Abstract
Abnormal alterations in the expression and biological function of retinoid X receptor alpha (RXRα) have a key role in the development of cancer. Potential modulators of RXRα as anticancer agents are explored in growing numbers of studies. A series of (4/3-(pyrimidin-2-ylamino)benzoyl)hydrazine-1-carboxamide/carbothioamide derivatives are synthesised and evaluated for anticancer activity as RXRα antagonists in this study. Among all synthesised compounds, 6A shows strong antagonist activity (half maximal effective concentration (EC50) = 1.68 ± 0.22 µM), potent anti-proliferative activity against human cancer cell lines HepG2 and A549 cells (50% inhibition of cell viability (IC50) values < 10 µM), and low cytotoxic property in normal cells such as LO2 and MRC-5 cells (IC50 values > 100 µM). Further bioassays indicate that 6A inhibits 9-cis-RA-induced activity in a dose-dependent manner, and selectively binds to RXRα-=LΒD with submicromolar affinity (Kd = 1.20 × 10−7 M). 6A induces time-and dose-dependent cleavage of poly ADP-ribose polymerase, and significantly stimulates caspase-3 activity, leading to RXRα-dependent apoptosis. Finally, molecular docking studies predict the binding modes for RXRα-LBD and 6A.
Graphic Abstract
Compound 6A with low cytotoxic property in normal cells acts as a selective RXR alpha ligand to promote TNF alpha-mediated apoptosis of cancer cells.
1. Introduction
Retinoid X receptor alpha (RXRα) is a ligand-dependent transcription factor of the nuclear receptor superfamily, and it is composed of A/B region (modulating N-terminal domain or AF-1 domain), DNA-binding domain (DBD), hinge region, and ligand-binding domain (LBD)Citation1–3. RXRα forms dimers with itself or other nuclear receptors such as the thyroid hormone receptor (TR), retinoic acid receptor (RAR), vitamin D receptor (VDR), liver X receptor (LXR), and peroxisome-proliferator-activated receptor (PPAR), to exert its transcriptional activities in the nucleus and act biologically in the cytoplasmCitation4–7. It’s worth noting that RXRα dimers are activated by endogenous ligand binding to its LBD, and then regulates gene expression in various biological processesCitation8,Citation9. Besides, dysregulations of RXRα expression and function are implicated in the initiation and development of many cancersCitation1,Citation10–12. So exogenous RXRα modulators may obstruct the development of various cancer and diseases. Experiments have shown that agonist can activate RXRα and upregulated the expression of CDKN1A (a target gene of RXR), resulting in p21WAF1/CIP1-dependent cell apoptosis and suppressing cell proliferationCitation13,Citation14, while synthetic antagonists can promote mitochondria-mediated apoptosis and inflammationCitation10,Citation15,Citation16, or induce TNFα-dependent apoptosis through suppressing PI3K/AKT-mediated cell survival pathwayCitation17,Citation18. In a word, previous studies have suggested a potential role for RXRα in cancer initiation and cancer therapy.
In recent years, increasing RXRα modulators including many agonists and few antagonists were identified based on virtual screening and structure-based designCitation19–21. Among them, only Bexarotene (an RXR full agonist) has been clinically used for the treatment of cancers such as Cutaneous T cell lymphomaCitation22. However, synthetic RXRα ligands reported are associated with unacceptable toxicity or undesirable side effectsCitation23. For example, Bexarotene has adverse effects such as elevation of blood triglyceride, weight gain, and hepatomegalyCitation24,Citation25. So, the current challenge is to develop a variety of novel RXRα modulator with more potent, high specific and low cytotoxic properties.
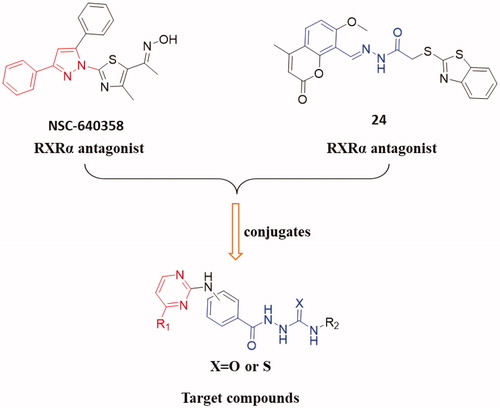
In this work, to explored novel RXRα antagonists, a series of new (3/4-(pyrimidin-2-ylamino)benzoyl)-based hydrazine-1-carboxamide/carbothioamide derivatives were designed based on the structures of compounds NSC-640358 and 24 (). First, we fused 4-substituted pyrimidine (replacing 3-phenyl-1H-pyrazole moiety in NSC-640358) to 2-benzoylhydrazine-1-carboxamide (as a substitute for (E)-N’-benzylideneacetohydrazide moiety in compound 24) to build the scaffold structure of target compounds. Second, to obtain potent RXRα antagonists, we modified the two moiety structures such as introducing the aryl group to the C-4 position (R1) of pyrimidine ring, introducing the alkyl/aryl/aralkyl group to the nitrogen atom of carboxamide (R2), and synthesising carboxamide or carbothioamide derivatives (X). Next, we synthesised target compounds and evaluated their biological activities such as in vitro antiproliferative activity and modulating RXRα activity. Then, we attempt to establish the preliminary structure − activity relationships, and the selected compound 6 A as a new high potent RXRa antagonist was further investigated for the RXRα-dependent apoptosis induction including inhibition of RXRα transactivation, physical binding of 6A to RXRα-LBD, and induction of cell apoptosis, and checking the level of cleaved poly ADP-ribose polymerase (c-PARP) and caspase-3. At last, a molecular docking study was performed to explore the binding nature of 6A to the ligand-binding pocket (LBP) of RXRα with antagonistic conformation (PDB: 3A9E).
