Abstract
Histamine H3 receptor (H3R), a kind of G-protein coupled receptor (GPCR), is expressed mainly in the central nervous system (CNS) and plays a vital role in homoeostatic control. This study describes the design and synthesis of a series of novel H3R antagonists based on the iso-flavone scaffold. The results of the bioactivity evaluation show that four compounds (1c, 2c, 2h, and 2o) possess significant H3R inhibitory activities. Molecular docking indicates that a salt bridge, π–π T-shape interactions, and hydrophobic interaction all contribute to the interaction between compound 2h and H3R.
Introduction
Histamine, a distinctly important neurotransmitter, exerts as a modulator in the brain and dominates several homoeostatic functions such as thermoregulation, fluid balance, and energy metabolismCitation1. Apart from that, histamine is also involved in numerous processes, for instance, circadian rhythms, the sleep–wake cycle, attention, memory, learning, and neuroendocrine regulationCitation2. According to recent studies, the biosynthesis and release of histamine in central nervous system (CNS) are modulated by four different G-protein coupled receptors (GPCRs) subtypes, namely histamine H1 receptor (H1R), histamine H2 receptor (H2R), histamine H3 receptor (H3R), histamine H4 receptor (H4R). Unlike H1R and H2R, H3R shows higher homology to H4RCitation3 and is highly expressed in brainCitation4, such as basal ganglia and globus pallidus, which could couple with G i/oα protein and then activate mitogen-activated protein kinase (MAPK) and phosphatidylinositol 3-kinase (PI3K) pathwaysCitation5. Subsequently, the phospholipase A2 (PLA2) is induced to recruit Ca2+ from intracellular storesCitation6, reduces cAMP formationCitation7, and enhances phosphorylationCitation2. Moreover, H3R is recognised as an auto- and hetero-receptor on non-histaminergic neurons controlling the release of many other important neurotransmittersCitation8,Citation9, such as acetylcholine, norepinephrine, dopamine, and serotoninCitation10. A clinical study revealed that neurotransmitters could trigger the postsynaptic signalling pathways bound to cognition which supported the hypothesis that H3R is a drug target for cognitive disordersCitation3,Citation6,Citation11,Citation12, especially for Alzheimer Disease (AD), schizophrenia and epilepsyCitation13–16. Because of the unique functions of H3R, a wide variety of selective H3R antagonists have been developed and some of them have shown promising effectsCitation4,Citation12,Citation17–21.
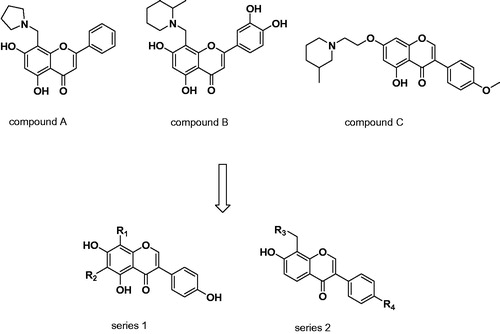
Flavone and iso-flavone, which are regarded as privileged structures, exhibit variety of pharmacological activities, such as anti-cancer, antimicrobial, anti-inflammatory, and also are used in neurodegenerative disorders, for example, Alzheimer’s diseaseCitation22–24. Our previous study had confirmed the iso-flavone and flavone compounds possessed moderate inhibitory activity against H3RCitation25. Particularly, the optimization at the 8-position of the flavones and 7-position of iso-flavone provided satisfactory bioactivity (compound A, B, and C, ), which enlightened us to modify 8-position of iso-flavone to enhance the H3R inhibitory effect. In addition, we also want to modify the 6-position of isoflavones to see whether compounds with better antagonistic activity can be obtained. In this current work, two series of novel iso-flavone derivatives were designed and synthesised based on our previous study. After screening the H3R inhibitory activities at a fixed concentration, compounds that possessed good H3R inhibitory activity were further tested to determine the IC50 values. In addition, molecular docking studies were performed to investigate the interaction between H3R and the most potent antagonist.
Materials and methods
Chemistry
Unless otherwise indicated, all solvents and organic reagents were obtained from commercially available sources and were used without further purification. The reaction process was monitored using thin layer chromatography (TLC) with silica gel plates (thickness = 0.20 mm, GF254) under UV light. Column chromatography was performed using a ZCX-II (200–300 mesh), to purify the final products. All final products were found to have purities ≥95% analysed by HPLC. Melting points were determined using a YRT-3 apparatus (Tian Jin Optical Instrument Factory, Tianjin, China) and were presented as uncorrected values. 1H NMR spectra were recorded on a Varian Mercury-300 MHz instrument, whereas 13 C NMR was recorded at 400 MHz on a Varian Mercury using DMSO-d6 as a solvent and tetramethylsilane (TMS) as an internal standard (1H NMR and 13 C NMR were recorded in different time). Mass spectra were obtained using a Waters Acquity UPLC-SQD mass spectrometer (Waters, Milford, MA). High-resolution mass spectra (HRMS) were recorded on an Agilent Technologies LC/MSD TOF spectrometer (Agilent Technologies Co. Ltd., Santa Clara, CA).
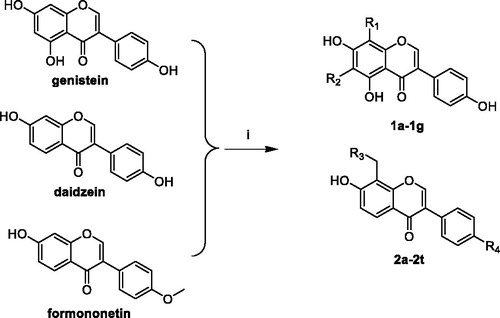
The synthetic route of novel compounds is depicted in Scheme 1. All title compounds were synthesised through Mannich reactions using iso-flavone, 37% formalin, and aliphatic amines as starting materials. Compounds 1a–1g, 2a–2i, and 2j–2t were synthesised from genistein, daidzein, and formononetin, respectively. The use of DMF-methanol as a solvent for formononetin and daidzein never resulted in the formation of 6-substituted products, but only 8-position substituted products were obtained.
