?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Cholinesterases (ChEs) are enzymes that break down neurotransmitters associated with cognitive function and memory. We isolated cinnamic acids (1 and 2), indolinones (3 and 4), and cycloartane triterpenoid derivatives (5–19) from the roots of Cimicifuga dahurica (Turcz.) Maxim. by chromatography. These compounds were evaluated for their inhibitory activity toward ChEs. Compound 1 was determined to have an IC50 value of 16.7 ± 1.9 μM, and to act as a competitive inhibitor of acetylcholinesterase (AChE). Compounds 3, 4 and 14 were found to be noncompetitive with IC50 values of 13.8 ± 1.5 and 6.5 ± 2.5 μM, and competitive with an IC50 value of 22.6 ± 0.4 μM, respectively, against butyrylcholinesterase (BuChE). Our molecular simulation suggested each key amino acid, Tyr337 of AChE and Asn228 of BuChE, which were corresponded with potential inhibitors 1, and 3 and 4, respectively. Compounds 1 and 4 were revealed to be promising compounds for inhibition of AChEs and BuChEs, respectively.
Introduction
Cholinesterases (ChEs), which are enzymes that hydrolyse choline esters, are classified as acetylcholinesterase (EC 3.1.1.7, AChE) and butyrylcholinesterase (EC 3.1.1.8, BuChE)Citation1. AChE is responsible for the conversion of acetylcholine (ACh) into choline and acetic acid in cholinergic synapses. AChE is formed as a tetramer of ∼70-kDa monomeric subunitsCitation1,Citation2. Its 3D structure was revealed by examining the enzyme of electric eelsCitation3. AChE has an active site with α-helix and β-sheet structures and a catalytic triad of serine, histidine, and glutamic acidCitation1,Citation3. BuChE, an enzyme that breaks down artificial butyrylcholine, is known to hydrolyse ACh and other ester derivatives in the bodyCitation4,Citation5. BuChE, which is a tetrameric serine esterase consisting of monomers of ∼90-kDa molecular mass, showed over 65% structural similarity to AChECitation4,Citation6. ACh is a neurotransmitter that is produced from the acetylation reaction of choline and acetyl-CoA by choline acetyltransferase, and is distributed in the central and peripheral nervous systemsCitation7. ACh plays a key role in nerve-nerve communication by binding to ACh receptorsCitation8. This molecule is associated with maintenance of cognitive function and memoryCitation5,Citation8. In particular, Alzheimer’s disease (AD) patients are characterised by a decline in ACh levelsCitation8. Two ChEs have been regarded as target enzymes for treatment of ADCitation1,Citation5,Citation8.
Cimicifuga dahurica (Turcz.) Maxim., in the family Ranunculaceae, is commonly called “shengma” and is distributed throughout Korea, Japan, China and RussiaCitation9. In China, the rhizomes of C. dahurica have been used as a traditional medicine to treat headaches and toothachesCitation10. Phytochemical studies of this plant indicated the presence of cycloartane triterpenoids and cinnamic acid derivativesCitation9. These compounds exhibit neuroprotective activity and enhance cell viability by eliminating H2O2 in PC12 cellsCitation9,Citation11. Cycloartane triterpenoids have anti-tumour activities including induction of apoptosis and G2/M cell cycle arrest in solid tumours, blood tumours and drug-resistant tumoursCitation10.
These findings led us to search for products that block the catalytic reaction of ChE. We isolated compounds 1–19 from the roots of C. dahurica using open column chromatography (CC). These compounds were tested for interactions with both AChE and BuChE in vitro. Through molecular simulation, the inhibitor- ChE complex structure was predicted visually using the Autodock 4.2 programme. The complex that was constructed considered the interaction between the inhibitor and ChE in terms of molecular dynamics (MDs).