2. Results and discussion
2.1. Chemistry
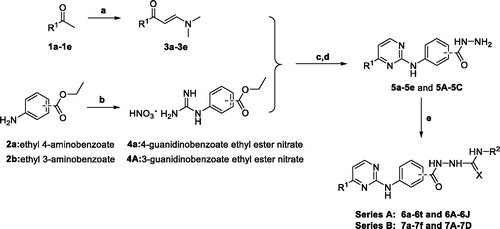
The synthetic strategy of target compounds (Series A andB) is depicted in Scheme 1. Commercially available ketones 1a–1e were reacted with N,N-dimethylformamide dimethyl acetal (DMF-DMA) in dry toluene at 110 °C for 18 h to give corresponding compounds 3a–3e. Ethyl 4-aminobenzoate (2a) and ethyl 3-aminobenzoate (2b) were refluxed with cyanamide in the presence of concentrated HCl for 24 h in ethanol, and then treated with ammonium nitrate to afford 4-guanidino benzoic acid ethyl ester nitrate (4a) and 3-guanidino benzoic acid ethyl ester nitrate (4b), respectively. Reactions of 4a with 3-(dimethylamino)-1-arylprop-2-en-1-ones (3a–3e) in the presence of sodium hydroxide were performed under reflux conditions in ethanol for 48 h to produce 4-((4-arylpyrimidin-2-yl)amino)benzoic acids, followed by refluxing with hydrazine hydrate in ethanol to get corresponding aroylhydrzides 5a–5e. Finally, target compounds of series A were prepared via reactions between aroylhydrzides 5a–5e with different isocyanates or isothiocyanates. On the other hand, target compounds of series B were synthesised with 4b instead of 4a in a similar method.
2.2. Biology activity
2.2.1. 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay and structure activity relationships (SARs) of the antiproliferative activity
To explore the antitumor activity of the target compounds, all synthesised target compounds were first assayed for in vitro antiproliferative activities against two human cancer cell lines (HepG2 and A549) by MTT method with Sorafenib as a reference. The concentrations of compounds required for 50% inhibition of cell viability (IC50) were determined and listed in and . According to these data, the preliminary SARs of these novel 2–(3/4-((-pyrimidin-2-yl)amino)benzoyl)hydrazine-1-carboxamide derivatives were summarised in : (1) First, the R1 substituent is crucial to the antitumor activities. As shown in , urea derivatives with 4-pyridyl at the R1-position showed stronger antiproliferative activity than those with 3-pyridyl and 2-pyridyl (6a vs. 6i and 6m, 6d vs. 6j and 6o, 6e vs. 6k, 6h vs. 6l, and 6b vs. 6n). The synthesised thiourea derivatives 6A–6J possessed a similar SAR, with the 4-pyridyl group to the C-4 position (R1) of pyrimidine ring being the better substitution. 4-Pyridyl substitution at R1 position was also better than 4-chlorophenyl and phenyl substitutions (6b vs. 6t, 6A vs. 6I, 6A vs. 6G, and 6B vs. 6H). (2) For the substituent R2 of compounds (6a∼6h) which contained 4-pyridyl group at R1 position, it was found that both compounds 6a–6d bearing alkyl groups (n-Butyl, t-Butyl, Cyclohexyl, and Cyclopentyl) and 6h having an aralkyl group (phenethyl) generally had good anti-proliferative properties, while the R2 analogues 6e (p-Tolyl), 6f (2,4-Difluorophenyl), and 6g (3-Chloro-4-methylphenyl) exhibited different cytotoxicity due to their different substituted phenyl ring at the R2-position. A comparison of the activities of compounds 6e and 6f shows that the p-Tolyl substitution is better than 2,4-Difluorophenyl, but when 3-Chloro-4-methylphenyl was introduced into the molecule, the biological activity of 6g was significantly reduced. Among the synthesised thiourea derivatives (6A–6J), compounds with one aralkyl group (benzyl) at R2-position displayed the better cytotoxicity against HepG2 cells than those with one aryl group (phenyl) such as 6B vs. 6A, 6E vs. 6D, and 6J vs 6I. (3) The various scaffold had a certain influence on antitumor activity. As shown in and , replacement of the 4-(pyrimidin-2-ylamino)benzohydrazide scaffold with the 3-(pyrimidin-2-ylamino)benzohydrazide tended to decrease the activity (7a vs. 6b, 7b vs. 6d, 7d vs. 6i, and 7f vs. 6m).
Figure 2. Summarised SARs of synthesised compounds (Series A and B) in Scheme 1.
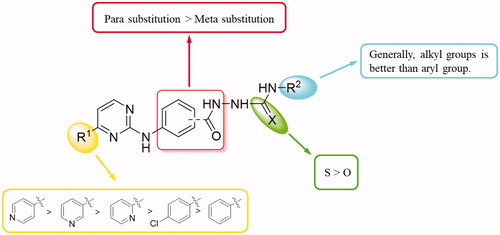
Table 1. In vitro antiproliferative activities of 2–(4-((-pyrimidin-2-yl)amino)benzoyl) hydrazine-1-carboxamide derivatives (Series A) on two selected cancer cell lines.a
Table 2. In vitro antiproliferative activities of 2–(3-((-pyrimidin-2-yl)amino)benzoyl) hydrazine-1-carboxamide derivatives (Series B) on two selected cancer cell lines. a
2.2.2. Identification of novel RXRα modulators from target compounds
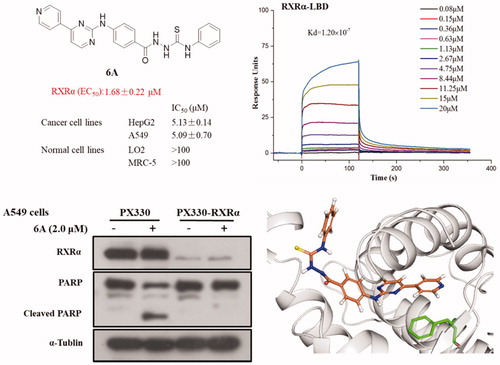
RXRα is an attractive target for the treatment of cancer and we’d like to discover new potential modulators of RXRα as anticancer agents. So, all target compounds were evaluated for their effect on regulating RXRα transcriptional function using a luciferase transcription assay in HEK293T cells with the Gal4- RXRα/LBD chimaera and Gal4 reporter system. When tested at 1.0 μM concentration with absence 9-cis-RA in this system, no compound exhibited increase transcriptional activity, suggesting no agonist effect for them. However, 11 compounds (6c, 6e, 6k, 6A, 6B, 6C, 6D, 6E, 6F, 6G, and 6H) of series A showed good antagonist effect on 9cis-RA-induced RXRα transactivation with an inhibition of >50% (), while target compounds of series B exhibited little antagonist effect on 9-cis-RA-induced reporter activity, indicating 4-(pyrimidin-2-ylamino)benzohydrazide scaffold being crucial to the transcriptional activity of RXRα. Among these candidate compounds, compound 6A showed the strongest inhibition activity 9-cis-RA-induced Gal4-DBD-RXRα-LBD transactivation at the tested concentration (1.0 μM). To confirm the antagonist effect of these 11 potential antagonists of RXRα, we next used a retinoid X receptor response element (RXRE)-luciferase reporter-based assay to evaluate their half maximal effective concentration (EC50) values of the inhibition on 9-cis-RA-induced RXRE-luciferase reporter activation. As shown in , many of them (6k, 6A, 6B, 6C, 6G, and 6H) showed good antagonism with EC50 less than 10.0 μM, while the EC50 values of several compounds (6c, 6e, 6D, and 6E) could not be calculated. Furthermore, we evaluated the cytoxic effect of the six potent RXRα antagonists (EC50 < 10.0 μM) on two normal cell lines to determine their safety (). Among the six tested compounds, 6A showed low cytotoxic property to both LO2 and MRC-5 with both IC50 values more than 100 μM.
