?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Carbonic anhydrases (CAs) are ubiquitous metalloenzymes, which started to be investigated in detail in pathogenic, as well as non-pathogenic species since their pivotal role is to accelerate the physiological CO2 hydration/dehydration reaction significantly. Here, we propose the marine unicellular diatom Phaeodactylum tricornutum as a model organism for testing the membrane penetrability of CA inhibitors (CAIs). Seven inhibitors belonging to the sulphonamide type and possessing a diverse scaffold have been explored for their in vitro inhibition of the whole diatom CAs and the in vivo inhibitory effect on the growth of P. tricornutum. Interesting, inhibition of growth was observed, in vivo, demonstrating that this diatom is a good model for testing the cell wall penetrability of this class of pharmacological agents. Considering that many pathogens are difficult and dangerous to grow in the laboratory, the growth inhibition of P. tricornutum with different such CAIs may be subsequently used to design inhibition studies of CAs from pathogenic organisms.
1. Introduction
The physiologic/biosynthetic processes requiring CO2 or (respiration, photosynthesis/gluconeogenesis, lipogenesis, ureagenesis, carboxylation) and biochemical pathways involving pH homeostasis, secretion of electrolytes, calcification, bone resorption, transport of CO2, and bicarbonate, etc., are connected with the interconversion of CO2 to bicarbonate and protons (CO2 + H2O ⇌
+ H+)Citation1,Citation2. In all living organism, the CO2 hydration/dehydration reaction is catalysed by a superfamily of ubiquitous metalloenzymes, known as carbonic anhydrases (CAs, EC 4.2.1.1)Citation3–10, which catalyse these reactions at very high rates, with a pseudo-first order kinetic constant (kcat) ranging from 104 to 106 s−1 for the CO2 hydrationCitation11,Citation12. At the intracellular concentrations of CO2, the uncatalysed CO2 hydration/dehydration reaction has a too low rate with an effective kcat of 0.15 s−1 for the hydration reaction, and a rate constant of 50 s−1 for the reverse reactionCitation11,Citation12. CAs have thus the physiologically pivotal role to accelerate the CO2 hydration/dehydration reaction significantly, in order to support the fast processes involving dissolved inorganic carbon, which otherwise would be impaired without these enzymesCitation13. In this context, CAs started to be investigated in detail in pathogenic, as well as non-pathogenic organisms recently, since it has been demonstrated that CAs are essential for the life cycle of bacteria, fungi and protozoaCitation4,Citation9,Citation13–20. For example, fungal β-CAs plays a critical role in the CO2-sensing and in the regulation of sexual development of many fungal organismsCitation20. Plasmodium falciparum, which is a protozoan, uses its η-CA for producing
, which is the substrate of the first enzyme involved in the pyrimidine pathway and necessary for DNA/RNA synthesis during the exponential growth and replication or the parasiteCitation4,Citation21. Again, the growth of two non-pathogenic bacteria Ralstonia eutropha (its genome encodes for one periplasmic ɑ-CA, two β-CAs, and one ɣCA) and Escherichia coli (its genome contains one β-CA and one ɣ-CA) is strictly correlated to the CA activityCitation22,Citation23. The two CAs (ɑ and β) encoded in the genome of the pathogen Helicobacter pylori, are essential for the acid acclimation and the survival of the pathogen within the human stomachCitation24–26; Vibrio cholerae uses its CAs (ɑ, β and ɣ) for producing sodium bicarbonate, which is an inducer of the cholera toxin gene expressionCitation27–32. The two pathogenic bacteria, Brucella suis and Mycobacterium tuberculosis, need functional CAs for growingCitation33–38. It is readily apparent that CAs from pathogens are potential drug targets and their inhibition leads to growth impairment or growth defects of the microorganism. Fortunately, many CA inhibitors (CAI) exists, which could be classified as inhibitors binding the metal ion (anion, sulphonamides and their bioisosteres, dithiocarbamates, xanthates); inhibitors anchoring to the water molecule/hydroxide ion coordinated to the metal (phenols, polyamines, thioxocoumarins, sulphocumarins); inhibitors occluding the active site entrance (coumarins and their isosteres) and inhibitors binding out of the active siteCitation39. As described in the literature, the principal drawback using CAIs as antiinfectives agents is the lack of selectivity towards the pathogenic versus human isoformsCitation40–43. For this reason, many research groups are continually involved in the synthesis of new CAIs or in the modification/optimisation of the existing inhibitors, which are commonly tested on CAs from mammalian and pathogenic organismsCitation11,Citation13–15,Citation20,Citation44–46. Another critical issue is related to the inhibitor penetrability into microbial cells. Many CAIs are very efficient inhibitors when tested in vitro on the purified enzymes (inhibitors with a nanomolar KI) but showed ineffective in vivo results when tested on the microorganismsCitation38,Citation47,Citation48. Since it is very complicated to obtain specific control measures and containment levels for activities with pathogenic organisms, in this article, we propose the marine unicellular diatom Phaeodactylum tricornutum as a model organism for testing the membrane penetrability of the CAIs. P. tricornutum is a eukaryotic organism characterised by fusiform cells with a cell wall poor in silicaCitation49,Citation50. The genome of the P. tricornutum encodes for nine CAs: five α-CAs confined in the matrices of the four-layered plastid membranes, two β-CAs (PtCA1 and PtCA2) located in the pyrenoid and two mitochondrial γ-CAsCitation49. Recently, in the lumen of the pyrenoid-penetrating thylakoid a new class of CAs, named θ-CA, has been identifiedCitation49. The CA inhibition of marine diatoms, as well as of other microalgae, lead to growth impairment or defects in the microalga because these enzymes have a pivotal role in ensuring the supply of inorganic carbon (Ci) to RuBisCO and phosphoenolpyruvate carboxylaseCitation49,Citation51.