Materials and methods
General experimental procedures
Optical rotations were measured using a JASCO P-2000 polarimeter (JASCO, Oklahoma, OK, USA). IR spectra were obtained on a Bruker TENSOR 37 FT-IR spectrometer (Bruker, Billerica, MA, USA). NMR spectra were recorded on JEOL JNM-AL 400 MHz and JEOL ECA 600 MHz spectrometer (JEOL, Peabody, MA, USA), chemical shift (δ) are expressed in ppm with reference to the TMS signals. Gas chromatography spectra were recorded on a Shimadzu-2010 spectrometer (Shimadzu, Kyoto, Japan), SPB-1 capillary (30 m × 0.25 mm and 30 m × 0.32 mm); Mightysil RP-18 GP, Kanto Chemical, 10 × 250 mm. The electrospray ionisation and the high-resolution electrospray ionisation mass spectrometer were operated in the positive-ion mode, with sodium iodide being used for mass calibration from an Agilent 6530 Accurate-Mass Q-TOF LC/MS system (Micromass, Wythenshawe, UK). CC was conducted using on 65–250 or 230–400 mesh silica gel (Sorbent Technologies, Atlanta, GA, USA), porous polymer gel (Diaion® HP-20, 20–60 mesh, Mitsubishi Chemical, Tokyo, Japan), Sephadex™ LH-20 (Supelco, Bellefonte, PA, USA), octadecyl silica (ODS, 50 μm, Cosmosil 140 C18-OPN, Nacalai Tesque), and YMC RP-C18 resins (30–50 μm, Fuji Silysia Chemical). Analytical thin layer chromatography (TLC) systems were performed on precoated silica gel 60 F254 (1.05554.0001, Merck) and RP-18 F254S plates (1.15685.0001, Merck) and compounds were visualised by spraying with 10% H2SO4 in water and then heating for 1.5–2 min. All procedures were carried out with solvents purchased from commercial sources that were used without further purification.
Chemicals and reagents
AChE (C3389), acetylthiocholine iodide (A5751), BuChE (C1057), butyrylthiocholine iodide (B3253) and 5,5-dithiobis(2-nitrobenzoic acid) (DTNB) were purchased from Sigma-Aldrich (St Louis, MO, USA).
Plant material
The roots (3.5-years old) of C. dahurica were purchased from a herbal company, Naemome Dah, Ulsan, Korea, in February 2016. This sample was identified by Prof. Y.H. Kim. A voucher specimen (CNU-16003) representing this collection has been deposited at the Herbarium of the College of Pharmacy, Chungnam National University, Daejeon, Korea.
Extraction and isolation
The roots of C. dahurica (2.5 kg) were extracted three times with 5.0 L of 95% ethanol at 40 °C. Concentrated ethanol extract (65.3 g) was suspended in distilled water and progressively fractioned with n-hexane (9.6 g), dichloromethane (15.2 g) and water (40.5 g) fractions.
Dichloromethane fraction was subjected to silica gel CC by using gradient solvent system of n-hexane and EtOAc (from 95:5 to 2.5:5) to achieve seven fractions (D1?D7). D3 fraction was chromatographed by Sephadex LH-20 CC with MeOH-H2O solvent (95:5) to isolate compounds 7 (10.2 mg) and 10 (8.9 mg). Compounds 5 (13.5 mg), 6 (4.0 mg), 9 (3.8 mg) and 13 (3.1 mg) were purified by silica gel CC with an isocratic solvent system of n-hexane and EtOAc (2:1) from D4 fraction. D5 fraction was chromatographed by RP-C18CC using mixture solvent system of acetone and H2O (1:4) to afford compounds 1 (3 mg), 8 (5.6 mg) and 12 (3.0 mg). D7 fraction was purified by over Sephadex LH-20 CC with MeOH solvent to give three fractions (D7.1?D7.3). D7.1 fraction was subjected to RP-C18 CC by using solvent system of MeOH and H2O (5.5:1) to obtain compounds 3 (20 mg) and 4 (5 mg). Compounds 2 (32.6 mg) and 11 (5 mg) were isolated by over silica gel CC with solvent system of CH2Cl2 and acetone (4:1) from D7 fraction. H2O fraction was subjected to a Diaion HP-20 CC by using gradient solvent system of MeOH and H2O (from 25:75 to 100:0) to give four fractions (H1–H4). H2 fraction was isolated as compounds 14 (11.7 mg), 15 (18.2 mg), 18 (15.2 mg) and 19 (10.1 mg) by RP-C18 CC with gradient solvent system of MeOH and H2O (from 35:65 to 100:0). H4 fraction was purified by over RP-C18 CC with solvent system of MeOH and H2O (35:65) to achieve compounds 16 (10.3 mg) and 17 (3.5 mg).