Figure 3. Identification of RXRα antagonists via a luciferase transcription assay in HEK293T cells with the Gal4-RXRα/LBD chimaera and Gal4 reporter system. The inhibition of target compounds at 1 μM on 9-cis-RA activating Gal4-DBD-RXRα-LBD transcriptional activity in HEK293T cells transfected with p-BIND-RXRα-LBD (50 ng) and pG5-luc (50 ng) were tested. Luciferase activities were measured 12 h after treatment using the dual-luciferase assay system kit and Renilla luciferase values were normalised to firefly luciferase activity and plotted as relative luciferase activity.
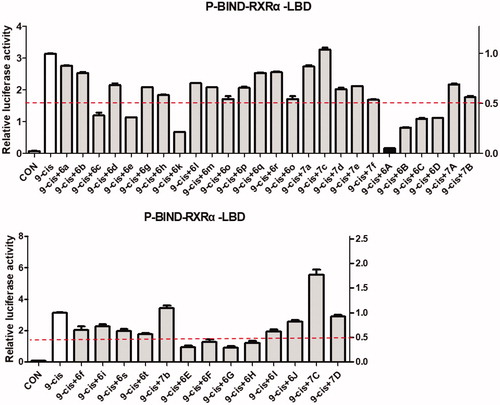
Table 3. The antagonist effect on RXRα (EC50) and cytotoxic effect on two normal cell lines (IC50) of the selected compounds.a
2.2.3. 6A binds to RXRα and acts as an antagonist
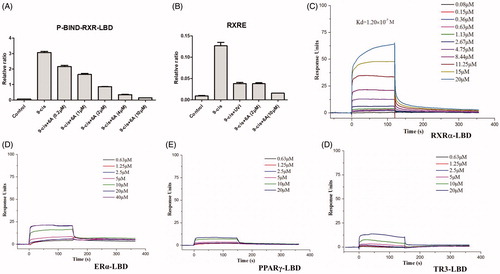
Compound 6A displayed low cytotoxic property, potent antiproliferative (IC50 < 5.30 μM) and high antagonistic (EC50 = 1.68 ± 0.22 μM) effects, so compound 6A was selected for further biological study. Compound 6A was assessed in a dose-dependent reporter gene assay. As shown in , compound 6A showed the dose-dependent effect on inhibiting the 9-cis-RA-induced Gal4-DBD-RXRα-LBD transactivation and 9-cis-RA-induced RXRE-luciferase reporter activation, and it at 2.0 μM displayed a similar inhibitory effect when compared with UVI3003 (1.0 μM), a known RXRα classical antagonist. In order to confirm compound 6A as RXRα ligand, we then performed a surface plasmon resonance (SPR) technology-based experiment to directly measure the physical binding of compound 6A to RXRα-LBD protein. As shown in , we found that 6 A dose dependently bound to RXRα-LBD with a Kd value of 1.20 × 10−7 M. Furthermore, we measured the physical binding of compound 6A to other nuclear receptors (ERα-LBD, PPARγ-LBD, and TR3-LBD). The results showed that compound 6A could not bound to these tested nuclear receptors well (). This implied that compound 6A was able to selectively interact with RXRα.
Figure 4. The antagonist effect of 6A by reporter gene assay (A,B) and the physical binding of 6A to RXRα-LBD (C), ERα-LBD (D), PPARγ-LBD (E), and TR3-LBD (F) by SPR assay. (A) Dose-dependent effect of 6A on inhibiting RXRα transactivation. HEK293T cells transfected with p-BIND-RXRα-LBD and pG5-luc were treated with indicated concentrations of 6A in the presence of 9-cis-RA (0.10 μM). (B) Inhibition of 9-cis-RA-induced RXRE-luciferase reporter activation by 6A. HEK293T cells transfected with RXRE (50 ng), full-length RXRα (50 ng), Renilla (1 ng) were treated with 9-cis-RA in the presence of indicated concentrations of 6A or UVI3003 (1 μM). (C) Surface plasmon resonance of 6A binding to RXRα-LBD. D. Surface plasmon resonance of 6A binding to ERα-LBD. E. Surface plasmon resonance of 6A binding to PPARγ-LBD. F. Surface plasmon resonance of 6A binding to TR3-LBD. (C–F) Gradient concentrations of 6A were respectively injected through chips immobilised with RXRα-LBD, ERα-LBD, PPARγ-LBD, and TR3-LBD.
2.2.4. 6A induces RXRα-dependent apoptosis of cancer cells
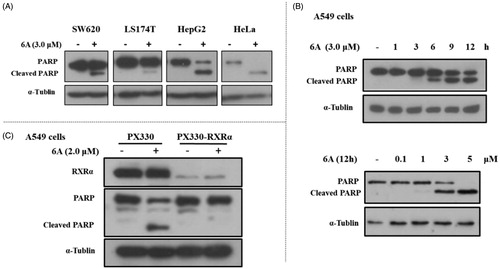
Previous studies showed RXRα antagonists with anticancer activity could induce apoptotic cell death. So, we studied whether the inhibitory effect of 6A on the growth of cancer cells was caused by cellular apoptosis. First, we examined the expression of c-PARP (an apoptosis-related protein) in several cancer cell lines treated with or without 6A. As shown in , the c-PARP increased markedly in HepG2 and Hela cells after treatment with 6A at 3.0 μM for 12 h, whereas 6A at the same concentration has a weak effect on PARP cleavage in SW620 and LS174T cells. A time- and dose- dependent increase of c-PARP, was also observed in A549 cells after treatment with 6A (). To address the role of RXRα, we then evaluated the effect of 6A on cleaved PARP in RXRα knockout A549 cells (PX330-KO). Comparison of the obvious effect of 6A in normal A549 cells (PX330), 6A showed little effect on inducing PARP cleavage in RXRα knockout A549 cells (PX330-KO; ). Thus, 6A could induce RXRα-dependent PARP cleavage in cancer cells.