General procedure for the synthesis of compounds 1a–1g
Genistein (0.50 g, 1.85 mmol), 37% formalin (0.30 g, 3.70 mmol), aliphatic amines (0.225 g, 2.780 mmol), and methanol (30 ml) were added into a three-necked flask (100 ml) and stirred at 25 °C for 24 h. After reactions completed monitored by TLC (DCM:MeOH = 10:1), the solvent was removed under reduced pressure. The residue was purified by column chromatography using a mixture of dichloromethane and methanol (30:1) as the eluent to give the target compounds in yields ranging from 41% to 91%.
The similar procedure was followed for the synthesis of compounds 2a–2t.
Title compounds were characterised as follows:
5,7-dihydroxy-3-(4-hydroxyphenyl)-8-(pyrrolidin-1-ylmethyl)-4H-chromen-4-one (1a)
White solid, yield: 24%; mp 218–220 °C; 1H NMR (300 MHz, DMSO-d6) δ 8.15 (s, 1H), 7.35 (d, J = 8.7 Hz, 2H), 6.80 (d, J = 8.4 Hz, 2H), 6.10 (s, 1H), 3.96 (s, 2H), 2.83 (m, 4H), 1.83 (m, 4H). 13 C NMR (100 MHz, DMSO-d6) δ 179.9, 170.4, 161.6, 159.5, 157.8, 153.5, 130.7, 123.8, 122.2, 115.5, 104.7, 102.2, 94.7, 53.3, 50.0, 23.6. HR-MS (ESI) Calcd for C20H19NO5 [M + H]+, 354.1341, found: 354.1368.
8-((4-benzylpiperazin-1-yl)methyl)-5,7-dihydroxy-3-(4-hydroxyphenyl)-4H-chromen-4-one (1b)
White solid, yield: 24%; mp 191–193 °C; 1H NMR (300 MHz, DMSO-d6) δ 8.33 (s, 1H), 7.35–7.28 (m, 7H), 6.81 (d, J = 8.4 Hz, 2H), 6.17 (s, 1H), 3.81 (s, 2H), 3.46 (s, 2H), 2.56 (m, 4H), 2.40 (m, 4H). 13 C NMR (100 MHz, DMSO-d6) δ 180.9, 165.5, 161.1, 157.9, 155.6, 154.2, 138.5, 130.7, 129.4, 128.7, 127.5, 122.7, 121.7, 115.6, 104.7, 100.1, 99.5, 62.3, 52.8, 52.5, 51.7. HR-MS (ESI) Calcd for C27H26N2O5 [M + H]+, 459.1920, found: 459.1939.
5,7-dihydroxy-3-(4-hydroxyphenyl)-8-((3-methylpiperidin-1-yl)methyl)-4H-chromen-4-one (1c)
White solid, yield: 18%; mp 209–211 °C; 1H NMR (300 MHz, DMSO-d6) δ 8.29 (s, 1H), 7.36 (d, J = 8.7 Hz, 2H), 6.81 (d, J = 8.4 Hz, 2H), 6.10 (s, 1H), 3.84 (s, 2H), 2.89 (brs, 2H), 2.15 (t, J = 10.8 Hz, 1H), 1.89 (t, J = 10.8 Hz, 1H), 1.65–1.48 (m, 4H), 0.94 (m, 1H), 0.82 (d, J = 6.6 Hz, 3H). 13 C NMR (100 MHz, DMSO-d6) δ 180.4, 168.0, 163.5, 157.9, 157.4, 154.1, 130.7, 122.5, 121.9, 115.6, 104.0, 103.5, 94.5, 60.0, 53.1, 52.7, 32.0, 30.9, 24.9, 19.6. HR-MS (ESI): Calcd for C22H23NO5 [M + H]+, 382.1654, found: 382.1682.
6-(((3R,5S)-3,5-dimethylmorpholino)methyl)-5,7-dihydroxy-3–(4-hydroxyphenyl)-4H-chromen-4-one (1d)
White solid, yield: 20%; mp >250 °C; 1H NMR (300 MHz, DMSO-d6) δ 13.02 (s, 1H), 8.36 (s, 1H), 7.37 (d, J = 8.4 Hz, 2H), 6.81 (d, J = 8.4 Hz, 2H), 6.22 (s, 1H), 3.73 (s, 2H), 3.54 (t, J = 9 Hz, 2H), 2.81(d, J = 12 Hz, 2H), 1.78 (t, J = 10.8 Hz, 2H), 1.04 (d, J = 6.3 Hz, 6H). 13 C NMR (100 MHz, DMSO-d6) δ 181.0, 177.9, 176.8, 164.5, 157.9, 155.8, 154.3, 130.7, 121.7, 115.6, 104.9, 100.5, 99.4, 71.5, 58.6, 51.2, 19.4. HR-MS (ESI) Calcd for C22H23NO6 [M + H]+, 39.1604, found: 398.1633.
5,7-dihydroxy-8-((4-(hydroxymethyl)piperidin-1-yl)methyl)-3–(4-hydroxyphenyl)-4H-chromen-4-one (1e)
White solid, yield: 19%; mp 223–225 °C; 1H NMR (300 MHz, DMSO-d6) δ 8.29 (s, 1H), 7.36 (d, J = 8.4 Hz, 2H), 6.81 (d, J = 8.7 Hz, 2H), 6.09 (s, 1H), 3.86 (s, 2H), 3.57 (brs, 2H), 2.84 (t, J = 6.6 Hz, 2H), 2.38 (t, J = 10.2 Hz, 2H), 1.68 (d, J = 12.9 Hz, 2H), 1.12 (brs, 1H), 1.23–1.11 (m, 2H). 13 C NMR (100 MHz, DMSO-d6) δ 180.6, 167.4, 161.4, 157.9, 155.4, 153.8, 130.7, 122.6, 121.8, 115.6, 104.1, 99.9, 99.2, 65.9, 52.7, 52.6, 38.1, 28.6. HR-MS (ESI) Calcd for C22H23NO6 [M + H]+:398.1604, found: 398.1584.
5,7-dihydroxy-3-(4-hydroxyphenyl)-6-(morpholinomethyl)-4H-chromen-4-one (1f).