Here, seven inhibitors belonging to the sulphonamide types, such as the AAZ, MZA, which are clinically used agents, and compounds 1–5 have been explored for their in vitro inhibition of the diatom CAs and in vivo inhibitory effect on the growth of the P. tricornutum cell. Our results demonstrate that the growth of the P. tricornutum cells is affected by the CAIs and the unicellular diatom represents a good model for verifying the CAIs membrane penetrability.
2. Material and methods
2.1. Chemistry
Compounds 3–5 used in the work were reported earlier by our groupsCitation52,Citation53. AAZ and MZA were commercially available from Sigma-Aldrich (Milan, Italy). All the chemicals and solvents were purchased from Sigma-Aldrich (Milan, Italy). All reactions involving air- or moisture-sensitive compounds were performed under a nitrogen atmosphere using dried glassware and syringes techniques to transfer solutions. Nuclear magnetic resonance (1H-NMR, 13C-NMR) spectra were recorded using a Bruker Advance III 400 MHz spectrometer in DMSO-d6. Chemical shifts are reported in parts per million (ppm) and the coupling constants (J) are expressed in Hertz (Hz). Splitting patterns are designated as follows: s, singlet; d, doublet; t, triplet; q, quadruplet; m, multiplet; bs, broad singlet; dd, double of doubles. The assignment of exchangeable protons was confirmed by the addition of D2O. Analytical thin-layer chromatography (TLC) was carried out on Sigma Aldrich silica gel F-254 plates. Flash chromatography purifications were performed on Sigma Aldrich Silica gel 60 (230–400 mesh ASTM) as the stationary phase and ethyl acetate/n-hexane or MeOH/DCM were used as eluents. Melting points (mp) were measured in open capillary tubes with a Gallenkamp MPD350.BM3.5 apparatus and are uncorrected. The solvents used in MS measures were acetone, acetonitrile (Chromasolv grade), purchased from Sigma-Aldrich (Milan-Italy), and mQ water 18 MΩ, obtained from Millipore's Simplicity system (Milan-Italy). The mass spectra were obtained using a Varian 1200 L triple quadrupole system (Palo Alto, CA) equipped by Electrospray Source (ESI) operating in both positive and negative ions. Stock solutions of analytes were prepared in acetone at 1.0 mg mL−1 and stored at 4 °C. Working solutions of each analyte were freshly prepared by diluting stock solutions in a mixture of mQ water:acetonitrile 1:1 (v/v) up to a concentration of 1.0 µg mL−1 The mass spectra of each analyte were acquired by introducing, via syringe pump at 10 µL min−1, of the its working solution. Raw-data were collected and processed by Varian Workstation Vers. 6.8 software.
2.1.1. Synthesis of 4-(t-butyl)-N-(3-methyl-5-sulphamoyl-1,3,4-thiadiazol-2(3H)-ylidene)benzamide 1 740
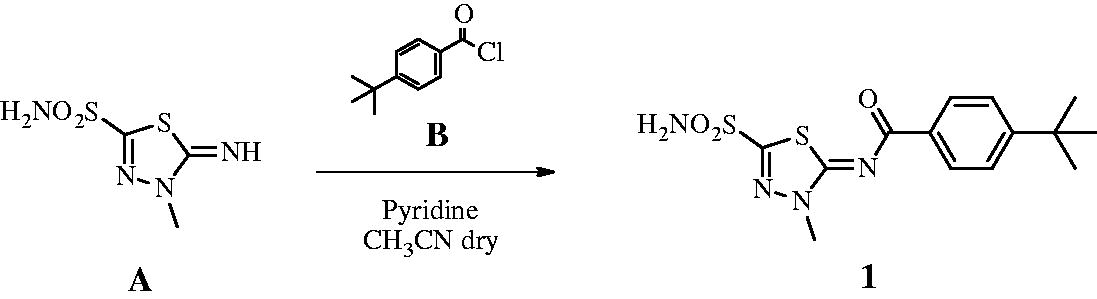
4-t-Butylbenzyl chloride B (1.2 eq) was added to a solution of 5-imino-4-methyl-4,5-dihydro-1,3,4-thiadiazole-2-sulphonamide A (0.3 g, 1.0 eq) and pyridine (3.0 eq) in dry acetonitrile (5 ml) at 0 °C under a nitrogen atmosphere. The solution was stirred at r.t. until the starting material was consumed (TLC monitoring), then quenched with HCl 1 M (15 ml). The formed precipitate was filtered-off and purified by silica gel column chromatography eluting with 40% EtOAc in n-hexane to afford the title compound 1 as a white solid. 52% yield; m.p. 295–6 °C; silica gel TLC Rf 0.47 (EtOAc/n-hexane 60% v/v); δH (400 MHz, DMSO-d6): 1.32 (s, 9H, 3 × CH3), 4.08 (s, 3H, NCH3), 7.56 (d, J = 8.8, 2H, Ar–H), 8.20 (d, J = 8.8, 2H, Ar–H), 8.46 (s, 2H, exchange with D2O, SO2NH2); δC (100 MHz, DMSO-d6): 31.6, 35.8, 39.3, 126.3, 130.1, 133.3, 156.7, 159.0, 166.3, 174.3; m/z (ESI negative) 353.0 [M-H]−.