Figure 5. 6A-induced PARP cleavage in cancer cells. (A) SW620, LS174T, HepG2, and Hela cells were treated with 6A for 12 h and cell lysates prepared were analysed by Western blotting. (B) A549 cells were time- and dose- dependently treated with 6A for 12 h and cell lysates prepared were analysed by Western blotting for PARP cleavage. (C) A549 and RXRα-/- A549 cells were treated with 6A for 12 h and cell lysates prepared were analysed by Western blotting for PARP cleavage.
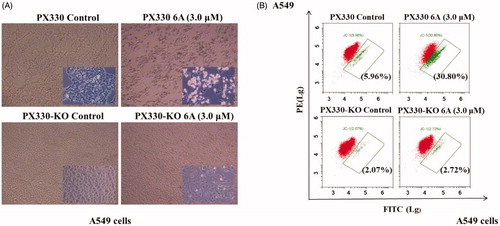
To further confirm the role of RXRα in the apoptosis induction by 6A, we evaluated the apoptosis effect of compound 6A in A549 and RXRα knockout A549 cells. As observed in , 6A-treated A549 cells exhibited obvious apoptosis characteristics with the accumulation of the nuclear convolution and fragmentation in membrane-enclosed bodies, but little apoptotic bodies were detected in treated RXRα knockout A549 cells. Meanwhile, showed that when A549 cells were incubated with 6A at 10.0 μM, the percentage of apoptotic cells was strikingly elevated to 30.8% from the control group (5.96%). However, the percentages of apoptotic cells in RXRα knockout A549 cells without and with 6A treatment were 2.07 and 2.72%, respectively. These results demonstrated that apoptosis cells increased significantly after the treatment of 6A in A549 cells, and the apoptosis induction of 6A was mainly through RXRα-dependent pathways. It indicates the expression of RXRα plays a role in the apoptosis induction by 6A.
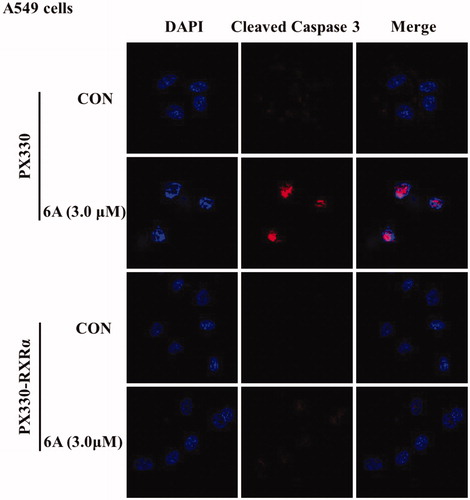
Figure 6. 6A-induced apoptotic effect. (A) The images of A549 and RXRα-/- A549 cells after treatment with or without 6A for 12 h; (B) Flow cytometry analysis of A549 and RXRα-/- A549 cells after treatment with or without 6A for 12 h.
Finally, to find out whether the apoptotic induction of 6A is dependent on caspase pathway, we next used immunofluorescence assay to evaluate the effect of 6A on the activation of caspase-3 a major executioner caspase known to cleave PARP. As shown in , 6A treatment of A549 cells resulted in the strong activation of caspase-3 and nuclear shrinkage compared with control cells, whereas there is no significant difference between treated and untreated RXR-/- A549 cells. This result was consistent with c-PARP. It suggests that compound 6A induces cell death in A549 cells via caspase-dependent pathway and RXRα dependent pathway.
2.3. Molecular docking studies
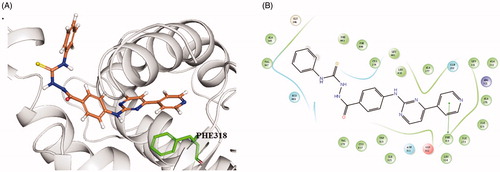
Based on the antagonist effect of 6A on RXRα, the molecular docking of 6A with antagonistic conformation of RXRα (PDB: 3A9E) was performed using Schrodinger 2015.1 Glide protocol to explore their binding mode. The docking study showed that 6A was well accommodated into LBP of RXRα (see and Supplementary movie). The docking results predict the binding features of 6A with RXRα. First, the right 4-(pyridin-4-yl)pyrimidine portion of compound 6A occupies the sub-pocket formed by hydrophobic aromatic amino acid residues LEU438, LEU331, ALA276, PHE318, and TRP310, the pyridine ring forming π–π interaction with PHE318. Second, the hydrazine-1-carbothioamide structure locates at the polar region involving HID440. Third, the left phenyl group of Compound 6A is directed to the protein-solvent interface and makes hydrophobic interactions with surrounding lipophilic amino acids such as VAL347 and PHE443. These findings may provide a direction for the further modifications of this new class of RXRα regulator.
Figure 8. Predicted binding modes of 6A with RXRα-LBP under antagonistic conformation (PDB ID: 3A9E). (A) Close-up view depiction (3 D model) of the superposition of RXRα LBP bound to 6A. The RXRα binding site is presented with a grey ribbon. The 6A is shown in stick with orange carbon, blue nitrogen, and yellow sulphur. The figure was prepared using PyMol (http://www.pymol.org/). (B) Two-dimensional diagram of interactions between compound 6A and RXRα. The π–π interactions are displayed as green solid line. Colour lines around 6A stand for the binding pocket and the residues in colours nearby established the pocket. The green colour denotes the hydrophobic nature of amino acids, the red colour denotes the acid amino acids, the purple denotes the alkalinity of amino acids, the cyan denotes the polar amino acids, and the grey points of ligand atoms denotes the solvent accessibility.
