,2,3-triazole-linked 5-amidinobenzimidazoles 7a–7e, 8a–8e, and 9a–9e were synthesised by a Cu(I)catalysed 1,3-dipolar cycloaddition reaction applying microwave and ultrasound irradiation, with subsequent formation of a benzimidazole moiety by oxidative coupling of o-phenylenediamines with benzaldehydes. It was found that the 7c–9c, 7d–9d, and 7e–9e sets of compounds non-covalently bound to ds-DNA/RNA. The small bathochromic shifts in UV–Vis titration spectra upon addition of ctDNA, modest thermal stabilisation effects, and strong positive ICD bands in CD titration experiments supported minor groove binding as the dominant binding mode of these compounds. Conversely, the appearance of negative ICD bands in CD titration experiments with polyA-polyU and polyC-polyG, and density of binding sites obtained from UV–Vis titrations, identified intercalation as the predominant binding mode.
Furthermore, SARs showed that the type of aromatic substituents at N-1 of 1,2,3-triazole had profound effects on anti-bacterial and anti-protozoal activities. Thus, results of anti-bacterial evaluations revealed that o-chlorophenyl-1,2,3-triazole and N-isopropylamidine moieties in 8c had a considerable impact on inhibitory activity against resistant Gram-positive bacteria, particularly the MRSA strain. On the other hand, non-substituted amidine and phenyl rings in 7a contributed to a strong inhibitory effect on an ESBL-producing E. coli strain, with the potency better than those of the reference antibiotics ceftazidime and ciprofloxacin Compounds 7 b, 9a, and 9 b that showed extremely low affinity to ctDNA had also negligible anti-microbial activity (MIC ≥128 µg/ml). Contrary to this, the 5-N-isopropylamidinobenzimidazole series 8a–8e, which had better binding affinity relative to other amidines, showed some selective activity (MIC = 8–32 µg/ml) against the MRSA strain. Notably, compound 7a emerged as the most promising candidate because of its higher potency (MIC = 4 µg/ml) against ESBL-producing E. coli. It had the highest affinity among the tested compounds to ctDNA ( and ).
Results of anti-trypanosomal evaluations showed that the o-chlorophenyl group in 7c–9c had a negative impact on activity, whereas the p-methoxyphenyl substituent in 7b–9b enhanced activity, with 8b (IC50 = 1.1 µM and IC90 = 3.5 µM) being more potent than nifurtimox. In contrast to the observed correlation between anti-microbial activity and DNA binding, the antiprotozoal effects of 8b did not correlate with its DNA affinity. Further investigations will, therefore, be required to clarify the mechanism of anti-protozoal activity.
The promising anti-bacterial activity of compounds 7a and 8c and the anti-trypanosomal potency of compound 8b suggest that further structural optimisation of the 1,2,3-triazole-linked 5-amidinobenzimidazole class could enhance the potential anti-HAT and anti-bacterial activity against resistant pathogenic microorganisms.
Supplemental Material
Download PDF (2 MB)Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Akhtar W, Khan MF, Verma G, et al. Therapeutic evolution of benzimidazole derivatives in the last quinquennial period. Eur J Med Chem 2017;126:705–53.
- Akhtar J, Khan AA, Ali Z, et al. Structure-activity relationship (SAR) study and design strategies of nitrogen-containing heterocyclic moieties for their anticancer activities. Eur J Med Chem 2017;125:143–89.
- Yadav G, Ganguly S. Structure activity relationship (SAR) study of benzimidazole scaffold for different biological activities: a mini-review. Eur J Med Chem 2015;97:419–43.
- Akhtar MJ, Siddiqui AA, Khan AA, et al. Design, synthesis, docking and QSAR study of substituted benzimidazole linked oxadiazole as cytotoxic agents, EGFR and erbB2 receptor inhibitors. Eur J Med Chem 2017;126:853–69.
- Bhattacharya S, Chaudhuri P. Medical implications of benzimidazole derivatives as drugs designed for targeting DNA and DNA associated processes. Curr Med Chem 2008;15:1762–77.