2.1.2. Synthesis of t-butyl (2–(4-sulphamoylbenzamido)ethyl)carbamate 2 484
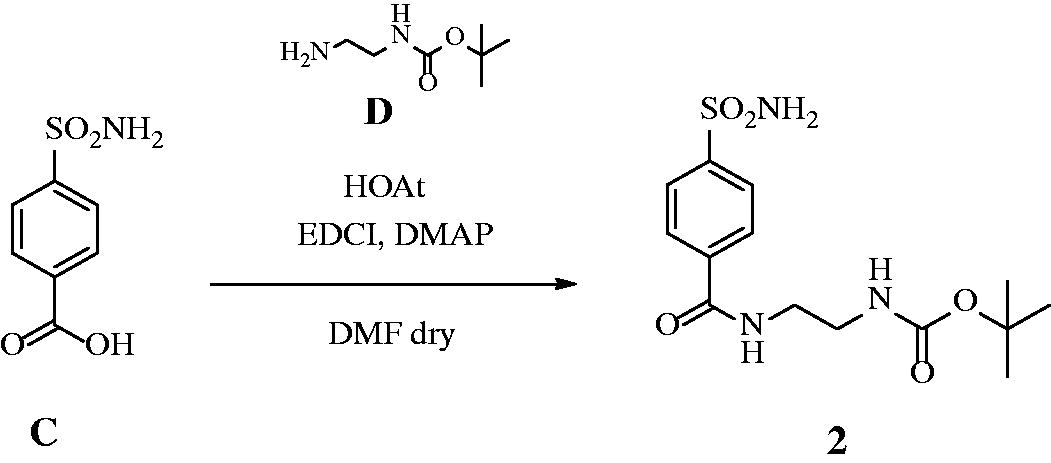
HOAt (1.2 eq) was added to a solution of 4-sulphamoylbenzoic acid C (0.2 g, 1.0 eq) and N-boc-ethylenediamine D (1.2 eq) in dry DMF (3 ml) under a nitrogen atmosphere, followed by DMAP (0.03 eq) and EDCI (1.2 eq). The solution was stirred at r.t. until the starting material was consumed (TLC monitoring), then quenched with slush (15 ml) and extracted with EtOAc (2 × 20 ml). The organic layers were washed with HCl 0.5 M (2 × 15 ml) and brine (2 × 15 ml), dried over Na2SO4, filtered-off and concentrated under vacuo. The obtained residue was purified by silica gel column chromatography eluting with 10% MeOH in DCM to afford the title compound 2 as a white solid. 73% yield; m.p. 198–199 °C; silica gel TLC Rf 0.35 (MeOH/DCM 10% v/v); δH (400 MHz, DMSO-d6): 1.41 (s, 9H, 3× CH3), 3.17 (q, J = 5.6, 2H, CH2), 3.34 (q, J = 5.6, 2H, CH2), 6.95 (m, 1H, exchange with D2O, CONH), 7.50 (s, 2H, exchange with D2O, SO2NH2), 7.93 (d, J = 8.4, 2H, Ar–H), 8.02 (d, J = 8.4, 2H, Ar–H), 8.66 (m, 1H, exchange with D2O, CONH); δC (100 MHz, DMSO-d6): 29.2, 40.4, 40.4, 78.7, 126.5, 128.8, 138.4, 147.2, 156.7, 166.3; m/z (ESI negative) 341.9 [M-H]−.
2.2. Cell culture
The CCMP632 strain of P. tricornutum (Pt1) Bohlin was obtained from the Provasoli-Guillard National Centre for Culture of Marine Phytoplankton. Cultures were grown in f/2-Si mediumCitation54 at 18 °C under white fluorescent lights (70 μmol m−1 s−1), 12 h:12 h dark–light cycle as described by De Riso and co-workersCitation55. Analyses of the wild-type Pt1 have been performed on cells in exponential phase of growth and collected 4 h after the beginning of the light period.
2.3. Spot test analysis
Different dilutions of wild-type Pt1 cells (0.5 × 107; 0.75 × 107; 1 × 107; 1.5 × 107 and 2 × 107 cells) were spotted on f/2-Si agar plates (volume of the spot 5µl). Cells were inoculated with CAIs (AAZ, MZA, 1–5) diluted with 10% DMSO at two different concentrations: 0.4 and 1.0 mM. Plates inoculated in an identical manner but only with 10% DMSO and plates inoculated only with f2/-Si were used as control. Cell survival was monitored after 5 days of growth at 18 °C under white fluorescent lights (70 μmol m–1 s–1), 12 h:12 h dark–light cycle.
2.4. Enzyme purification
All the purification steps were carried out at a temperature of 4 °C. Approximately, 10 g of pelleted diatom culture were homogenised in 20 ml of 20 mM Tris-HCl buffer pH 8.3 containing 10−3 M PMSF, 10−3 M benzamidine and 2 × 10−3 M EDTA. The homogenate was centrifuged twice for 30 min at 12,000×g, and the resulting supernatant was centrifuged again for 45 min at 100,000×g. The hydratase activity was detected in the supernatant fraction. This fraction was dialysed against 20 mM Tris–HCl buffer pH 7.5 and further purified by affinity chromatography on a p-aminomethylbenzenesulphonamide agarose resin (pAMBS; Sigma-Aldrich). 1 ml of pAMBS resin was applied to an empty column (BioRad) and equilibrated with 0.1 M Tris-HCl, pH 7.5 buffer containing 0.2 M K2SO4, 0.5 mM EDTA. The sample containing about 1 mg of total protein was loaded on the p-AMBS column equilibrated as aforementioned. Unbound proteins were removed by washing extensively with the same buffer. The bound carbonic anhydrase was eluted using 0.4 M KSCN dissolved in 0.1 M Tris–HCl, pH 7.5 buffer. The CA-containing fractions were pooled, dialysed and concentrated by ultrafiltration. The CA-containing sample was subject to SDS-PAGE.