White solid, yield: 16%; mp 210–212 °C; 1H NMR (300 MHz, DMSO-d6) δ 13.03 (s, 1H), 8.36 (s, 1H), 7.38 (d, J = 8.4 Hz, 2H), 6.82 (d, J = 8.4 Hz, 2H), 6.24 (s, 1H), 3.74 (s, 2H), 3.58 (m, 4H), 2.49 (m, 4H). 13C NMR (100 MHz, DMSO-d6) δ 180.9, 164.6, 161.4, 157.9, 154.4, 143.3, 130.7, 122.7, 121.7, 115.6, 104.9, 100.7, 99.3, 66.6, 53.14, 51.4. HR-MS (ESI) Calcd for C20H19NO6 [M + H]+, 370.1291, found: 370.1320.
5,7-dihydroxy-3-(4-hydroxyphenyl)-8-((4-methylpiperazin-1-yl)methyl)-4H-chromen-4-one (1g)
White solid, yield: 19%; mp 231–233 °C; 1H NMR (300 MHz, DMSO-d6) δ 8.34 (s, 1H), 7.37 (d, J = 8.4 Hz, 2H), 6.82 (d, J = 8.7 Hz, 2H), 6.18(s, 1H), 3.80(s, 2H), 3.55(m, 4H), 2.35 (m, 4H), 2.07(s, 3H). 13 C NMR (100 MHz, DMSO-d6) δ 180.7, 166.4, 161.5, 158.0, 155.6, 154.2, 130.7, 122.7, 121.7, 115.6, 104.7, 100.1, 99.6, 54.9, 52.4, 51.7, 46.0. HR-MS (ESI) Calcd for C21H22N2O5 [M + H]+, 383.1607, found: 383.1609.
7-hydroxy-8-((4-(2-hydroxyethyl)piperazin-1-yl)methyl)-3-(4-hydroxyphenyl)-4H-chromen-4-one (2a)
White solid, yield: 20%; mp 230–232 °C; 1H NMR (300 MHz, DMSO-d6) δ 8.30 (s, 1H), 7.91 (d, J = 8.7 Hz, 1H), 7.37(d, J = 8.7 Hz, 2H), 6.88 (d, J = 9.0 Hz, 1H), 6.77 (d, J = 8.4 Hz, 2H), 3.95 (s, 2H), 3.49 (t, J = 6.3 Hz, 2H), 2.57–2.48 (m, 8H), 2.38 (t, J = 6.3 Hz, 2H). 13 C NMR (100 MHz, DMSO-d6) δ 175.3, 163.4, 157.7, 155.5, 153.0, 130.6, 126.3, 123.8, 123.0, 116.9, 115.5, 115.5, 108.7, 60.5, 59.0, 53.4, 52.7, 52.4. HR-MS (ESI) Calcd for C22H24N2O5 [M + H]+, 397.1763, found: 397.1767.
7-hydroxy-8-((4-(hydroxymethyl)piperidin-1-yl)methyl)-3-(4-hydroxyphenyl)-4H-chromen-4-one (2b)
White solid, yield: 24%; mp 244–246 °C; 1H NMR (300 MHz, DMSO-d6) δ 8.28 (s, 1H), 7.89 (d, J = 8.7 Hz, 1H), 7.37 (d, J = 8.4 Hz, 2H), 6.84–6.77 (m, 3H), 3.98 (s, 2H), 3.26 (d, J = 6.3 Hz, 2H), 2.95 (d, J = 11.1 Hz, 2H), 2.21 (t, J = 11.1 Hz, 2H), 1.73 (d, J = 13.2 Hz, 2H), 1.43 (brs, 1H), 1.19 (m, 2H). 13 C NMR (100 MHz, DMSO-d6) δ 175.3, 164.2, 157.7, 155.5, 152.9, 130.6, 126.1, 123.8, 123.0, 116.6, 115.7, 115.5, 108.4, 66.00, 53.3, 52.9, 38.2, 28.9. HR-MS (ESI) Calcd for C22H23NO5 [M + H]+, 382.1654, found: 382.1648.
7-hydroxy-3-(4-hydroxyphenyl)-8-((4-methylpiperidin-1-yl)methyl)-4H-chromen-4-one (2c)
White solid, yield: 14%; mp 250–252 °C; 1H NMR (300 MHz, DMSO-d6) δ 8.28 (s, 1H), 7.89 (d, J = 8.7 Hz, 1H), 7.37 (d, J = 8.7 Hz, 2H), 6.84–6.77 (m, 3H), 3.97 (s, 2H), 2.95 (d, J = 11.1 Hz, 2H), 2.22 (t, J = 11.4 Hz, 2H), 1.68 (d, J = 13.2 Hz, 2H), 1.42 (brs, 1H), 1.17 (m, 2H), 0.91 (d, J = 6.6 Hz, 3H). 13 C NMR (100 MHz, DMSO-d6) δ 175.3, 164.2, 157.7, 155.5, 152.9, 130.6, 126.1, 123.8, 123.0, 116.6, 115.7, 115.5, 108.5, 53.1, 34.1, 30.2, 22.0. HR-MS (ESI): Calcd for C22H23NO4 [M + H]+, 366.1705, found: 366.1749.
8-(((3R,5S)-3,5-dimethylmorpholino)methyl)-7-hydroxy-3–(4-hydroxyphenyl)-4H-chromen-4-one (2d)
White solid, yield: 27%; mp 230–232 °C; 1H NMR (300 MHz, DMSO-d6) δ 8.32 (s, 1H), 7.92 (d, J = 9 Hz, 1H), 7.38 (d, J = 9 Hz, 2H), 6.93 (d, J = 9 Hz, 1H), 6.80 (d, J = 8.4 Hz, 2H), 3.88 (s, 2H), 3.56 (t, J = 8.4 Hz, 2H), 2.83 (d, J = 10.8 Hz, 2H), 1.90 (t, J = 11.1 Hz, 2H), 1.05 (d, J = 6.3 Hz, 6H). 13 C NMR (100 MHz, DMSO-d6) δ 175.4, 162.8, 157.8, 155.8, 153.1, 130.6, 126.4, 123.8, 123.00, 117.1, 115.5, 115.4, 109.1, 71.5, 58.7, 51.8, 19.4. HR-MS (ESI) Calcd for C22H23NO5 [M + H]+, 382.1654, found: 382.1664.