- Sontakke VA, Lawande PP, Kate AN, et al. Antiproliferative activity of bicyclic benzimidazole nucleosides: synthesis, DNA-binding and cell cycle analysis. Org Biomol Chem 2016;14:4136–45.
- Hegde M, Kumar KSS, Thomas E, et al. A novel benzimidazole derivative binds to the DNA minor groove and induces apoptosis in leukemic cells. RSC Adv 2015;5:93194–208.
- Singla P, Luxami V, Paul K. Triazine–benzimidazole conjugates: synthesis, spectroscopic and molecular modelling studies for interaction with calf thymus DNA. RSC Adv 2016;6:14741–50.
- Chaudhuri P, Ganguly B, Bhattacharya S. An experimental and computational analysis on the differential role of the positional isomers of symmetric bis-2-(pyridyl)-1H-benzimidazoles as DNA binding agents. J Org Chem 2007;72:1912–23.
- Thomas JR, Hergenrother PJ. Targeting RNA with small molecules. Chem Rev 2008;108:1171–224.
- Guan L, Disney MD. Recent advances in developing small molecules targeting RNA. ACS Chem Biol 2012;7:73–86.
- Zheng Y, Cai X, Bradley JE. MicroRNAs in parasites and parasite infection. RNA Biol 2013;10:371–9.
- Cheng G, Dai M, Ahmed S, et al. Antimicrobial drugs in fighting against antimicrobial resistance. Front Microbiol 2016;7:470.
- Cassir N, Rolain JM, Brouqui P. A new strategy to fight antimicrobial resistance: the revival of old antibiotics. Front Microbiol 2014;5:551.
- Yang SW, Pan J, Yang C, et al. Benzimidazole analogs as WTA biosynthesis inhibitors targeting methicillin resistant Staphylococcus aureus. Bioorg Med Chem Lett 2016;26:4743–7.
- Hu L, Kully ML, Boykin DW, Abood N. Optimization of the central linker of dicationic bis-benzimidazole anti-MRSA and anti-VRE agents. Bioorg Med Chem Lett 2009;19:3374–7.
- Rodrigo‐Troyano A, Sibila O. The respiratory threat posed by multidrug resistant Gram‐negative bacteria. Respirology 2017;22:1288–99.
- Ansari KF, Lal C. Synthesis and evaluation of some new benzimidazole derivatives as potential antimicrobial agents. Eur J Med Chem 2009;44:2294–9.
- Sharma D, Narasimhan B, Kumar P, Jalbout A. Synthesis and QSAR evaluation of 2-(substituted phenyl)-1H-benzimidazoles and [2-(substituted phenyl)-benzimidazol-1-yl]-pyridin-3-yl-methanones. Eur J Med Chem 2009;44:1119–27.
- Ates-Alagoz Z. Antimicrobial activities of 1-H-benzimidazole-based molecules. Curr Top Med Chem 2016;16:2953–62.
- Song D, Ma S. Recent development of benzimidazole‐containing antibacterial agents. Chem Med Chem 2016;11:646–59.
- Desai N, Trivedi A, Pandit U, et al. Hybrid bioactive heterocycles as potential antimicrobial agents: a review. Mini Rev Med Chem 2016;16:1500–26.
- El-Gohary NS, Shaaban MI. Synthesis and biological evaluation of a new series of benzimidazole derivatives as antimicrobial, antiquorum-sensing and antitumor agents. Eur J Med Chem 2017;131:255–62.
- Klimesová V, Kocí J, Pour M, et al. Synthesis and preliminary evaluation of benzimidazole derivatives as antimicrobial agents. Eur J Med Chem 2002;37:409–18.
- Gibney KA, Sovadinova I, Lopez AI, et al. Poly(ethylene imine)s as antimicrobial agents with selective activity. Macromol Biosci 2012;12:1279–89.