2.5. SDS-PAGE
Sodium dodecyl sulphate (SDS)-polyacrylamide gel electrophoresis (PAGE) was carried out according to LaemmliCitation56. Samples were dissolved in buffer with 5% b-mercaptoethanol. Gel was stained with Coomassive blue.
2.6. Colourimetric carbonic anhydrase assay
CA activity assay was a modification of the procedure described by Capasso et al.Citation57. Briefly, the hydratase assay was performed at 0 °C using CO2 as substrate following the pH variation due to the catalysed conversion of CO2 to bicarbonate. Bromothymol blue was used as pH indicator. The production of hydrogen ions during the CO2 hydration reaction lowers the pH of the solution leading to a colour transition of the dye. The time required for the colour change is inversely proportional to the amount of CA present in the sample. The Wilbur-Anderson units (WAU) were calculated according to the following definition: one WAU of CA activity is defined as the ratio (T0 − T)/T, where T0 (the time needed for the pH indicator colour change for the uncatalysed reaction) and T (the time needed for the pH indicator colour change for the catalysed reaction) are recorded as the time (in seconds) required for the pH to drop from 8.3 to the transition point of the dye (pH 6.8) in a control buffer and the presence of enzyme, respectively.
2.7. CA inhibition determination
An Applied Photophysics stopped-flow instrument was used for assaying the total CAs catalysed CO2 hydration activity of the algal homogenates. Phenol red was used as pH indicator (0.2 mM) in 20 mM Hepes as buffer (pH 7.5) working at the absorbance maximum of 557 nm or, alternatively, bromothymol blue (0.2 mM in 20 mM Tris, pH 8.3) working at an absorbance maximum of 602 nm. 20 mM Na2SO4 was added to maintain constant the ionic strength in both media. The initial rates of the CA-catalysed CO2 hydration reaction were followed for a period of 10–100 s. For each inhibitor at least six traces of the initial 5–10% of the reaction are used for determining the initial velocity. The uncatalysed rates are determined in the same manner and subtracted from the total observed rates. Stock solutions of inhibitor (0.1 mM) were prepared in distilled-deionised water and dilutions up to 0.01 nM were done thereafter with the assay buffer. The inhibitor and enzyme solutions were pre-incubated together at room temperature prior to assay, in order to allow for the formation of the E-I complex. The rates of the CA-catalysed reaction were followed by measuring the absorbance at the λmax of the proper pH indicator, with the inhibition constants obtained by non-linear least-squares methods using PRISM 3 and the Cheng–Prusoff equation as the mean from at least three different determinations.
2.8. Lipophilicity determination
Prediction of logP is a method for obtaining information on the partition coefficient of a compound. The platform SwissADME (http://www.swissadme.ch/) was used to calculate the logP of the sulphonamide compoundsCitation58.
3. Results and discussion
3.1. Lipophilicity of the sulphonamide CAIs
Sulphonamides inhibitors with the sulphamoyl group have been extensively studied as CAIs because of their ability to coordinate the metal ions of CA with high affinities (e.g. 106–109 M−1 for the human isoform, hCA II)Citation39. The zinc is located at the bottom of a deep conical cavity and is coordinated by three histidine ligands (e.g., α-CAs) or two cysteines and one histidine (e.g., β-CAs) and a hydroxide ion with tetrahedral geometry ()Citation39. The sulphonamide inhibitor nitrogen atom displaces a zinc-bound hydroxide from the active site to form a stable enzyme-inhibitor complex, as elucidated through numerous solved crystallographic structures of various CA-inhibitor adductsCitation39. For an optimal in vivo activity, a balanced hydrosolubility and liposolubility of the sulphonamide inhibitors are necessary, even if some sulphonamides characterised by low lipid solubility, such as acetazolamide (AAZ), are used as effective drugs for a long period. In the present paper, seven sulphonamides inhibitors were investigated for their lipophilicity and effects on the growth of P. tricornutum: acetazolamide (AAZ), methazolamide (MZA), and compounds 1–5 ()Citation52,Citation53. The partition coefficient (P) in the system octanol/water, which is one of the essential factors for evaluating the drug penetrability through a biological membrane was calculated by using various commercially available programmes. The seven CAIs showed a lipophilicity consensus ranging from −0.28 to 2.55 (). A negative consensus LogP value means the compound has a higher affinity for the aqueous phase; when the consensus is 0, the compound is equally partitioned between the lipid and aqueous phases; a positive consensus LogP value denotes a higher concentration in the lipid phase (the compound is more lipophilic). The lipophilicity increases as the consensus LogP values increase (). From , it is possible to note that only three of the investigated sulphonamides have low lipid solubility (AAZ, MZA and 5), which all incorporate predominantly hydrophilic moieties. The remaining sulphonamides also possess various highly lipophilic moieties (tert-Bu-phenyl; phenylthio, T-Bu, trifluromethyl-phenyl, etc) which lead to positive LogP values and thus an enhanced liposolubility over AAZ and MZA ().
Figure 1. (A) The active site of hCA II as representative of hCA isoforms. The catalytic triad is indicated with the residues: H94, H96 and H119 and (B) The catalytic site of the β-CA from P. tricornutum (PtLCIB4). The metal (yellow sphere) is coordinated by two Cys (C46 e C126) and one His (H102) residues and one water molecule (red sphere).
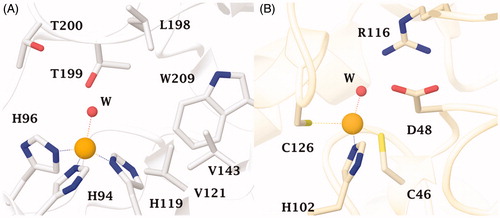
Figure 2. Sulphonamide compounds investigated for their effect on the growth of P. tricornutum cells.