3. Conclusion
In summary, 2-(4-(pyrimidin-2-yl)amino)benzoyl)hydrazine-1-carboxamide/carbothioamide (Series A) and 2-(3-(pyrimidin-2-yl)amino)benzoyl) hydrazine-1-carboxamide/carbothioamide (Series B) derivatives were first synthesised and evaluated as potential anti-tumour agents targeting RXRα. Compounds 6c, 6e, 6h, 6A, and 6J exhibited potent antiproliferative activity with IC50 values of lower than 15 μM against two human tumour cell lines, while compounds 6k, 6A, 6B, 6C, 6G, and 6H were identified as good antagonists of RXRα with EC50 values of lower than 10.0 μM at RXRE-luciferase reporter-based assay. Compound 6A with the low cytotoxity of normal cells was identified as the most potent anticancer agent targeting RXRα. Moreover, it was found that the representative compound 6A effectively induced RXRα-dependent apoptosis of cancer cells in a time- and concentration- dependent manner. SPR technology-based experiment confirmed that compound 6A could selectively bind to RXRα-LBD with a Kd value of 1.20 × 10−7 M, and molecular docking analysis showed that compound 6A could bind to the LBP of RXRα like the classical RXRα antagonists. Taken together, this class of synthetic compounds may be served as a good starting point to design and synthesise new RXRα antagonists, and further studies of compound 6A on activity in vivo would be considered in the future.
4. Experimental section
4.1. Chemistry
All reagents and solvents were purchased from commercial sources and were used without further purification. Oxygen or water sensitive reactions, which required the use of nitrogen atmosphere. All reactions were magnetically stirred and monitored by thin-layer chromatography (TLC) on (Qingdao Haiyang Chemical, China) silica gel 60 F-254 by fluorescence. 1H-NMR and 13C-NMR spectra were obtained using a Bruker AV2 600 Ultra shield spectrometer at 600 and 150 MHz, respectively. Chemical shifts were given in ppm (δ) relative to tetramethylsilane as the internal standard. Spin multiplicities were described as s (singlet), d (doublet), dd (double doublet), t (triplet), br (broad signal), and orm (multiplet). High-resolution mass spectra (HRMS) data were acquired using a quadrupole-orbitrap (Q-Exactive) mass spectrometer (Thermo Scientific, Hemel Hempstead, UK) equipped with a heated electrospray ionisation source (HESI-II). Melting points were measured on SGW X-4 micro-melting point spectrometer and were uncorrected. The high-performance liquid chromatography (HPLC) analysis was performed on an Agilent Technologies 1100 Series HPLC system (Palo Alto, CA). The mobile phase was (A) methanol and (B) water in a linear gradient mode as follows: (A) from 5 to 100% and (B) from 95 to 0% during 0–30 min. The flow rate was 1 ml/min, and the detection wavelengths were 254 and 280 nm.
4.1.1. General procedure for the synthesis of compounds 3a–3e
To a solution of appropriate 1a–1e (0.041 mol) in toluene (50 ml), DMF-DMA (0.082 mol) was added, the reaction mixture was stirred at 110 °C for 18 h. Then, the mixture was cooled to room temperature, the solvent was removed by reduced pressure distillation. Then 20 ml ethyl acetate was added. The resulting solution was stirred at room temperature for 1 h, and filtered under suction, washed with (ethyl acetate: n-hexane = 3:1) to afford the compounds 3a–3e.
4.1.1.1. (E)-3-(dimethylamino)-1-(pyridin-4-yl)prop-2-en-1-one (3a)
According to the general procedure, compound 3a was obtained by using 4-acetylpyridine, yellow solid, 4.2 g, yield: 58.3%. Melting point (mp): 84–85 °C. 1H NMR (600 MHz, DMSO-d6) δ 8.67–8.69 (m, 2H), 7.79 (d, J = 12.1 Hz, 1H), 7.76 (d, J = 5.5 Hz, 2H), 5.84 (d, J = 12.1 Hz, 1H), 3.18 (s, 3H), 2.95 (s, 3H); 13C NMR (150 MHz, DMSO-d6) δ 155.7, 150.6 (2C), 147.2, 121.5 (2C), 91.2, 45.2, 37.8 (2C).
4.1.1.2. (E)-3-(dimethylamino)-1-(pyridin-3-yl)prop-2-en-1-one (3b)
According to the general procedure, compound 3b was obtained by using 3-acetylpyridine, yellow solid, 3.2 g, yield: 49.5%. mp: 83–85 °C. 1H NMR (600 MHz, DMSO-d6) δ 9.07 (d, J = 1.7 Hz, 1H), 8.66 (dd, J = 4.8, 1.7 Hz, 1H), 8.22 (td, J = 8.0, 1.7 Hz, 1H), 7.77 (d, J = 12.1 Hz, 1H), 7.47–7.51 (m, 1H), 5.87 (d, J = 12.1 Hz, 1H), 3.17 (s, 3H), 2.94 (s, 3H); 13C NMR (150 MHz, DMSO-d6) δ 155.1, 151.8, 149.0, 135.6, 135.1, 123.9, 91.4, 45.1, 37.7 (2C).
4.1.1.3. (E)-3-(dimethylamino)-1-(pyridin-2-yl)prop-2-en-1-one (3c)
According to the general procedure, compound 3c was obtained by using 2-acetylpyridine, Yellow solid, 3.5 g, yield: 53%. mp: 78–79 °C. 1H NMR (600 MHz, DMSO-d6) δ ppm 8.62−8.64 (m, 1 H) 7.99 (d, J = 7.7 Hz, 1 H) 7.91 (td, J = 7.7, 1.7 Hz, 1 H) 7.80 (d, J = 12.0 Hz, 1 H) 7.50 (m, 1 H) 6.38 (d, J = 12.0 Hz, 1 H) 3.18 (s, 3 H) 2.92 (s, 3 H); 13C NMR (150 MHz, DMSO-d6) δ 156.2, 154.8, 148.9, 137.5, 126.1, 121.6, 90.5, 45.1, 37.6 (2C).
4.1.1.4. (E)-3-(dimethylamino)-1-phenylprop-2-en-1-one (3d)
According to the general procedure, compound 3d was obtained by using acetophenone, Yellow solid, 2.6 g, yield: 60%. mp: 95–96 °C. 1H NMR (600 MHz, DMSO-d6) δ 7.87–7.89 (m, 2 H) 7.71 (d, J = 12.3 Hz, 1 H), 7.46–7.50 (m, 1 H), 7.41–7.45 (m, 2 H), 5.82 (d, J = 12.3 Hz, 1 H), 3.14 (s, 3 H), 2.91 (s, 3 H); 13C NMR (150 MHz, DMSO-d6) δ 154.6, 140.7, 131.2, 128.6 (2C), 127.6 (2C), 91.4, 45.0, 37.6 (2C).