7-hydroxy-3-(4-hydroxyphenyl)-8-((3-hydroxypiperidin-1-yl)methyl)-4H-chromen-4-one (2e)
White solid, yield: 15%; mp 226–228 °C; 1H NMR (300 MHz, DMSO-d6) δ 8.28 (s, 1H), 7.89 (d, J = 9 Hz, 1H), 7.37 (d, J = 8.4 Hz, 2H), 6.84–6.77 (m, 3H), 3.96 (s, 2H), 2.87 (d, J = 7.2 Hz, 2H), 2.13 (t, J = 11.1 Hz, 1H), 1.87 (t, J = 10.8 Hz, 1H), 1.69–1.42 (m, 4H), 0.90 (m, 1H), 0.84 (d, J = 6.3 Hz, 3H). 13 C NMR (100 MHz, DMSO-d6) δ 175.3, 164.2, 157.6, 155.4, 152.9, 130.6, 126.2, 123.8, 123.0, 116.6, 115.7, 115.5, 108.3, 60.6, 53.4, 53.2, 32.3, 31.2, 25.2, 19.7. HR-MS (ESI) Calcd for C22H23NO4 [M + H]+, 366.1705, found: 366.1716.
7-hydroxy-3-(4-hydroxyphenyl)-8-(pyrrolidin-1-ylmethyl)-4H-chromen-4-one (2f).
White solid, yield: 23%; mp 177–179 °C; 1H NMR (300 MHz, DMSO-d6) δ 8.27 (s, 1H), 7.88 (d, J = 8.7 Hz, 1H), 7.37 (d, J = 8.4 Hz, 2H), 6.84–6.77 (m, 3H), 4.08 (s, 2H), 2.67 (m, 4H), 1.77 (m, 4H). 13C NMR (100 MHz, DMSO-d6) δ 175.3, 164.7, 157.7, 155.4, 152.8, 130.6, 126.2, 123.8, 123.1, 116.1, 115.9, 115.5, 109.1, 53.6, 50.0, 23.7. HR-MS (ESI) Calcd for C20H19NO4 [M + H]+, 338.1392, found: 338.1413.
(S)-7-hydroxy-8-((2-(hydroxymethyl)pyrrolidin-1-yl)methyl)-3–(4-hydroxyphenyl)-4H-chromen-4-one (2g)
White solid, yield: 35%; mp 205–207 °C; 1H NMR (300 MHz, DMSO-d6) δ 8.27 (s, 1H), 7.88 (d, J = 8.7 Hz, 1H), 7.37 (d, J = 8.7 Hz, 2H), 6.85–6.77 (m, 3H), 4.34–4.01 (s, 2H), 3.51 (brs, 2H), 2.92–2.83 (d, J = 27.6 Hz, 2H), 2.40 (d, J = 8.1 Hz, 1H), 1.89 (m, 1H), 1.67 (m, 3H). 13 C NMR (100 MHz, DMSO-d6) δ 175.3, 172.8, 157.8, 155.2, 152.6, 138.2, 130.6, 126.1, 123.8, 123.0, 120.0, 115.5, 109.6, 65.6, 62.8, 54.6, 49.5, 27.6, 23.1. HR-MS (ESI) Calcd for C21H21NO5 [M + H]+, 368.1498, found: 368.1482.
7-hydroxy-3-(4-hydroxyphenyl)-8-((2-methylpiperidin-1-yl)methyl)-4H-chromen-4-one (2h)
White solid, yield: 12%; mp 228–230 °C; 1H NMR (300 MHz, DMSO-d6) δ 8.28 (s, 1H),7.89 (d, J = 8.7 Hz, 1H), 7.37 (d, J = 8.4 Hz, 2H), 6.79–6.77 (m, 3H), 4.32–4.27 (d, J = 15, 1H), 3.9–3.85 (d, J = 15 Hz, 1H), 2.83 (d, J = 12 Hz, 1H), 2.66 (brs, 1H), 2.30 (t, J = 9.3 Hz, 1H), 1.62–1.35 (m, 6H), 1.15 (d, J = 6.3 Hz, 3H). 13 C NMR (100 MHz, DMSO-d6) δ 175.3, 164.6, 157.7, 155.2, 152.9, 130.6, 125.9, 123.8, 123.1, 116.4, 115.9, 115.4, 108.7, 57.8, 56.3, 50.4, 36.3, 33.8, 25.6, 22.5. HR-MS (ESI) Calcd for C22H23NO4 [M + H]+, 366.1705, found: 366.1731.
7-hydroxy-3-(4-hydroxyphenyl)-8-((4-methylpiperazin-1-yl)methyl)-4H-chromen-4-one (2i)
White solid, yield: 18%; mp 215–217 °C; 1H NMR (300 MHz, DMSO-d6) δ 8.28 (s, 1H), 7.90 (d, J = 8.7 Hz, 1H), 7.37 (d, J = 8.7 Hz, 2H), 6.88–6.77 (m, 3H), 3.93(s, 2H), 2.56 (m, 4H), 2.34 (m, 4H), 2.15 (s, 3H). 13 C NMR (100 MHz, DMSO-d6) δ 175.4, 163.3, 157.7, 155.6, 153.1, 137.2, 130.6, 126.3, 123.9, 123.0, 116.9, 115.5, 108.9, 55.0, 52.6, 52.3, 46.1. HR-MS (ESI) Calcd for C21H22N2O4 [M + H]+, 367.1658, found: 367.1646.
7-hydroxy-3-(4-methoxyphenyl)-8-((4-methylpiperazin-1-yl)methyl)-4H-chromen-4-one (2j)
White solid, yield: 23%; mp 202–204 °C; 1H NMR (300 MHz, DMSO-d6) δ 8.36 (s,1H), 7.92 (d, J = 8.7 Hz, 1H), 7.50 (d, J = 9 Hz, 2H), 6.99 (d, J = 8.7 Hz, 2H), 6.87 (d, J = 9 Hz, 1H), 3.95 (s, 2H), 3.77 (s, 3H), 2.57 (m, 4H), 2.36 (m, 4H), 2.16 (s, 3H). 13C NMR (100 MHz, DMSO-d6) δ 175.3, 163.2, 159.5, 155.6, 153.4, 130.6, 126.3, 124.7, 123.5, 116.9, 115.5, 114.1, 109.0, 55.7, 55.0, 52.6, 52.3, 46.1. HR-MS (ESI) Calcd for C22H24N2O4 [M + H]+, 381.1814, found: 381.1814.