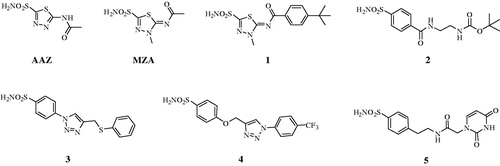
Table 1. The lipophilicity of the seven sulphonamides used in the present study.
3.2. CAs from P. tricornutum
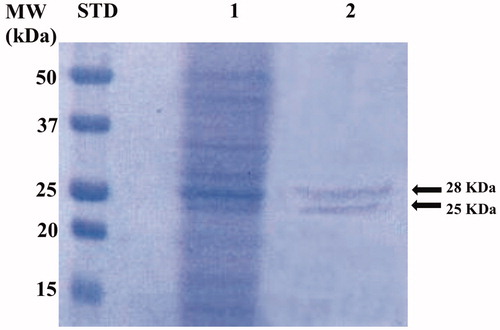
The genome of P. tricornutum encodes for nine CAs belonging to the α-, β-, γ-, and θ-CA classesCitation49. The native P. tricornutum CAs (PtrCAs) were extracted and purified from 10 g of pelleted diatom culture. Using CO2 as a substrate, the activity of PtrCAs in the extract was determined. Most of the CA activity was recovered in the soluble fraction of cellular extract after centrifugation and the calculated specific activity was 200 Wilbur-Anderson units (WAU) mg−1 of protein. The extract containing the diatom hydratase activity was further purified by p-aminomethylbenzenesulphonamide (pAMBS) affinity chromatography because most of the diatom CA-classes show affinity for this resin as described in the literatureCitation59. In fact, the resin consists of a cyanogen bromide-activated agarose matrix to which the primary amino group of the pAMBS has been attached. In , the SDS-PAGE acquired after the diatom culture homogenisation and column affinity chromatography is shown. The whole native CAs were purified to apparent homogeneity from the other proteins, as indicated by the two single bands evidenced on the SDS-PAGE. The molecular weight estimated by SDS-PAGE was of about 28.0 and 25.0 kDa under reducing conditions. Interesting, the determination of the theoretical molecular weight of the different diatom CA classes, calculated from their amino acid sequences, showed that most of them had a Mw corresponding to 28 and 25 kDa. The diatom CAs were indicated with the acronym PtrCA and are mainly represented by these two bands, as shown by SDS-PAGE ().
3.3. In vitro activity/inhibition of the diatom CAs
The sulphonamide inhibitors described in the previously paragraph, acetazolamide (AAZ) and methazolamide (MZA), which are clinically used agents, and compounds 1–5, were used for determining the IC50 values relative to the inhibition of the PtrCAs purified from the marine diatom P. tricornutum as described above (). These values were compared with those obtained for the two human CA isoforms (hCA I and hCA II). All the inhibitors considered resulted very efficient versus PtrCAs, with IC50 in a range of 8.6–96.6 nM, and the human isoform hCA II (IC50 = 1.2–42.1 nM). These inhibitors were less efficient for the human isoform hCA I, and two of them resulted in inefficient inhibitors (IC50 1 = 1458.9 or IC50 4 > 10,000). It is interesting to note that among the three CAIs with a low lipophilicity (see ), only the AAZ has an IC50 of 8.6 nM, which resulted ten times lower respect to those of the two inhibitors with a negative consensus (MZA and compound 5) and the four inhibitors with a good lipophilicity (compounds 1, 2, 3 and 4) ().
Table 2. Inhibition data with the sulphonamides reported here of the two human isoforms, hCA I and hCA II, and the diatom CAs, PtrCAs.
3.4. In vivo inhibition of growth in the presence of sulphonamides
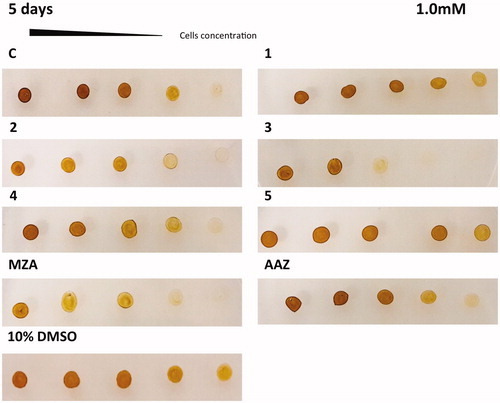
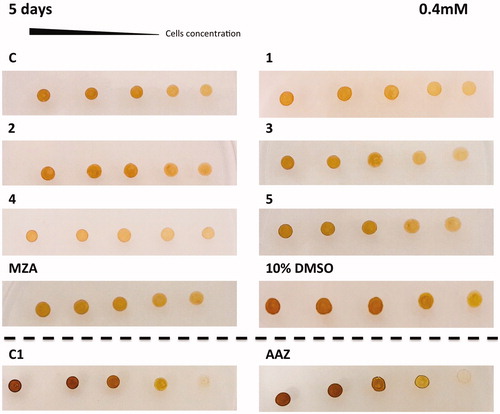
The in vivo cell wall penetrability of the sulphonamide inhibitors AAZ, MZA, 1–5 cannot be easily quantified, and thus we tested their effects on the cellular growth of P. tricornutum. Different numbers of diatom cells were spotted on the agar plates containing two different concentrations of each inhibitor (0.4 and 1 mM) ( and ). The agar plate free of the inhibitor and the plate inoculated with the solvent (10% DMSO) were used as controls. Diatom cell survival was monitored for five days under normal light conditions, at 18 °C. Under laboratory conditions, the effect of the inhibitors on the diatom growth is evidenced by the disappearance of the diatom colonies. As evidenced from and , all the sulphonamides CAIs, which were characterised in vitro for their ability to inhibit PtrCAs (), impaired the growth of P. tricornutum cells when the plates were supplemented with these sulphonamides at concentrations of 0.4 and 1.0 mM. Compounds 4, 3, 1 and 5 showed a noticeable effect on the cellular growth at 0.4 mM concentrations, compared to the controls, and this effect was more pronounced at a low number of cells (). At higher concentration of inhibitors (1 mM) it is readily apparent that MZA, 2 and 3 resulted to be more effective than the other inhibitors investigated here (). However, except for compound 3, these compounds showed different behaviour when tested at the two different concentrations. For example, MZA seems to be not efficient at 0.4 mM, but it interferes efficiently with the diatom growth when used at 1.0 mM ( and ). The stronger effect at a higher concentration of some of these inhibitors