4.1.1.5. (E)-1-(4-chlorophenyl)-3-(dimethylamino)prop-2-en-1-one (3e)
According to the general procedure, compound 3e was obtained by using 4-chloroacetophenone, yellow solid, 2.6 g, yield: 56%. mp: 101–102 °C. 1H NMR (600 MHz, DMSO-d6) δ 7.89–7.94 (m, 2H), 7.73 (d, J = 12.1 Hz, 1H), 7.49–7.51 (m, 1H), 7.47–7.48 (m, 1H), 5.82 (d, J = 12.1 Hz, 1H), 3.15 (s, 3H), 2.92 (s, 3H); 13C NMR (150 MHz, DMSO-d6) δ 184.7, 155.0, 139.4, 136.0, 129.5 (2C), 128.6 (2C), 91.1, 45.0, 37.6.
4.1.2. 4-Guanidinobenzoate ethyl ester nitrate (4a) and 3-guanidinobenzoate ethyl ester nitrate (4A)
4.1.2.1. 4-guanidinobenzoate ethyl ester nitrate (4a)
To a solution of ethyl p-aminobenzoate (2a; 5.0 g, 0.030 mol) in ethanol (20 ml), cyanamide (50.0% w/w, 7.6 g, 0.091 mol) was added, Concentrated hydrochloric acid (3.8 ml, 0.045 mol) was added dropwise with stirring at 0 °C. After the completion of the dropwise addition, the temperature was raised to reflux for 24 h. The reaction mixture was concentrated under reduced pressure. The residue was diluted with 50 ml of water, and then an aqueous solution of ammonium nitrate (4.8 g, 0.061 mol) was added dropwise at 0 °C. After the completion of the dropwise addition, the mixture was kept for 1 h, then filtered to give 4a.
White solid, 6.3 g, yield: 78%. mp: 175–176 °C. 1H NMR (600 MHz, DMSO-d6): δ 9.96 (br s, 1H), 8.00 (d, J = 8.6 Hz, 2H), 7.65 (s, 3H), 7.36 (d, J = 8.6 Hz, 2H), 4.32 (q, J = 7.1 Hz, 2H), 1.30–1.34 (m, 3H); 13C NMR (150 MHz, DMSO-d6): δ 165.6, 155.8, 140.9, 131.2(2C), 127.2, 123.4(2C), 61.2, 14.6.
4.1.2.2. 3-guanidinobenzoate ethyl ester nitrate (4A)
According to the synthetic procedure of 4a, compound 4A was obtained by using ethyl 3-aminobenzoate (2b).
White solid, 6.0 g, yield: 74%. mp: 177–178 °C. 1H NMR (600 MHz, DMSO-d6): δ 7.85 (m, 1H), 7.78 (t, J = 1.8 Hz, 1H), 7.58–7.62 (m, 1H), 7.51–7.56 (m, 4H), 4.34 (q, J = 7.0 Hz, 2H), 1.33 (t, J = 7.0 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 165.6, 156.3, 136.5, 131.7, 130.7, 129.6, 127.3, 125.3, 61.5, 14.6.
4.1.3. General procedure for the synthesis of compounds 5a–5e
To a solution of (E)-3-(dimethylamino)-1-(pyridin-4-yl)prop-2-en-1-one (1.3 g, 7.4 mmol) and ethyl 4-guanidinobenzoate nitrate (4a;2.0 g, 7.4 mmol), sodium hydroxide (355 mg, 8.9 mmol) was added, and the mixture was heated to reflux for 48 h. After the completion of the reaction, the mixture was concentrated and purified by column chromatography using appropriate mixtures of dichloromethane and methanol (MeOH) to afford ethyl 4-((4-(pyridin-4-yl)pyrimidin-2-yl)amino)benzoate, then to a solution of appropriate compound. To a solution of ethyl 4-((4-(pyridin-4-yl)pyrimidin-2-yl)amino)benzoate (1.00 eq) in ethanol, hydrazine hydrate (1.00 eq) was added, and the reaction mixture was stirred at 87 °C for 20 h. The reaction mixture was cooled to room temperature, filtered and the concentrated filter cake was dried to obtain 5a. Yield: 84.3%.
Compounds 5b–5e were synthesised following the general procedure as described above with the yield of 52–68%.
4.1.3.1. 4-((4-(pyridin-4-yl)pyrimidin-2-yl)amino)benzohydrazide (5a)
Yellow solid 1.1 g, yield: 58%. mp: 298–300 °C. 1H NMR (600 MHz, DMSO-d6): δ 10.11 (s, 1H), 9.62 (s, 1H), 8.81 (dd, J = 4.4, 1.6 Hz, 2H), 8.72 (d, J = 5.1 Hz, 1 H), 8.11 (dd, J = 4.4, 1.6 Hz, 2H), 7.92 (dt, J = 8.0, 1.8 Hz, 2H), 7.83 (dt, J = 8.0, 1.8 Hz, 2H), 7.60 (d, J = 5.1 Hz, 1H), 4.46 (br s, 2H); 13C NMR (150 MHz, DMSO-d6): δ 166.3, 162.0, 160.5, 160.4, 151.1, 144.2(2C), 143.4, 128.2 (2C), 126.5, 121.4 (2C), 118.4 (2C), 109.7.
4.1.3.2. 4-((4-(pyridin-3-yl)pyrimidin-2-yl)amino)benzohydrazide (5b)
White solid, 1.0 g, yield: 53%. mp: 285–287 °C. 1H NMR (600 MHz, DMSO-d6): δ 10.05 (s, 1H), 9.61 (s, 1H), 9.36–9.37 (m, 1H), 8.75 (m, 1H), 8.66 (d, J = 5.1 Hz, 1H), 8.52 (dt, J = 8.01.8 Hz, 1H), 7.90 (dt, J = 8.0, 1.8 Hz, 2H), 7.82 (dt, J = 8.0, 1.8 Hz, 2H), 7.59–7.63 (m, 1 H), 7.58 (d, J = 5.1 Hz, 1H), 4.45 (br s, 2H); 13C NMR (150 MHz, DMSO-d6): δ 166.3, 162.2, 160.4, 159.9, 152.1, 148.7, 143.5, 134.9, 132.6, 128.2, 126.4, 124.4 (2C), 118.3 (2C), 109.4.