ChE assay
AChE and BuChE inhibition assays were performed as described by Othman et al.Citation12 with some modifications. Briefly, each 130 μL of AChE (∼0.05 U/mL) and BuChE (∼0.05 U/mL) in 50 mM phosphate buffer (pH 7.4) was added to 96-well plates containing 20 μL of MeOH or sample dissolved in MeOH. 25 μL acetylthiocholine iodide (5 mM) or butyrylthiocholine iodide (5 mM), and 25 μL DTNB (1 mM) were added into the mixture in order. After initiating ChE reaction at 37 °C, the products were scanned at 475 nm UV-Vis photometer for 20 min. The inhibition activity was calculated using the following equation:
Where control and sample were the intensity of control and inhibitor after 20 min, respectively.
The ChE inhibitory activity of each sample was expressed as IC50 (µM required to inhibit the hydrolysis of the ChE substrates by 50%) determined from the log-dose inhibition curve.
Molecular docking of inhibitor with ChE
Molecular docking was performed as previously described using the Autodock 4.2 programme (La Jolla, CA, USA)Citation12. Single bond of ligand was flexibly assigned by using torsion tree of Autodocktools. Each pdb files of AChE (pdb ID: 1C2B) and BuChE (pdb ID: 4BDS) were downloaded from RCSB protein data bank. Achieved protein was added in hydrogens, and then this was assigned with compute gasteiger charges. For the docking, the grid containing activity site or all protein was set. Ligand was docked into that with default values of genetic algorithm parameters (number of GA runs: 50, maximun number of evals: 25,000,000). The result was presented with Ligplot (Cambridge, UK) and Chimaera (San Francisco, CA, USA).
Molecular simulation of inhibitor with ChE
MDs were performed to simulate the complex of ligand with protein by the Gromacs version 4.6.5 package. Itp and gro files of ligand were built at Prodrg server. Gro and topology files of ChE were generated by pdbgmx utility. These were edited to add ligand files. The ligand with protein was dissolved in water molecules of a cubic box with a size of 12 × 12 × 12 containing six sodium ions (1.0 Å distance). Moreover, then this complex was minimised until it reach the maximal force of 10 kJ/mol. Each NVT and NPT was simulated at 300 K temperature and 1 bar pressure for 100 ps in the order, respectively. Lastly, equilibrated complex was subjected to MD simulation for 10,000 ps.
Statistical analysis
Statistical significance was determined using a one-way analysis of variance and Students t-test (Systat Inc., Evanston, IL, USA). A p value <0.01 was considered significant. All results are presented as the mean ± SEM.
Results and discussion
Isolation and identification
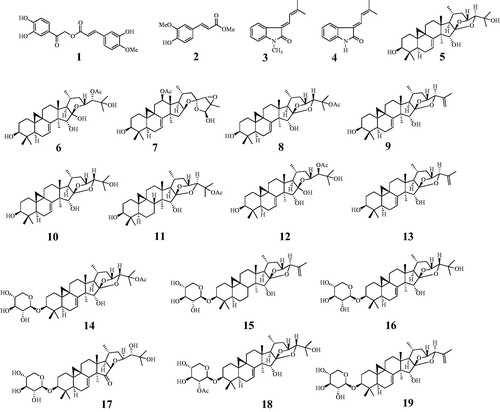
An ethanol extract of the roots of C. dahurica was progressively partitioned into n-hexane, dichloromethane, and water fractions. The dichloromethane and water fractions were subjected to various CC methods to obtain compounds 1–13 and 14–19, respectively. These compounds were investigated based on spectroscopic data and comparison with previous reports. The nineteen extracted compounds were identified as cimiciphenone (1)Citation13, ferulic acid methyl ester (2)Citation13, (E)-3‐(3′-methyl-2′-butenylidene)-1-methyl-2-indolinone (3)Citation13, (E)-3‐(3′-methyl-2′-butenylidene)-2-indolinone (4)Citation13, 7,8-didehydrocimigenol (5), 24-epi-24-O-acetyl-7,8-didehydroshengmanol (6)Citation14, 25-triepoxy-12β-acetoxy-3β,26-dihydroxy-9,19-cyclolanost-7-ene (7)Citation15, 25-O-acetyl-7,8-didehydrocimigenol (8)Citation16, 25-anhydro-7,8-didehydrocimigenol (9)Citation15, 24-epi-7,8-didehydrocimigenol (10)Citation16, 25-O-acetylcimigenol (11)Citation15, 24-epi-24-O-acetyl-7,8-didehydroshengmanol (12)Citation14, 25-anhydrocimigenol (13)Citation15, 25-O-acetyl-7,8-didehydrocimigenol 3-O-β-D-xylopyranoside (14)Citation14, 25-anhydrocimigenol-3-O-β-D-xylopyranoside (15)Citation17, 24-epi-7,8-didehydrocimigenol 3-O-β-D-xylopyranoside (16)Citation14, 3-O-β-D-xylopyranosyl-24S,25-dihydroxy-15-oxo-acta-(16R,23R)-16,23-monoxoside (17)Citation18, cimiricaside A(18)Citation19, and 7,8-didehydro-25-anhydrocimigenol-3-O-β-D-xylopyranoside (19)Citation17 ().