7-hydroxy-3-(4-methoxyphenyl)-8-(morpholinomethyl)-4H-chromen-4-one (2k).
White solid, yield: 28%; mp 235–237 °C; 1H NMR (300 MHz, DMSO-d6) δ 8.31 (s, 1H), 7.92 (d, J = 8.7 Hz, 1H), 7.38 (d, J = 8.7 Hz, 2H), 6.93 (d, J = 9 Hz, 1H), 6.77 (d, J = 8.4 Hz, 2H), 3.88 (s, 3H), 3.59 (m, 6H), 2.48 (m, 4H). 13C NMR (100 MHz, DMSO-d6) δ 175.4, 162.6, 157.7, 155.9, 153.2, 130.6, 126.4, 123.8, 123.0, 117.1, 115.5, 115.3, 109.3, 66.6, 53.2, 52.0. HR-MS (ESI) Calcd for C20H19NO5 [M + H]+, 354.1341, found: 354.1315.
7-hydroxy-3-(4-methoxyphenyl)-8-((4-methylpiperidin-1-yl)methyl)-4H-chromen-4-one (2l)
White solid, yield: 21%; mp 208–210 °C; 1H NMR (300 MHz, DMSO-d6) δ 8.33 (s, 1H), 7.90 (d, J = 8.7 Hz, 1H), 7.50 (d, J = 9 Hz, 2H), 6.95 (d, J = 11.7 Hz, 2H), 6.82 (d, J = 9 Hz, 1H), 3.98 (s, 2H), 3.77 (s, 3H), 2.96 (d, J = 11.4 Hz, 2H), 2.22 (t, J = 10.8 Hz, 2H), 1.68 (d, J = 12.3 Hz, 2H), 1.42 (brs, 1H), 1.17 (m, 2H), 0.91 (d, J = 6.6 Hz, 3H). 13C NMR (100 MHz, DMSO-d6) δ 175.2, 164.3, 159.5, 155.5, 153.2, 130.6, 126.2, 124.7, 123.5, 116.6, 115.8, 114.1, 108.4, 55.7, 53.2, 53.1, 34.1, 30.2, 22.0. HR-MS (ESI) Calcd for C23H25NO4 [M + H]+, 380.1862, found: 380.1881.
7-hydroxy-3-(4-methoxyphenyl)-8-(pyrrolidin-1-ylmethyl)-4H-chromen-4-one (2m)
White solid, yield: 12%; mp 173–175 °C; 1H NMR (300 MHz, DMSO-d6) δ 8.32 (s, 1H),7.89 (d, J = 8.7 Hz, 1H), 7.50 (d, J = 8.7 Hz, 2H), 6.98 (d, J = 8.7 Hz, 2H),6.82 (d, J = 8.7 Hz, 1H), 4.09 (s, 2H), 3.77 (s, 3H), 2.68 (m, 4H), 1.77 (m, 4H). 13 C NMR (100 MHz, DMSO-d6) δ 175.2, 164.5, 159.5, 155.4, 153.1, 130.6, 126.2, 124.8, 123.4, 116.2, 115.9, 114.1, 109.3, 55.6, 53.6, 49.9, 23.7. HR-MS (ESI) Calcd for C21H21NO4 [M + H]+, 352.1549, found: 352.1568.
7-hydroxy-8-((4-hydroxypiperidin-1-yl)methyl)-3–(4-methoxyphenyl)-4H-chromen-4-one (2n)
White solid, yield: 21%; mp 205–207 °C; 1H NMR (300 MHz, DMSO-d6) δ 8.33 (s, 1H), 7.90 (d, J = 8.7 Hz, 1H), 7.50 (d, J = 8.7 Hz, 2H), 6.98 (d, J = 9 Hz, 2H), 6.83 (d, J = 9 Hz, 1H), 3.96 (s, 2H), 3.77 (s, 3H), 3.56 (brs, 1H), 2.80 (t, J = 7.2 Hz, 2H), 2.35 (t, J = 10.8 Hz, 2H), 1.75 (d, J = 12.9 Hz, 2H), 1.45 (q, J = 6.9 Hz, 2H). 13 C NMR (100 MHz, DMSO-d6) δ 175.2, 164.1, 159.5, 155.4, 153.2, 131.5, 125.9, 124.3, 123.4, 116.5, 115.3, 113.9, 109.4, 55.6, 55.4, 52.9, 50.6, 34.4. HR-MS (ESI) Calcd for C22H23NO5 [M + H]+, 382.1654, found: 382.1669.
7-hydroxy-3-(4-methoxyphenyl)-8-((3-methylpiperidin-1-yl)methyl)-4H-chromen-4-one (2o)
White solid, yield: 21%; mp 165–167 °C; 1H NMR (300 MHz, DMSO-d6) δ 8.33 (s, 1H), 7.90 (d, J = 9 Hz, 1H), 7.50 (d, J = 8.7 Hz, 2H), 6.98 (d, J = 9 Hz, 2H), 6.82 (d, J = 9 Hz, 1H), 3.96 (s, 2H), 3.77 (s, 3H), 2.87 (d, J = 7.5 Hz, 2H), 2.17(t, J = 11.7 Hz, 1H), 1.83 (t, J = 10.8 Hz, 1H), 1.65–1.49 (m, 4H), 0.93 (m, 1H), 0.82 (d, J = 6.6 Hz, 3H). 13C NMR (100 MHz, DMSO-d6) δ 175.2, 164.3, 159.5, 155.5, 153.2, 130.6, 126.2, 124.7, 123.5, 116.6, 115.7, 114.1, 108.4, 60.6, 55.7, 53.4, 53.2, 32.3, 31.2, 25.2, 19.7. HR-MS (ESI) Calcd for C23H25NO4 [M + H]+, 380.1862, found: 380.1897.