4.1.3.3. 4-((4-(pyridin-2-yl)pyrimidin-2-yl)amino)benzohydrazide (5c)
White solid, 0.8 g, yield: 68%. mp: 286–288 °C. 1H NMR (600 MHz, DMSO-d6): δ 10.06 (s, 1H), 9.62 (s, 1H), 8.76–8.78 (m, 1 H), 8.70 (d, J = 5.1 Hz, 1H), 8.43 (d, J = 7.9 Hz, 1H), 8.07 (td, J = 7.9, 1.8 Hz, 1H), 7.92 (dt, J = 8.0, 1.8 Hz, 2H), 7.84 (dt, J = 8.0, 1.8 Hz, 2H), 7.79 (d, J = 5.1 Hz, 1H), 7.59 (m, 1H), 4.46 (br s, 2 H); 13C NMR (150 MHz, DMSO-d6): δ 166.3, 163.4, 160.3, 160.1, 154.0, 150.1, 143.6, 138.1, 128.2, 126.3, 126.3, 121.6 (2C), 118.3 (2C), 109.1.
4.1.3.4. 4-((4-phenylpyrimidin-2-yl)amino)benzohydrazide (5d)
White solid, 0.6 g, yield: 67%. mp: 303–305 °C. 1H NMR (600 MHz, DMSO-d6): δ 9.98 (s, 1H), 9.60 (s, 1 H), 8.59–8.63 (m, 1H), 8.18–8.20 (m, 2H), 7.93 (d, J = 8.8 Hz, 2H), 7.82 (d, J = 8.8 Hz, 2H), 7.56–7.59 (m, 3H), 7.49 (d, J = 5.1 Hz, 1H), 4.44 (s, 2H); 13C NMR (150 MHz, DMSO-d6): δ 166.3, 164.2, 160.4, 159.6, 143.7, 137.0, 131.5 (2C), 129.4 (2C), 128.2, 127.4(2C), 126.2, 118.2 (2C), 109.1
4.1.3.5. 4-((4-(4-chlorophenyl)pyrimidin-2-yl)amino)benzohydrazide (5e)
Yellow solid, 1.1 g, yield: 58%. mp: 300–302 °C. 1H NMR (600 MHz, DMSO-d6): δ 9.98 (s, 1 H), 9.60 (s, 1H), 8.61 (d, J = 5.1 Hz, 1H), 8.18–8.20 (m, 2H), 7.93 (d, J = 8.8 Hz, 2H), 7.82 (d, J = 8.8 Hz, 2H), 7.57–7.58 (m, 2H), 7.49 (d, J = 5.1 Hz, 1H), 4.44 (s, 2H); 13C NMR (150 MHz, DMSO-d6): δ 166.3, 163.0, 160.4, 159.8, 143.6, 136.3, 135.8, 129.5(2C), 129.2 (2C), 128.2 (2C), 126.3, 118.3 (2C), 109.0.
4.1.4. General procedure for the synthesis of compounds 5A–5C
According to the synthesis of compounds 5a–5c, compounds 5A–5C were synthesised with 4A instead of 4a in a similar method.
4.1.4.1. 3-((4-(pyridin-4-yl)pyrimidin-2-yl)amino)benzohydrazide (5A)
Yellow solid, 0.5 g, yield: 63%. mp: 302–304 °C. 1H NMR (600 MHz, DMSO-d6) δ 9.98 (s, 1H), 9.73 (s, 1H), 8.79–8.80 (m, 1H), 8.79–8.79 (m, 1H), 8.69 (d, J = 5.1 Hz, 1H), 8.45 (s, 1H), 8.13–8.17 (m, 2H), 7.87–7.91 (m, 1H), 7.58 (d, J = 5.1 Hz, 1H), 7.41–7.44 (m, 1H), 7.38–7.41 (m, 1H), 4.54 (br s, 2H); 13C NMR (150 MHz, DMSO-d6) δ 166.8, 161.8, 160.7, 160.4, 151.1 (2C), 144.2, 140.9, 134.5, 128.9, 122.0, 121.4 (2C), 120.2, 118.8, 109.2.
4.1.4.2. 3-((4-(pyridin-3-yl)pyrimidin-2-yl)amino)benzohydrazide (5B)
Yellow solid, 1.2 g, yield: 79%. mp: 306–308 °C. 1H NMR (600 MHz, DMSO-d6): δ 9.92 (s, 1H), 9.72 (s, 1H), 9.38 (dd, J = 2.2, 0.7 Hz, 1H), 8.74 (dd, J = 4.7, 1.6, Hz, 1H), 8.64 (d, J = 5.1 Hz, 1H), 8.57 (dt, J = 8.1, 1.9 Hz, 1H), 8.41 (s, 1H), 7.89–7.92 (m, 1H), 7.60 (m, 1H), 7.55 (d, J = 5.1 Hz, 1H), 7.41–7.45 (m, 1H), 7.34–7.40 (m, 1H), 4.49–4.54 (m, 2H); 13C NMR (150 MHz, DMSO-d6): δ 166.8, 162.1, 160.6, 159.9, 152.0, 148.7, 141.0, 134.9, 134.5, 132.6, 128.9, 124.4, 121.9, 120.1, 118.7, 108.9.
4.1.4.3. 3-((4-(Pyridine-2-yl)pyrimidin-2-yl)amino)benzohydrazide (5C)
Yellow solid, 1.1 g, yield: 68%. Mp: 311–312 °C. 1H NMR (600 MHz, DMSO-d6): δ 9.92 (s, 1H), 9.72 (s, 1H), 8.76 (td, J = 4.6, 1.1 Hz, 1H), 8.67 (d, J = 5.1 Hz, 1H), 8.51 (d, J = 7.7 Hz, 1H), 8.48 (s, 1H), 8.05 (dt, J = 7.7, 1.7, Hz, 1H), 7.90 (td, J = 7.7, 1.7 Hz, 1H), 7.76 (d, J = 5.1 Hz, 1H), 7.58 (ddd, J = 7.7, 4.6, 1.1 Hz, 1H), 7.41–7.43 (m, 1H), 7.37–7.41 (m, 1H), 4.52 (br s, 2H); 13C NMR (150 MHz, DMSO-d6): δ 166.9, 163.3, 160.5, 160.1, 154.0, 150.1, 141.1, 138.1, 134.5, 128.9, 126.2, 121.8, 121.7, 120.0, 118.6, 108.7.
4.1.5. General procedure for the synthesis of target compounds 6a–6t and 6A–6J (Series A)
Compounds 5a–5e (1.00 eq) were separately used to react with an appropriate isocyanate or isothiocyanate (1.00 eq) in ethanol at 70 °C for 4 h, then cooled to room temperature, filtered to get crude products of compounds 6a-6t and 6A-6J, which were further purified by silica gel column chromatography (DCM:MeOH = 50:1).