may be due to the toxicity problems caused by the off-targeting of other proteins than CAs. Interesting, 10% DMSO, which was the solvent of the inhibitors, had no effects on the cellular diatom growth ( and ). Besides, the results obtained using a concentration of inhibitors of 0.4 mM reflect those of the lipophilicity. AAZ, MZA and 5, which have low lipophilicity, were the less effective inhibitors of the diatom cell growth because of difficulties to penetrate through the membrane. AAZ has a LogP of −0.70 and is an excellent in vitro inhibitor when tested on the whole diatom CAs. This inhibitor failed to show growth impairment of the cells when used at 0.4 mM, while MZA and 5, with lipophilicity values higher than AAZ, showed a better inhibition of growth at 0.4 mM ().
Figure 4. Diatom cells were spotted on the agar plates containing 0.4 mM of each sulphonamide inhibitors. Panel containing the agar with the inhibitor has been indicated with the acronym of the inhibitor used. The panel C1 and AAZ are separated by the other groups because obtained from a different set of experiments. Legend: C and C1, panels with the plate free of the inhibitor; 10% DMSO, panel with the plate inoculated with 10% DMSO; Panels 1, 2, 3, 4, 5, MZA, AAZ, plates with the sulphonamide inhibitors; Number of cells going from left to right: 2.0 × 107; 1.5 × 107; 1.0 × 107; 0.75 × 107 and 0.5 × 107. Diatom cell survival was monitored after five days under normal light conditions and 18 °C. The disappearance of the diatom colonies evidences the inhibitor effect of the CAIs.
4. Conclusions
In this study, we proposed the marine unicellular diatom Phaeodactylum tricornutum as a model organism for testing the membrane penetrability of CAIs and their effects on the growth of the organism. P. tricornutum is an eukaryotic organism characterised by fusiform cells with a cell wall poor in silicaCitation49,Citation50. The scarcity of silica in the composition of the cell wall makes possible to consider these diatoms as a good model for testing the membrane penetrability of small molecules, such as the CA sulphonamide inhibitors. The seven sulphonamides inhibitors AAZ, MZA, and compounds 1–5, resulted to be effective inhibitors of the diatom CAs when tested in vitro (IC50–s in the range of 8.6–96 nM). Some of these inhibitors probably can cross the membrane of P. tricornutum possessing both hydrophobic and hydrophilic moieties in their molecule. The most efficient in vivo inhibitors were compounds 4, 3, 1 and 5 with a noticeable inhibitory growth effect at a low number of cells. Furthermore, these sulphonamides inhibitors showed a different behaviour when tested at the concentrations of 0.4 and 1.0 mM, which is difficult to rationalise. For example, MZA was not efficient at 0.4 mM, but it interfered efficiently with the diatom growth when used at 1.0 mM. Probably this effect might be due to the off-targeting of other proteins than the CAs considered here. In general, these results demonstrate that P. tricornutum might be considered as an excellent and rather simple model for testing the membrane penetrability of new CAIs and their effects on the growth of the organism. Considering that many pathogens are difficult and dangerous to grow in the laboratory, the preliminary results obtained for the growth inhibition of P. tricornutum with different such CAIs may be subsequently used to design inhibition studies of CAs from pathogenic organisms, which may pave the way to novel anti-infectives with a diverse mechanism of action.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Nishimori I, Onishi S, Takeuchi H, et al. The alpha and beta classes carbonic anhydrases from Helicobacter pylori as novel drug targets. Curr Pharm Des 2008;14:622–30.
- Morishita S, Nishimori I, Minakuchi T, et al. Cloning, polymorphism, and inhibition of beta-carbonic anhydrase of Helicobacter pylori. J Gastroenterol 2008;43:849–57.
- Del Prete S, De Luca V, De Simone G, et al. Cloning, expression and purification of the complete domain of the eta-carbonic anhydrase from Plasmodium falciparum. J Enzyme Inhib Med Chem 2016;31:54–9.
- Del Prete S, Vullo D, De Luca V, et al. Cloning, expression, purification and sulfonamide inhibition profile of the complete domain of the eta-carbonic anhydrase from Plasmodium falciparum. Bioorg Med Chem Lett 2016;26:4184–90.
- Del Prete S, Vullo D, De Luca V, et al. Anion inhibition profiles of the complete domain of the eta-carbonic anhydrase from Plasmodium falciparum. Bioorg Med Chem 2016;24:4410–14.
- Annunziato G, Angeli A, D'Alba F, et al. Discovery of new potential anti-infective compounds based on carbonic anhydrase inhibitors by rational target-focused repurposing approaches. Chem Med Chem 2016;11:1904–14.