7-hydroxy-8-((3-hydroxypiperidin-1-yl)methyl)-3–(4-methoxyphenyl)-4H-chromen-4-one (2p)
White solid, yield: 25%; mp 188–190 °C; 1H NMR (300 MHz, DMSO-d6) δ 8.33 (s, 1H), 7.91 (d, J = 9 Hz, 1H), 7.50 (d, J = 8.7 Hz, 2H), 6.98 (d, J = 9 Hz, 2H), 6.84 (d, J = 8.7 Hz, 1H), 3.95 (s, 2H), 3.77 (s, 3H), 3.58 (brs, 1H), 2.85 (d, J = 9.3 Hz, 1H), 2.66 (d, J = 11.1 Hz, 1H), 2.27–2.13 (m, 2H), 1.71 (d, J = 10.5 Hz, 2H), 1.45 (m, 1H), 1.24 (m, 1H). 13C NMR (100 MHz, DMSO-d6) δ 175.2, 164.0, 159.5, 155.5, 153.2, 130.6, 126.2, 124.7, 123.5, 116.7, 115.7, 114.1, 108.6, 65.8, 60.4, 55.6, 53.0, 52.9, 32.6, 22.7. HR-MS (ESI) Calcd for C22H23NO5 [M + H]+, 382.1654, found:382.1718.
8-((4-benzylpiperazin-1-yl)methyl)-7-hydroxy-3-(4-methoxyphenyl)-4H-chromen-4-one (2q)
White solid, yield: 27%; mp 220–222 °C; 1H NMR (300 MHz, DMSO-d6) δ 8.36 (s,1H), 7.92 (d, J = 8.7 Hz, 1H), 7.50 (d, J = 8.7 Hz, 2H), 7.29 (m, 5H), 6.98 (d, J = 8.7 Hz, 2H), 6.87 (d, J = 8.7 Hz, 1H), 3.96 (s, 2H), 3.77 (s, 3H), 3.47 (s, 2H), 2.59–2.49 (m, 8H). 13C NMR (100 MHz, DMSO-d6) δ 175.3, 159.7, 159.5, 157.4, 155.7, 142.5, 138.5, 130.6, 129.4, 128.7, 127.3, 124.4, 123.3, 116.7, 115.5, 114.1, 108.6, 69.1, 62.4, 55.8, 52.9, 52.6. HR-MS (ESI) Calcd for C28H28N2O4 [M + H]+, 451.2127, found: 457.2113.
7-hydroxy-8-((4-(hydroxymethyl)piperidin-1-yl)methyl)-3-(4-methoxyphenyl)-4H-chromen-4-one (2r)
White solid, yield: 27%; mp 195–197 °C; 1H NMR (300 MHz, DMSO-d6) δ 8.33 (s, 1H), 7.90 (d, J = 8.7 Hz, 1H), 7.50 (d, J = 8.7 Hz, 2H), 6.98 (d, J = 9 Hz, 2H), 6.82 (d, J = 9 Hz, 1H), 3.98 (s, 2H), 3.77 (s, 3H), 3.27 (d, J = 6 Hz, 2H), 2.99 (d, J = 11.1 Hz, 2H), 2.25 (t, J = 11.4 Hz, 2H), 1.73 (d, J = 12.9 Hz, 2H), 1.23 (brs, 1H), 1.11 (m, 2H). 13 C NMR (100 MHz, DMSO-d6) δ 175.2, 164.3, 159.5, 155.5, 153.2, 130.6, 126.2, 124.7, 123.5, 116.5, 115.8, 114.1, 108.4, 66.00, 55.7, 53.3, 52.9, 38.2, 28.9. HR-MS (ESI) Calcd for C23H25NO5 [M + H]+, 396.1811, found: 396.1806.
7-hydroxy-8-((4-(2-hydroxyethyl)piperazin-1-yl)methyl)-3-(4-methoxyphenyl)-4H-chromen-4-one (2s)
White solid, yield: 27%; mp 196–198 °C; 1H NMR (300 MHz, DMSO-d6) δ 8.35 (s, 1H), 7.91 (d, J = 8.7 Hz, 1H), 7.50 (d, J = 8.7 Hz, 2H), 6.98 (d, J = 9 Hz, 2H), 6.89 (d, J = 9 Hz, 1H), 3.94 (s, 2H), 3.77 (s, 3H), 3.49 (t, J = 6.3 Hz, 3H), 2.57–2.40 (m, 8H), 2.36 (t, J = 7.2 Hz, 2H). 13C NMR (100 MHz, DMSO-d6) δ 175.2, 163.4, 159.5, 155.6, 153.3, 130.6, 126.3, 124.7, 123.3, 116.9, 115.5, 114.1, 108.8, 60.5, 59.0, 55.7, 53.5, 52.7, 52.4. HR-MS (ESI) Calcd for C23H26N2O5 [M + H]+, 411.1920, found: 411.1904.
7-hydroxy-3-(4-methoxyphenyl)-8-((2-methylpiperidin-1-yl)methyl)-4H-chromen-4-one (2t)
White solid, yield: 17%; mp 141–143 °C; 1H NMR (300 MHz, DMSO-d6) δ 8.32 (s, 1H), 7.87 (d, J = 9.9 Hz, 1H), 7.50 (d, J = 9.9 Hz, 2H), 6.98 (d, J = 9 Hz, 2H), 6.80 (d, J = 9.9 Hz, 1H), 4.31–4.26 (d, J = 15 Hz, 1H), 3.90–3.85 (d, J = 15 Hz, 1H), 3.77 (s, 3H), 2.83 (d, J = 12.3 Hz, 1H), 2.66 (brs, 1H), 2.33 (t, J = 9.6 Hz, 1H), 1.48–1.35 (m, 6H), 1.15 (d, J = 6.3 Hz, 3H). 13C NMR (100 MHz, DMSO-d6) δ 175.1, 164.8, 159.5, 155.2, 153.1, 130.6, 125.9, 124.8, 123.5, 116.4, 115.9, 114.1, 108.6, 56.6, 55.6, 51.7, 50.4, 33.8, 26.9, 25.7, 22.6. HR-MS (ESI) Calcd for C23H25NO4 [M + H]+, 380.1862, found: 380.2130.