ChE assay
To screen for the ability of the isolated compounds 1–19 to block catalytic reaction of ChE, they were analysed in vitro at 100 μM concentration using a UV-spectrophotometer. As shown in and , compounds 1–4 and 6–8, and compounds 2–6, 9 and 14–18 exhibited over 50% inhibitory activity against AChE and BuChE, respectively. To calculate their IC50 values, these compounds were subjected to an enzyme assay at a variety of concentrations. They caused decreases in activities of the two ChEs, with gradual or sharp slopes in activity curves in a dose-dependent manner (, ). These results showed that compounds 1– 4 and 6 – 8 had IC50 values ranging from 16.7 ± 1.9 to 95.8 ± 5.1 μM against AChE. In particular, compound 1 had an IC50 value of 16.7 ± 1.9 μM. Compounds 2–6, 9 and 14–18 were revealed to have IC50 values ranging from 6.5 ± 2.5 to 90.9 ± 6.0 μM against BuChE. Compounds 3, 4 and 14 exhibited greater inhibitory activity against BuChE than the other compounds tested, with IC50 values of 13.8 ± 1.2, 6.5 ± 2.5 and 22.6 ± 0.4 μM.
Figure 2. Inhibitory activity of compounds 1–19 at 100 μM on AChE and BuChE (A). IC50 values of them on AChE (B) and BuChE (C). Lineweaver-Burk plots of compound 1 on AChE (D) and of compounds 3, 4, and 14 on BuChE (E-G). Secondary plot of compounds 1, 3, 4 and 14 (H).
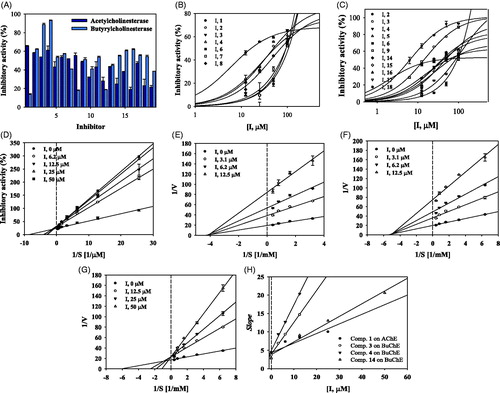
Table 1. Inhibitory activity of compounds 1–19 on two ChEs.
Enzyme kinetics on AChE and BuChE
As indicated in , compound 1 was competitive inhibitor which observed to have same Vmax value, and different Km values at 6.2–50 μM concentration on AChE. Compounds 3 and 4 were confirmed as noncompetitive mode due to various Vmax values and a Km value according to respective concentrations on BuChE (,F)). Whereas, compound 14 was revealed to take the binding into activity site by competing with substrate (). Additionally, these results were calculated with Ki values of the potential inhibitors using secondary replot. Compound 1 was calculated to be 16.2 ± 0.9 μM on AChE. Compounds 3, 4 and 14 were solved to be 4.9 ± 2.1, 3.5 ± 1.5 and 10.7 ± 1.3 μM on BuChE (, ).
Table 2. Enzyme kinetics of compounds 1, 3, 4 and 14 against two ChEs.