- Sharma V, Chitranshi N, Agarwal AK. Significance and biological importance of pyrimidine in the microbial world. Int J Med Chem 2014;2014:202784.
- Anthony NG, Breen D, Clarke J, et al. Antimicrobial lexitropsins containing amide, amidine, and alkene linking groups. J Med Chem 2007;50:6116–25.
- Bonandi E, Christodoulou MS, Fumagalli G, et al. The 1,2,3-triazole ring as a bioisostere in medicinal chemistry. Drug Discov Today 2017;22:1572–81.
- Dheer D, Singh V, Shankar R. Medicinal attributes of 1,2,3-triazoles: current developments. Bioorg Chem 2017;71:30–54.
- Tiwari VK, Mishra BB, Mishra KB, et al. Cu-catalyzed click reaction in carbohydrate chemistry. Chem Rev 2016;116:3086–240.
- DeSimone RW, Currie KS, Mitchell SA, et al. Privileged structures: applications in drug discovery. Comb Chem High Throughput Screening 2004;7:473–93.
- Gaba M, Singh S, Mohan C. Benzimidazole: an emerging scaffold for analgesic and anti-inflammatory agents. Eur J Med Chem 2014;76:494–505.
- Jadhav GR, Shaikh MU, Kale RP, et al. SAR study of clubbed [1,2,4]-triazolyl with fluorobenzimidazoles as antimicrobial and antituberculosis agents. Eur J Med Chem 2009;44:2930–5.
- Kühler TC, Swanson M, Christenson B, et al. Novel structures derived from 2-[[(2-pyridyl)methyl]thio]-1H-benzimidazole as anti-Helicobacter pylori agents, Part 1. J Med Chem 2002;45:4282–99.
- Maračić S, Kraljević TG, Paljetak HČ, et al. 1,2,3-Triazole pharmacophore-based benzofused nitrogen/sulfur heterocycles with potential anti-Moraxella catarrhalis activity. Bioorg Med Chem 2015;23:7448–63.
- Ranjan N, Story S, Fulcrand G, et al. Selective inhibition of Escherichia coli RNA and DNA topoisomerase I by Hoechst 33258 derived mono- and bisbenzimidazoles. J Med Chem 2017;60:4904–22.
- Grillot AL, Tiran AL, Shannon D, et al. Second-generation antibacterial benzimidazole ureas: discovery of a preclinical candidate with reduced metabolic liability. J Med Chem 2014;57:8792–816.
- Janupally R, Jeankumar VU, Bobesh KA, et al. Structure-guided design and development of novel benzimidazole class of compounds targeting DNA gyrase B enzyme of Staphylococcus aureus. Bioorg Med Chem 2014;22:5970–87.
- Lam T, Hilgers MT, Cunningham ML, et al. Structure-based design of new dihydrofolate reductase antibacterial agents: 7-(benzimidazol-1-yl)-2,4-diaminoquinazolines. J Med Chem 2014;57:651–68.
- Whalen KL, Chau AC, Spies MA. In silico Optimization of a Fragment‐Based Hit Yields Biologically Active, High‐Efficiency Inhibitors for Glutamate Racemase. ChemMedChem 2013;8:1681–1689.
- Pan W, Fan M, Wu H, et al. A new small molecule inhibits Streptococcus mutans biofilms in vitro and in vivo. J Appl Microbiol 2015;119:1403–11.
- Luo G, Spellberg B, Gebremariam T, et al. Combination therapy with iron chelation and vancomycin in treating murine staphylococcemia. Eur J Clin Microbiol Infect Dis 2014;33:845–51.
- Nimesh H, Sur S, Sinha D, et al. Synthesis and biological evaluation of novel bisbenzimidazoles as Escherichia coli topoisomerase IA inhibitors and potential antibacterial agents. J Med Chem 2014;57:5238–57.
- Mann J, Taylor PW, Dorgan CR, et al. The discovery of a novel antibiotic for the treatment of Clostridium difficile infections: a story of an effective academic–industrial partnership. MedChemComm 2015;6:1420–6.