Bioassay studies
Cell lines and cell culture
The cell-based histamine receptor 3 (H3R) assay was carried out based on β-lactamase complementation technology. The H3-bla U2OS cells (Invitrogen, Invitrogen, Waltham, Massachusetts) stably expressed two fusion proteins, as well as a β-lactamase reporter gene under the control of a UAS response element. The first fusion protein was human H3R linked to a Gal4-VP16 transcription factor through the TEV protease site, and the other was the β-arrestin/TEV protease fusion protein. H3-bla U2OS cells were cultured in McCoy’s 5 A Medium supplemented with 10% foetal bovine serum (FBS; Gibco, Shanghai, China) at 37 °C in a humidified atmosphere with 5% CO2. To each well in a 384-well plate was seeded exponentially growing cells in a density of 6.5 × 103 cells/mL in 32 μL of media. The plate was incubated at 37 °C, 18–24 h, 5% CO2 for cell adherence.
Fluorescent H3R assay
Stock solutions of test compounds (10 mM) were prepared in DMSO and then diluted 100 times in media. Cells were exposed to 4 μL of test compounds and the control compound thioperamide (Sigma-Aldrich, St. Louis, Missouri) for 30 min and then stimulated with 4 μL of methylhistamine at 400 nM (Sigma-Aldrich) for 5 h. Then, 8 μL of LiveBLAzer-FRET B/G Substrate (CCF4-AM; Invitrogen) was added and incubation continued for 2 h. Plates were subjected to the fluorescence reading with a Spectra Max M5 microplate reader (BioTek, Winooski, Vermont); equipped with 410 nm excitation and 460 nm and 530 nm emission filters. The inhibition percentage was calculated based on the fluorescence according to the following equation: % inhibition = (ModelResponse ratio–CompoundResponse ratio)/ModelResponse ratio. And IC50 values were determined from log concentration − inhibition curves. At least three separate tests were carried out.
Molecular docking
We chose the most active compounds for molecular docking studies to predict how molecules and proteins work. A homology modelling of H3R was built as our previous reportCitation25. The 3D structure of compound 2h was built using DS MODELER (Discovery Studio 2016, BIOVIA Inc, San Diego, CA) and evaluated the model according to the PDF Total Energy and the Profile-3D procedure. Flexible Docking was used for the docking procedure. The 3D model of H3R with the lowest PDF Total Energy was chosen for docking. Water and the cognate ligand (doxepin) were removed from the model, and hydrogen atoms were added to amino acid residues. The binding mode was shown by DS visualizer.
Results and discussion
Structure–activity relationship
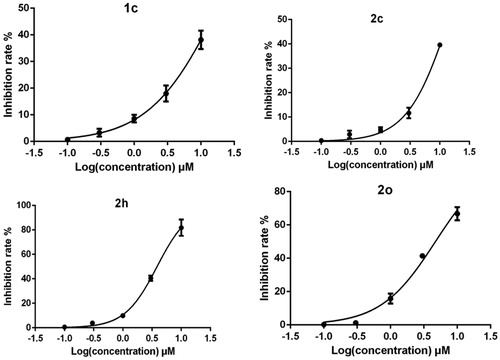
The compounds were initially evaluated for inhibition rate on H3R at a fixed concentration of 10 μM ( and ). Of the 27 compounds evaluated, four compounds (1c, 2c, 2h, 2o)performed satisfactory inhibitory effect (). According to reports in the literature, H3R inhibitory activities were increased by the introduction of pyrrolidine and piperidine to the iso-flavone scaffoldCitation10. Thus, we introduced various pyrrolidine, piperidine, piperazine and morpholine moieties onto 6- or 8-position of iso-flavone. The results for series 1 are shown in . The advantage of piperidine groups outweighed pyrrolidine moieties. As for substituted piperazine and morpholine moieties, the subsequent data did not give satisfactory results. Then, we modified daidzein and formononetin with substituted piperidine and pyrrolidine fragments. It should be noted that further steric modification on piperidine was detrimental for the inhibitory activities. For example, 4-hydroxymethyl, 3-hydroxy piperidine (compound 2b, 2e) attached to the structure of daidzein led the inhibitory activity to decrease. However, the 2-methyl piperidine group (compound 2h) showed very strong inhibition. Interestingly, for formononetin, 3-methyl piperidine (compound 2o) and pyrrolidine (compound 2m) fragments showed unexpected inhibitory effect. Structurally, substituted piperidine (such as methyl- and hydroxyl-) or pyrrolidine groups could improve bioactivity but bulky substitutions may hinder binding H3 pockets, namely, binding affinity would lossCitation10. Comparing different iso-flavone structures, even though 4′-hydroxy or 4′-methoxy benzene ring in 4-position of iso-flavone scaffold showed significant fluctuation in bioactivity level according to the data shown in , in most cases, daidzein derivatives have advantages over formononetin as H3R antagonists, for example, compound 2c vs 2l; 2h vs 2t.
Table 1. Structures and activities of compounds 1a–1g.
Table 2. Structures and activities of compounds 2a–2t.
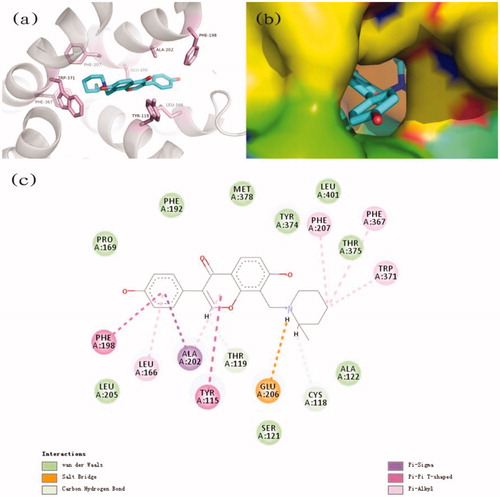
Binding modes of compound 2h
The results showed that compound 2h bound with H3R through multiple sites (). The protonated amine of the pyridine group interacted with Glu206 through a salt bridge. The Tyr-115 and Phe-198 bound to the aromatic ring structural on one side of compound 2h by π–π T-shape interactions. In addition to this, compound 2h also formed hydrophobic interaction, π–sigma and π–alkyl interaction with the protein.