Molecular docking of inhibitors with AChE and BuChE
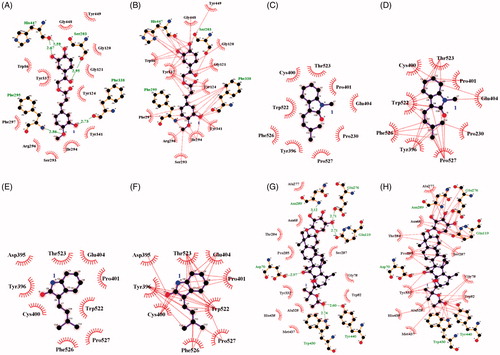
These findings suggest that compounds 1, 3, 4 and 14 may bind with either AChE or BuChE. An inhibitor (1) was found to dock into the active site of AChE, thus acting as a competitive inhibitor of this enzyme. This inhibitor was fitted into the binding site in a stable position with an Autodock score of –9.42 kcal/mol. Compound 1 formed five hydrogen bonds (Ser203: 2.95 Å; Phe295: 2.86 Å; Phe338: 2.73 Å; His447: 2.59 Å and 2.67 Å) with four amino acids in AChE and had a hydrophobic interaction with amino acids surrounding the active site (). Compounds 3, 4 and 14 exhibited molecular docking with BuChE. The noncompetitive inhibitors (3 and 4) were simulated in a blind docking test to search for possible binding with BuChE. The competitive inhibitor (14) docked into BuChE in the method described earlier. As shown in , compounds 3 and 4 were confirmed to have hydrophobic interactions with amino acids, but not to form hydrogen bonds. The predicted binding site was proposed as the location where the inhibitor clustered with a low Autodock score. In particular, compound 4 was stably placed in the active site, with the top five positions scoring from –7.25 to 7.26 kcal/mol (Supplementary Figure S1). Enzyme kinetic results showed that compound 4 preferentially bound to the allosteric site. Therefore, the catalytic site was excluded as a binding site for this compound (4). Our results predicted the binding site as that with the lowest Autodock score for the next cluster, similar to the results for compound 3. In addition, compound 14 exhibited hydrophobic interactions with seventeen residues and formed six hydrogen bonds (Asp70: 2.97Å; Glu119: 2.71 Å; Glu276: 2.71 Å; Asn289: 3.12 Å; Trp430: 2.74 Å; Tyr440: 2.60 Å) with six amino acids in the active site of BuChE ().
MDs of inhibitors with AChE and BuChE
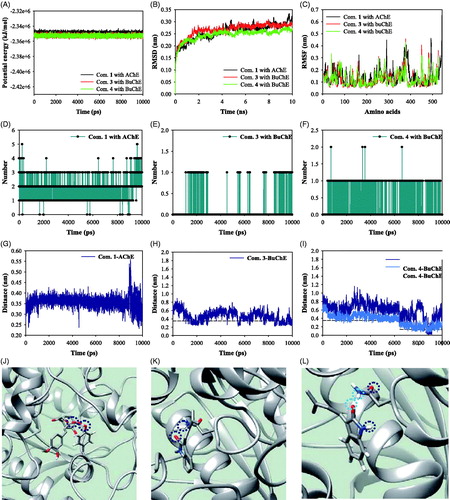
We performed MD simulations to study the stability of the inhibitor-ChE complex in solution at 300 K under 1 bar of pressure. The complexes of AChE with compound 1, and BuChE with compounds 3 and 4 were simulated stably with potential energies of about –2.35 × 106 kJ/mol (). As indicated in , each enzyme exhibited root mean square derivation (RMSD) values below 3.5 Å distance and root mean square fluctuations (RMSF) below 5.0 Å distance. Compound 1 formed about 1–3 hydrogen bonds with the active site of AChE for 10 ns (). Compounds 3 and 4 created 0–1 hydrogen bonds in the allosteric site of BuChE (,F)). Furthermore, these results indicated the key amino acids interacting with inhibitors during MD simulation. Analysis of complex formation during a 1-ns interval of MD simulation showed that compound 1 participated in hydrogen bonding with Tyr337 in the active site of AChE (). Compounds 3 and 4 were located within 3.5 Å of Asn228 in the predicted allosteric site of BuChE (). Compound 1 maintained a 3.5 Å distance with Tyr337 for 10 ns. Compounds 3 and 4 occasionally approached Asn228 at a 3.5 Å distance.