- Del Prete S, Vullo D, De Luca V, et al. Anion inhibition profiles of alpha-, beta- and gamma-carbonic anhydrases from the pathogenic bacterium Vibrio cholerae. Bioorg Med Chem 2016;24:3413–17.
- Abdel Gawad NM, Amin NH, Elsaadi MT, et al. Synthesis of 4-(thiazol-2-ylamino)-benzenesulfonamides with carbonic anhydrase I, II and IX inhibitory activity and cytotoxic effects against breast cancer cell lines. Bioorg Med Chem 2016;24:3043–51.
- Capasso C, Supuran CT. An overview of the carbonic anhydrases from two pathogens of the oral cavity: Streptococcus mutans and Porphyromonas gingivalis. Curr Top Med Chem 2016;16:2359–68.
- Del Prete S, Vullo D, De Luca V, et al. Comparison of the sulfonamide inhibition profiles of the alpha-, beta- and gamma-carbonic anhydrases from the pathogenic bacterium Vibrio cholerae. Bioorg Med Chem Lett 2016;26:1941–6.
- Supuran CT. Advances in structure-based drug discovery of carbonic anhydrase inhibitors. Expert Opin Drug Discov 2017;12:61–88.
- Supuran CT. Structure and function of carbonic anhydrases. Biochem J 2016;473:2023–32.
- Supuran CT, Capasso C. An overview of the bacterial carbonic anhydrases. Metabolites 2017;7:56–74.
- Supuran CT, Capasso C. Carbonic anhydrase from Porphyromonas gingivalis as a drug target. Pathogens 2017;6:30–42.
- Del Prete S, Vullo D, Osman SM, et al. Sulfonamide inhibition profiles of the beta-carbonic anhydrase from the pathogenic bacterium Francisella tularensis responsible of the febrile illness tularemia. Bioorg Med Chem 2017;25:3555–61.
- Del Prete S, Vullo D, Osman SM, et al. Anion inhibitors of the beta-carbonic anhydrase from the pathogenic bacterium responsible of tularemia, Francisella tularensis. Bioorg Med Chem 2017;25:4800–4.
- Capasso C, Supuran CT. Inhibition of bacterial carbonic anhydrases as a novel approach to escape drug resistance. Curr Top Med Chem 2017;17:1237–48.
- Capasso C, Supuran CT. An overview of the alpha-, beta- and gamma-carbonic anhydrases from Bacteria: can bacterial carbonic anhydrases shed new light on evolution of bacteria? J Enzyme Inhib Med Chem 2015;30:325–32.
- Capasso C, Supuran CT. An overview of the selectivity and efficiency of the bacterial carbonic anhydrase inhibitors. Curr Med Chem 2015;22:2130–9.
- Capasso C, Supuran CT. Bacterial, fungal and protozoan carbonic anhydrases as drug targets. Expert Opin Ther Targets 2015;19:1689–704.
- Supuran CT, Capasso C. The eta-class carbonic anhydrases as drug targets for antimalarial agents. Expert Opin Ther Targets 2015;19:551–63.
- Kusian B, Sultemeyer D, Bowien B. Carbonic anhydrase is essential for growth of Ralstonia eutropha at ambient CO(2) concentrations. J Bacteriol 2002;184:5018–26.
- Watson SK, Kan E. Effects of novel auto-inducible medium on growth, activity and CO(2) capture capacity of Escherichia coli expressing carbonic anhydrase. J Microbiol Method 2015;117:139–43.
- Compostella ME, Berto P, Vallese F, et al. Structure of alpha-carbonic anhydrase from the human pathogen Helicobacter pylori. Acta Crystallogr F Struct Biol Commun 2015;71:1005–11.
- Buzas GM, Supuran CT. The history and rationale of using carbonic anhydrase inhibitors in the treatment of peptic ulcers. In memoriam Ioan Puscas (1932-2015). J Enzyme Inhib Med Chem 2016;31:527–33.
- Modak JK, Liu YC, Machuca MA, et al. Structural basis for the inhibition of Helicobacter pylori alpha-carbonic anhydrase by sulfonamides. PLoS One 2015;10:e0127149.
- De Vita D, Angeli A, Pandolfi F, et al. Inhibition of the alpha-carbonic anhydrase from Vibrio cholerae with amides and sulfonamides incorporating imidazole moieties. J Enzyme Inhib Med Chem 2017;32:798–804.
- Del Prete S, Vullo D, De Luca V, et al. Sulfonamide inhibition studies of the beta-carbonic anhydrase from the pathogenic bacterium Vibrio cholerae. Bioorg Med Chem 2016;24:1115–20.
- Ferraroni M, Del Prete S, Vullo D, et al. Crystal structure and kinetic studies of a tetrameric type II beta-carbonic anhydrase from the pathogenic bacterium Vibrio cholerae. Acta Crystallogr D Biol Crystallogr 2015;71:2449–56.
- Ceruso M, Del Prete S, Alothman Z, et al. Sulfonamides with potent inhibitory action and selectivity against the alpha-carbonic anhydrase from Vibrio cholerae. ACS Med Chem Lett 2014;5:826–30.
- Vullo D, Isik S, Del Prete S, et al. Anion inhibition studies of the alpha-carbonic anhydrase from the pathogenic bacterium Vibrio cholerae. Bioorg Med Chem Lett 2013;23:1636–8.
- Abuaita BH, Withey JH. Bicarbonate induces Vibrio cholerae virulence gene expression by enhancing ToxT activity. Infect Immun 2009;77:4111–20.
- Monti SM, Meccariello A, Ceruso M, et al. Inhibition studies of Brucella suis beta-carbonic anhydrases with a series of 4-substituted pyridine-3-sulphonamides. J Enzyme Inhib Med Chem 2018;33:255–9.