Conclusions
In this work, two series of iso-flavone derivatives were synthesised and evaluated for their H3R inhibitory activity. Ultimately, we identified compound 1c, 2c, 2h, 2o which possessed favourable H3R inhibitory activity. The structure–activity relationship (SAR) study identified the piperazine group in the 8-position of iso-flavone was essential for the H3R inhibitory activity (compound 2h). Molecular docking showed 2′-methyl piperidine substituent of 2h formed a salt bridge and hydrophobic interactions with the protein. In this paper, we creatively modified the iso-flavone derivatives and determined this scaffold possessing the potential H3R inhibitory activity. Moreover, these results also provided clues for the development of novel H3R antagonists.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Sadek B, Saad A, Sadeq A, et al. Histamine H3 receptor as a potential target for cognitive symptoms in neuropsychiatric diseases. Behav Brain Res 2016;312:415–30.
- Ellenbroek BA, Ghiabi B. The other side of the histamine H3 receptor. Trends Neurosci 2014;37:191–9.
- Berlin M, Boyce CW, Ruiz Mde L. Histamine H3 receptor as a drug discovery target. J Med Chem 2011;54:26–53.
- Vanhanen J, Kinnunen M, Nuutinen S, Panula P. Histamine H3 receptor antogonist JNJ-39220675 modulates locomotor responses but not place conditioning by dopaminergic drugs. Psychopharmacology 2015;232:1143–53.
- Leurs R, Bakker RA, Timmerman H, de Esch IJ. The histamine H3 receptor: from gene cloning to H3 receptor drugs. Nat Rev Drug Discov 2005;4:107–20.
- Banuelos-Cabrera I, Cuéllar-Herrera M, Velasco AL, et al. Pharmacoresistant temporal lobe epilepsy modifies histamine turnover and H3 receptor function in the human hippocampus and temporal neocortex. Epilepsia 2016;57:e76–80.
- Rouleau A, Ligneau X, Tardivel-Lacombe J, et al. Histamine H3-receptor-mediated [35S]GTPγ[S] binding: evidence for constitutive activity of the recombinant and native rat and human H3 receptors. Br J Pharmacol 2002;135:383–92.
- Brown RE, Stevens DR, Haas HL. The physiology of brain histamine. Prog Neurobiol 2001;63:637–72.
- Riddy DM, Cook AE, Diepenhorst NA, et al. Isoform-specific biased agonism of histamine H3 receptor agonists. Mol Pharmacol 2017;91:87–99.
- Bordi F, Rivara S, Dallaturca E, et al. Dibasic biphenyl H3 receptor antagonists: steric tolerance for a lipophilic side chain. Eur J Med Chem 2012;48:214–30.
- Wager TT, Pettersen BA, Schmidt AW, et al. Discovery of two clinical histamine H3 receptor antagonists: trans-N-ethyl-3-fluoro-3- [3-fluoro-4-(pyrrolidinylmethyl)phenyl] cyclobutanecarbox amide (PF-03654746) andtrans- 3 -fluoro-3-[3-fluoro-4-(pyrrolidin-1-ylmethyl) phenyl]-N-(2-methylpropyl)cyc lobutanecarboxamide (PF-03654764). J Med Chem 2011;54:7602–20.
- Delay-Goyet P, Blanchard V, Schussler N, et al. SAR110894, a potent histamine H3-receptor antagonist, displays disease-modifying activity in a transgenic mouse model of tauopathy. Alzheimers Dement 2016;2:267–80.
- Sadek B, Schwed JS, Subramanian D, et al. Non-imidazole histamine H3 receptor ligands incorporating antiepileptic moieties. Eur J Med Chem 2014;77:269–79.
- Pierson PD, Fettes A, Freichel C, et al. 5-Hydroxyindole-2-carboxylic acid amides: novel histamine-3 receptor inverse agonists for the treatment of obesity. J Med Chem 2009; 52:3855–68.
- Ishikawa M, Watanabe T, Kudo T, et al. Investigation of the histamine H3 receptor binding site. Design and synthesis of hybrid agonists with a lipophilic side chain. J Med Chem 2010;53:6445–56.
- Hudkins RL, Raddatz R, Tao M, et al. Discovery and characterization of 6-{4-[3-(R)-2-methylpyrrolidin-1-yl) propoxy]phenyl}-2H-pyridazin-3-one (CEP-26401, irdabisant): a potent, selective histamine H3 receptor inverse agonist. J Med Chem 2011;54:4781–92.
- Hagenow S, Stasiak A, Ramsay RR, Stark H. Ciproxifan, a histamine H3 receptor antagonist, reversibly inhibits monoamine oxidase A and B. Sci Rep 2017;7:40541.
- Provensi G, Costa A, Passani MB, Blandina P. Donepezil, an acetylcholine esterase inhibitor, and ABT-239, a histamine H3 receptor antagonist/inverse agonist, require the integrity of brain histamine system to exert biochemical and procognitive effects in the mouse. Neuropharmacology 2016;109:139–47.
- Patnaik R, Sharma A, Skaper SD, et al. Histamine H3 inverse agonist BF 2649 or antagonist with partial H4 agonist activity clobenpropit reduces amyloid beta peptide-induced brain pathology in Alzheimer’s disease. Mol Neurobiol 2018;55:312–21.
- Iida T, Yoshikawa T, Karpati A, et al. JNJ10181457, a histamine H3 receptor inverse agonist, regulates in vivo microglial functions and improves depression-like behaviours in mice. Biochem Biophys Res Commun 2017;488:534–40.
- Tao M, Aimone LD, Huang Z, et al. Optimization of 5-pyridazin-3-one phenoxypropylamines as potent, selective histamine H3 receptor antagonists with potent cognition enhancing activity. J Med Chem 2012;55:414–23.
- Singh M, Kaur M, Silakari O. Flavones: an important scaffold for medicinal chemistry. Eur J Med Chem 2014; 84:206–39.
- Feng B, Li X, Xia J, Wu S. Discovery of novel isoflavone derivatives as AChE/BuChE dual-targeted inhibitors: synthesis, biological evaluation and molecular modelling. J Enzyme Inhib Med Chem 2017; 32:968–77.
- Carmela S, Stefania M, Gian LR. Anti-inflammatory effects of flavonoids in neurodegenerative disorders. Eur J Med Chem 2018;153:105–15.
- Wen G, Liu Q, Hu H, et al. Design, synthesis, biological evaluation, and molecular docking of novel flavones as H3 R inhibitors. Chem Biol Drug Des 2017;90:580–9.