Conclusions
AD is a neurodegenerative disease caused by destruction of neurons in the central nervous systemCitation20. Cycloartane triterpenoids and cinnamic acid derivatives from the roots of C. dahurica have been reported to have a neuroprotective effect on PC12 cellsCitation9,Citation11. The cholinergic hypothesis of AD is supported by increased memory and cognition function after binding of ACh to ACh receptors in the brainCitation8,Citation21. Therefore, AChE and BuChE are considered promising target enzymes for treating AD disease due to their effect of decreasing ACh levelsCitation4,Citation8.
Our study led to isolation of cinnamic acids (1 and 2), indolinones (3 and 4), and triterpenoid derivatives (5–19) from the roots of C. dahurica. We analysed these compounds to evaluate their inhibitory activity on both AChE and BuChE. Compound 1 has an IC50 value of 16.7 ± 1.9 μM against AChE, while compounds 3 and 4 were determined to have IC50 values of 13.8 ± 1.2 and 6.5 ± 2.5 μM against BuChE, respectively. According to BuChE assay results, triterpenoid glycosides showed more potent inhibitory activities than those of their aglycones except for compound 19. Above all, indolinone derivatives (3 and 4) were highly potential inhibitors compared to the others. In reported studies, alkaloid derivatives, such as atherosperminine, (+)-N-methylisococlaurine, berberine, 9-amino-1,2,3,4-tetrahydro acridine, and rivastigmine, were found to be famous ChEs inhibitorCitation12,Citation20,Citation21. Moreover, blood brain barrier (BBB) plays a role to keep neuronal cells from neurotoxic substances of outside. However, BBB transporters of glucose, phenylalanine, araginine and lactate are responsible for transporting small molecules, such as deoxyglucose, galactose, lysine, pyruvate and guanosine, into brainCitation22,Citation23. Especially, compound 4 having 199 Da alkaloid may overcome the block of BBB and potentially invade into brain.
Based on the enzyme kinetic study, compound 1 was shown to block catalytic reaction by interacting with the active site of AChE. Compounds 3 and 4 were revealed to have affinity for the allosteric site of BuChE. Their binding positions were predicted for the active or allosteric sites using the Autodock 4.2 package. Moreover, MD analysis led us to propose the key amino acid involved in ligand-receptor interactions. As a result, the ketone form of the ester in cimiciphenone (1) exhibited hydrogen bonding with the aromatic hydroxyl group of Tyr337 in the active site of AChE during simulation. To develop a new cinnamic acid moiety of AChE, chemists should consider compounds that are capable of interaction with Tyr337. (E)-3–(3′-methyl-2′-butenylidene)-1-methyl-2-indolinone (3) and (E)-3–(3′-methyl-2′-butenylid-ene)-2-indolinone (4) participated in hydrogen bonding with Asn228 located at its predicted binding site on BuChE. It is necessary to develop a prenyl indolinone derivative as a noncompetitive inhibitor to enhance hydrogen bonding with this polar amino acid (Asn228). In our research, we identified the key amino acids, which could not be found through molecular docking, using MD analysis. Among cycloartane triterpenoids and cinnamic acid derivatives that show neuroprotective activityCitation9,Citation11, cimiciphenone (1), (E)-3–(3′-methyl-2′-butenylidene)-1-methyl-2-indolinone (3) and (E)-3–(3′-methyl-2′-butenylidene)-2-indolinone (4) showed promise as potential inhibitors of AChE and BuChE, respectively. Compound 1 was determined to be the optimal compound for development as a competitive inhibitor of AChE, while compounds 3 and 4 may provide a new skeleton for noncompetitive inhibitors of BuChE.
Supplemental Material
Download PDF (417.2 KB)Disclosure statement
The authors have declared no conflict of interest.
Additional information
Funding
References
- Orhan IE, Senol FS, Shekfeh S, et al. Pteryxin-A promising butyrylcholinesterase-inhibiting coumarin derivative from Mutellina purpurea. Food Chem Toxicol 2017;109:970–4.
- Zhang X-J, Greenberg D. Acetylcholinesterase involvement in apoptosis. Front Mol Neurosci 2012;5:1–6.
- Dvir H, Silman I, Harel M, et al. Acetylcholinesterase: From 3D structure to function. Chem Biol Interact 2010;187:10–22.
- Ali R, Sheikh IA, Jabir NR, Kamal MA. Comparative review of decade’s research on cholinesterase inhibition. Am J Neuroprot Neuroregen 2012;4:136–8.