- Farahat AA, Bennett-Vaughn C, Mineva EM, et al. Synthesis, DNA binding and antitrypanosomal activity of benzimidazole analogues of DAPI. Bioorg Med Chem Lett 2016;26:5907–10.
- Karaaslan C, Kaiser M, Brun R, Göker H. Synthesis and potent antiprotozoal activity of mono/di amidino 2-anilinobenzimidazoles versus Plasmodium falciparum and Trypanosoma brucei rhodesiense. Bioorg Med Chem 2016;24:4038–44.
- Ismail MA, Batista-Parra A, Miao Y, et al. Dicationic near-linear biphenyl benzimidazole derivatives as DNA-targeted antiprotozoal agents. Bioorg Med Chem 2005;13:6718–26.
- Rice CA, Colon BL, Alp M, et al. Bis-benzimidazole hits against Naegleria fowleri discovered with new high-throughput screens. Antimicrob Agents Chemother 2015;59:2037–44.
- Alp M, Göker H, Brun R, Yildiz S. Synthesis and antiparasitic and antifungal evaluation of 2′-arylsubstituted-1H,1′H-[2,5']bisbenzimidazolyl-5-carboxamidines. Eur J Med Chem 2009;44:2002–8.
- Sola I, Artigas A, Taylor MC, et al. Synthesis and biological evaluation of N-cyanoalkyl-, N-aminoalkyl-, and N-guanidinoalkyl-substituted 4-aminoquinoline derivatives as potent, selective, brain permeable antitrypanosomal agents. Bioorg Med Chem 2016;24:5162–71.
- Pai S, Alam AS, Kapoor JN. Pentamidine salts useful in the treatment of pneumocystis carinii pneumonia, US. Patent Application US/07383, 243. US5084480 A, Fujisawa USA Inc., 1992.
- Jensch H4. 4'-Diamidino-diazoaminobenzene, a new agent in the treatment of trypanosomiasis and babesiasis. Arzneimitt Forsch 1955;5:634–635.
- Ansede JH, Anbazhagan M, Brun R, et al. As indicators of in vivo efficacy in a mouse model of Trypanosoma brucei rhodesiense infection. J Med Chem 2004;47:4335–8.
- Bakunov SA, Bakunova SM, Wenzler T, et al. Synthesis and antiprotozoal activity of cationic 1,4-diphenyl-1H-1,2,3-triazoles. J Med Chem 2010;53:254–72.
- Bistrović A, Krstulović L, Harej A, et al. Design, synthesis and biological evaluation of novel benzimidazole amidines as potent multi-target inhibitors for the treatment of non-small cell lung cancer. Eur J Med Chem 2018;143:1616–34. 2017
- Stolić I, Paljetak HČ, Perić M, et al. Synthesis and structure–activity relationship of amidine derivatives of 3,4-ethylenedioxythiophene as novel antibacterial agents. Eur J Med Chem 2015;90:68–81.
- Krstulović L, Stolić I, Jukić M, et al. New quinoline-arylamidine hybrids: synthesis, DNA/RNA binding and antitumor activity. Eur J Med Chem 2017;137:196–210.
- Hranjec M, Starčević K, Zamola B, et al. New amidino-benzimidazolyl derivatives of tylosin and desmycosin. J Antibiot 2002;55:308–14.
- Chandrika NT, Shrestha SK, Ngo HX, Garneau-Tsodikova S. Synthesis and investigation of novel benzimidazole derivatives as antifungal agents. Bioorg Med Chem 2016;24:3680–6.
- Kumar R, Arora J, Prasad AK, et al. Synthesis and antimicrobial activity of pyrimidine chalcones. Med Chem Res 2013;22:5624–32.
- Harbeck RJ, McCarter YS, Ortez JH, et al. Manual of antimicrobial susceptibility testing, Washington: American Society for Microbiology; 2005.
- Stolić I, Mišković K, Magdaleno A, et al. Effect of 3,4-ethylenedioxy-extension of thiophene core on the DNA/RNA binding properties and biological activity of bisbenzimidazole amidines. Bioorg Med Chem 2009;17:2544–54.