- Kohler S, Ouahrani-Bettache S, Winum JY. Brucella suis carbonic anhydrases and their inhibitors: towards alternative antibiotics? J Enzyme Inhib Med Chem 2017;32:683–7.
- Wani TV, Bua S, Khude PS, et al. Evaluation of sulphonamide derivatives acting as inhibitors of human carbonic anhydrase isoforms I, II and Mycobacterium tuberculosis beta-class enzyme Rv3273. J Enzyme Inhib Med Chem 2018;33:962–71.
- Angeli A, Del Prete S, Osman SM, et al. Activation studies with amines and amino acids of the beta-carbonic anhydrase encoded by the Rv3273 gene from the pathogenic bacterium Mycobacterium tuberculosis. J Enzyme Inhib Med Chem 2018;33:364–9.
- Johnson BK, Colvin CJ, Needle DB, et al. The carbonic anhydrase inhibitor ethoxzolamide inhibits the Mycobacterium tuberculosis PhoPR regulon and Esx-1 secretion and attenuates virulence. Antimicrob Agents Chemother 2015;59:4436–45.
- Nishimori I, Minakuchi T, Maresca A, et al. The β-carbonic anhydrases from Mycobacterium tuberculosis as drug targets. Curr Pharm Des 2010;16:3300–9.
- Supuran CT. How many carbonic anhydrase inhibition mechanisms exist? J Enzyme Inhib Med Chem 2016;31:345–60.
- Annunziato G, Giovati L, Angeli A, et al. Discovering a new class of antifungal agents that selectively inhibits microbial carbonic anhydrases. J Enzyme Inhib Med Chem 2018;33:1537–44.
- Fisher GM, Bua S, Del Prete S, et al. Investigating the antiplasmodial activity of primary sulfonamide compounds identified in open source malaria data. Int J Parasitol Drugs Drug Resist 2017;7:61–70.
- Kalinin S, Supuran CT, Krasavin M. Multicomponent chemistry in the synthesis of carbonic anhydrase inhibitors. J Enzyme Inhib Med Chem 2016;31:185–99.
- Carradori S, Secci D, De Monte C, et al. A novel library of saccharin and acesulfame derivatives as potent and selective inhibitors of carbonic anhydrase IX and XII isoforms. Bioorg Med Chem 2016;24:1095–105.
- Supuran CT. Bortezomib inhibits mammalian carbonic anhydrases. Bioorg Med Chem 2017;25:5064–7.
- Burghout P, Vullo D, Scozzafava A, et al. Inhibition of the beta-carbonic anhydrase from Streptococcus pneumoniae by inorganic anions and small molecules: toward innovative drug design of antiinfectives? Bioorg Med Chem 2011;19:243–8.
- Maresca A, Carta F, Vullo D, et al. Carbonic anhydrase inhibitors. Inhibition of the Rv1284 and Rv3273 beta-carbonic anhydrases from Mycobacterium tuberculosis with diazenylbenzenesulfonamides. Bioorg Med Chem Lett 2009;19:4929–32.
- Guzel-Akdemir O, Akdemir A, Pan P, et al. A class of sulfonamides with strong inhibitory action against the alpha-carbonic anhydrase from Trypanosoma cruzi. J Med Chem 2013;56:5773–81.
- Supuran CT, Scozzafava A. Benzolamide is not a membrane-impermeant carbonic anhydrase inhibitor. J Enzyme Inhib Med Chem 2004;19:269–73.
- Kikutani S, Nakajima K, Nagasato C, et al. Thylakoid luminal theta-carbonic anhydrase critical for growth and photosynthesis in the marine diatom Phaeodactylum tricornutum. Proc Natl Acad Sci USA 2016;113:9828–33.
- Harada H, Nakatsuma D, Ishida M, et al. Regulation of the expression of intracellular beta-carbonic anhydrase in response to CO2 and light in the marine diatom Phaeodactylum tricornutum. Plant Physiol 2005;139:1041–50.
- Hopkinson BM. A chloroplast pump model for the CO2 concentrating mechanism in the diatom Phaeodactylum tricornutum. Photosynth Res 2014;121:223–33.
- Nocentini A, Bua S, Lomelino CL, et al. Discovery of new sulfonamide carbonic anhydrase IX inhibitors incorporating nitrogenous bases. ACS Med Chem Lett 2017;8:1314–19.
- Nocentini A, Ferraroni M, Carta F, et al. Benzenesulfonamides incorporating flexible triazole moieties are highly effective carbonic anhydrase inhibitors: synthesis and kinetic, crystallographic, computational, and intraocular pressure lowering investigations. J Med Chem 2016;59:10692–704.
- Guillard R. Culture of phytoplankton for feeding marine invertebrates. In: Smith W, Chanley M, editors. Culture of marine invertebrate animals. Boston, MA: Springer, 1975.
- De Riso V, Raniello R, Maumus F, et al. Gene silencing in the marine diatom Phaeodactylum tricornutum. Nucleic Acids Res 2009;37:e96.
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970;227:680–5.
- Capasso C, De Luca V, Carginale V, et al. Biochemical properties of a novel and highly thermostable bacterial alpha-carbonic anhydrase from Sulfurihydrogenibium yellowstonense YO3AOP1. J Enzyme Inhib Med Chem 2012;27:892–7.
- Admire B, Yalkowsky SH. Predicting the octanol solubility of organic compounds. J Pharm Sci 2013;102:2112–9.
- Szabo E, Colman B. Isolation and characterization of carbonic anhydrases from the marine diatom Phaeodactylum tricornum. Physiologia Plantarum 2007;129:484–92.