- Nguyen VTN, Zhao BT, Seong SH, et al. Inhibitory effects of serratene-type triterpenoids from Lycopodium complanatum on cholinesterases and β-secretase 1. Chem Biol Interact 2017;274:150–7.
- Huang Y-J, Huang Y, Baldassarre H, et al. Recombinant human butyrylcholinesterase from milk of transgenic animals to protect against organophosphate poisoing. Pro Natl Acad Sci USA 2007;104:13603–8.
- de Souza LG, Rennã MN, Figueroa-Villar JD. Coumarins as cholinesterase inhibitors: A review. Chem Biol Interact 2016;254:11–23.
- Islam MM, Rohman MA, Gurung AB, et al. Correlation of cholinergic drug induced quenching of acetylcholinesterase bound thioflavon-T fluorescence with their inhibition activity. Spectrochim Acta A 2018;189:250–7.
- Lv C, Yang F, Qin R, et al. Bioactivity-guided isolation of chemical constituents against H2O2-induced neurotoxicity on PC12 from Cimicifuga dahurica (Turcz.) Maxim. Bioorg Med Chem Lett 2017;27:3305–9.
- Tian Z, Si J, Chang Q, et al. Antitumor activity and mechanisms of action of total glycosides from aerial part of Cimicifuga dahurica targeted against hepatoma. BMC Cancer 2007;7:7–10.
- Qin R, Zhao Y, Zhao Y, et al. Polyphenolic compounds with antioxidant potential and neuro-protective effect from Cimicifuga dahurica (Turcz.) Maxim. Fitoterapia 2016;115:52–6.
- Othman WNNW, Liew SY, Khaw KY, et al. Cholinesterase inhibitory activity of isoquinoline alkaloids from three Cryptocarya species (Lauraceae). Bioorg Med Chem 2016;24:4464–9.
- Thao NP, Luyen BTT, Lee JS, et al. Soluble epoxide hydrolase inhibitors of indolinone alkaloids and phenolic derivatives from Cimicifuga dahurica (Turcz.) Maxim. Bioorg Med Chem Lett 2017;27:1874–9.
- Kusano A, Takahira M, Shibano M, et al. Studies on the Constituents of Cimicifuga Species. XXVI. Twelve New Cyclolanostanol Glycosides from the Underground Parts of Cimicifuga simplex WORMSK. Chem Pharm Bull 1999;47:511–6.
- Thao NP, Kim JH, Luyen BTT, et al. In silico investigation of cycloartane triterpene derivatives from Cimicifuga dahurica (Turcz.) Maxim roots for the development of potent soluble epoxide hydrolase inhibitors. Inter J Biol Macromole 2017;98:526–34.
- Li JX, Kadota S, Hattori M, et al. Constituents of Cimicifugae Rhizoma. I. Isolation and Characterization of Ten New Cycloartenol Triterpenes from Cimicifuga heracleifolia KOMAROV. Chem Pharm Bull 1993;41:832–41.
- Thao NP, Luyen BT, Lee JS, et al. Inhibition potential of cycloartane-type glycosides from the roots of Cimicifuga dahurica against soluble epoxide hydrolase. J Nat Prod 2017;80:1867–75.
- Imai A, Lankin DC, Nikolić D, et al. Cycloartane triterpenes from the aerial parts of Actaea racemosa. J Nat Prod 2016;79:541–54.
- Yoshimitsu H, Nishida M, Sakaguchi M, Nohara I. Two new 15-deoxycimigenol-type and three new 24-epi-cimigenol-type glycosides from Cimicifuga rhizome. Chem Pharm Bull 2006;54:1322–5.
- Groner E, Ashani Y, Schorer-Apelbaum D, et al. The kinetics of Inhibition of Human acetylcholinesterase and butyrylcholinesterase by two series of novel carbamates. Mol Pharmacol 2007;71:1610–7.
- Jiang Y, Gao H, Turdu G. Traditional Chinese medicinal herbs as potential AChE inhibitors for anti-Alzheimer’s disease: A review. Bioorg Chem 2017;75:50–61.
- Pardridge WM. Drug transport across the blood-brain barrier. J Cereb Blood Flow Metab 2012;32:1959–72.
- Grabrucker AM, Ruozi B, Belletti D, et al. Nanoparticle transport across the blood brain barrier. Tissue Barriers 2016; 4:e1153568–19.