- Scatchard G. The attractions of proteins for small molecules and ions. Ann NY Acad Sci 1949;51:660–72.
- McGhee JD, von Hippel PH. Theoretical aspects of DNA-protein interactions: co-operative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice. J Mol Biol 1974 1974;86:469–89.
- Clinical and Laboratory Standards Institute. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. M7-A7, Wayne (PA): CLSI; 2006.
- Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. M100-S17, Wayne (PA): CLSI; 2007.
- Taylor MC, Lewis MD, Fortes Francisco A, et al. The Trypanosoma cruzi vitamin C dependent peroxidase confers protection against oxidative stress but is not a determinant of virulence. PLoS Negl Trop Dis 2015;9:e0003707.
- Kappe CO, Van der Eycken E. Click chemistry under non-classical reaction conditions. Chem Soc Rev 2010;39:1280–90.
- Ohemeng KA, Nguyen VN. Ortho -Mcneil Pharmaceutical. 2-substituted phenylbenzimidazole antibacterial agents, US. Patent Application 08/924,558, 1999, WO1999011627A1.
- Rehman SU, Sarwar T, Husain MA, et al. Studying non-covalent drug–DNA interactions. Arch Biochem Biophys 2015;576:49–60.
- Arjmand F, Parveen S, Afzal M, et al. Molecular drug design, synthesis and crystal structure determination of Cu II–Sn IV heterobimetallic core: DNA binding and cleavage studies. Eur J Med Chem 2012;49:141–50.
- Banerjee M, Farahat AA, Kumar A, et al. Synthesis, DNA binding and antileishmanial activity of low molecular weight bis-arylimidamides. Eur J Med Chem 2012;55:449–54.
- Wilson WD, Tanious FA, Fernandez-Saiz M, Rigl CT. Evaluation of drug-nucleic acid interactions by thermal melting curves. Methods Mol Biol 1997;90:219–40.
- Mergny JL, Lacroix L. Analysis of thermal melting curves. Oligonucleotides 2003;13:515–37.
- Husain MA, Yaseen Z, Rehman SU, et al. Naproxen intercalates with DNA and causes photocleavage through ROS generation. Febs J 2013;280:6569–80.
- Rodger A. Circular dichroism and linear dichroism. Encyclopedia of Analytical Chemistry. John Hoboken (NJ): Wiley & Sons, Ltd. 2014.
- Liu Y, Kumar A, Boykin DW, Wilson WD. Sequence and length dependent thermodynamic differences in heterocyclic diamidine interactions at AT base pairs in the DNA minor groove. Biophys Chem 2007;131:1–14.
- O'Sullivan P, Rozas I. Understanding the guanidine-like cationic moiety for optimal binding into the DNA minor groove . ChemMedChem 2014;9:2065–73.
- Nagle PS, Quinn SJ, Kelly JM, et al. Understanding the DNA binding of novel non-symmetrical guanidinium/2-aminoimidazolinium derivatives. Org Biomol Chem 2010;8:5558–67.
- Nguyen B, Tardy C, Bailly C, et al. Influence of compound structure on affinity, sequence selectivity, and mode of binding to DNA for unfused aromatic dications related to furamidine. Biopolymers 2002;63:281–97.
- Dyatkina NB, Roberts CD, Keicher JD, et al. Minor groove DNA binders as antimicrobial agents. 1. Pyrrole tetraamides are potent antibacterials against vancomycin resistant Enteroccoci and methicillin resistant Staphylococcus aureus. J Med Chem 2002;45:805–17.
- Khalaf AI. Minor groove binders: some recent research in drug development. Curr Trends Med Chem 2009;6:53–63.
- Havrylyuk D, Zimenkovsky B, Karpenko O, et al. Synthesis of pyrazoline–thiazolidinone hybrids with trypanocidal activity. Eur J Med Chem 2014;85:245–54.