4.1.5.1. N-butyl-2-(4-((4-(pyridin-4-yl)pyrimidin-2-yl)amino)benzoyl)hydrazine-1-carboxamide (6a)
Yellow solid (94.52%), 0.056 g, yield: 58%, mp: 213–215 °C. 1H NMR (600 MHz, DMSO-d6): δ 10.16 (s, 1H), 9.96 (br s, 1H), 8.82 (dd, J = 4.4, 1.4 Hz, 2H), 8.73 (d, J = 5.1 Hz, 1H), 8.13 (dd, J = 4.4, 1.4 Hz, 2H), 7.94 (d, J = 9.0 Hz, 2H), 7.89 (d, J = 9.0 Hz, 2H), 7.74 (s, 1H), 7.62 (d, J = 5.1 Hz, 1H), 6.45 (br s, 1H), 3.03 (q, J = 6.7 Hz, 2H), 1.38 (quin, J = 7.3 Hz, 2H), 1.28 (sxt, J = 7.3 Hz, 2H), 0.87 (t, J = 7.3 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 166.5, 161.9, 160.5, 159.0, 150.9 (2C), 144.4, 143.9 (2C), 128.9 (2C), 125.8, 121.5 (2C), 118.3 (2C), 109.8, 39.3, 32.5, 19.9, 14.2; HRMS (ESI, m/z): calcd for C21H24N7O2+ [M + H]+ 406.1986, found 406.1985.
4.1.5.2. N-(tert-butyl)-2-(4-((4-(pyridin-4-yl)pyrimidin-2-yl)amino)benzoyl) hydrazine-1-carboxamide (6b)
Yellow solid (99.24%), 0.054 g, yield: 60%, mp: 171–172 °C. 1H NMR (600 MHz, DMSO-d6): δ 10.16 (s, 1H), 9.95 (s, 1H), 8.80 (dd, J = 4.4, 1.4 Hz, 2H), 8.73 (d, J = 5.1 Hz, 1H), 8.11 (dd, J = 4.4, 1.4 Hz, 2H), 7.93 (d, J = 9.0 Hz, 2H), 7.88 (d, J = 9.0 Hz, 2H), 7.61 (s, 1H), 7.60 (d, J = 5.1 Hz, 1H)6.08 (br s, 1H), 1.27 (s, 9H); 13C NMR (150 MHz, DMSO- d6): δ 166.3, 162.0, 160.5, 157.8, 151.1(2C), 144.2, 143.9 (2C), 128.7 (2C), 125.7, 121.4 (2C), 118.3 (2C), 109.8, 49.9, 29.6 (3 C); HRMS (ESI, m/z): calcd for C21H24N7O2+ [M + H]+ 406.1986, found 406.1983, calcd for C21H23N7O2Na+ [M + Na]+ 428.1805, found 428.1803.
4.1.5.3. N-cyclohexyl-2-(4-((4-(pyridin-4-yl)pyrimidin-2-yl)amino)benzoyl) hydrazine-1-carboxamide (6c)
White solid (93.67%), 0.055 g, yield: 58%, mp: 181–183 °C. 1H NMR (600 MHz, DMSO-d6): δ 10.15 (s, 1H), 9.96 (s, 1H), 8.80 (dd, J = 4.4, 1.4 Hz, 2H), 8.72 (d, J = 5.1 Hz, 1H), 8.10 (dd, J = 4.4, 1.4 Hz, 2H), 7.92 (d, J = 9.0 Hz, 2H), 7.88 (d, J = 9.0 Hz, 2H), 7.69 (s, 1H), 7.60 (d, J = 5.1 Hz, 1H), 6.26 (d, J = 8.0 Hz, 1H), 3.38–3.43 (m, 1H), 1.75 (dd, J = 9.3, 3.8 Hz, 2H), 1.64 (dd, J = 9.3, 3.8 Hz, 2H), 1.53 (dd, J = 9.3, 3.8 Hz, 1H), 1.22–1.30 (m, 2H), 1.11–1.20 (m, 3H); 13C NMR (150 MHz, DMSO-d6): δ 166.4, 162.0, 160.4, 158.2, 151.1(2C), 144.2, 143.9 (2C), 128.8 (2C), 125.7, 121.4 (2C), 118.3(2C), 109.8, 48.6, 33.5 (2C), 25.7, 25.1 (2C); HRMS (ESI, m/z): calcd for C23H26N7O2+ [M + H]+ 432.2142, found 432.2141, calcd for C23H25N7O2Na+ [M + Na]+ 454.1962, found 454.1960.
4.1.5.4. N-cyclopentyl-2-(4-((4-(pyridin-4-yl)pyrimidin-2-yl)amino)benzoyl) hydrazine-1-carboxamide (6d)
Yellow solid (95.45%), 0.054 g, yield: 57%, mp: 238–239 °C. 1H NMR (600 MHz, DMSO-d6): δ 10.15 (s, 1H), 9.95 (s, 1H), 8.80 (dd, J = 4.4, 1.4 Hz Hz, 2H), 8.72 (d, J = 5.1 Hz, 1H), 8.11 (dd, J = 4.4, 1.4 Hz, 2H), 7.92–7.95 (m, 2H), 7.88 (d, J = 9.0 Hz, 2H), 7.67 (s, 1H), 7.60 (d, J = 5.1 Hz, 1H), 6.36 (d, J = 7.5 Hz, 1H), 3.90 (m, 1H), 1.76–1.83 (m, 2H), 1.58–1.65 (m, 2H), 1.45–1.53 (m, 2H), 1.33–1.41 (m, 2H); 13C NMR (150 MHz, DMSO-d6): δ 166.4, 162.0, 160.4, 158.5, 151.1(2C), 144.2, 143.9 (2C), 128.8 (2C), 125.8, 121.4 (2C), 118.3 (2C), 109.8, 51.6, 33.1 (2C), 23.7 (2C); HRMS (ESI, m/z): calcd for C22H24N7O2+ [M + H]+ 418.1986, found 418.1984, calcd for C22H23N7O2Na+ [M + Na]+ 440.1805, found 440.1804.
4.1.5.5. 2-(4-((4-(pyridin-4-yl)pyrimidin-2-yl)amino)benzoyl)-N-(p-tolyl)hydrazine-1-carboxamide (6e)
Yellow solid (99.58%), 0.061 g, yield: 69%, mp: 230–231 °C. 1H NMR (600 MHz, DMSO-d6): δ 10.17 (s, 1H), 10.14 (s, 1H), 8.80 (dd, J = 4.4, 1.4 Hz, 2H), 8.76 (br s, 1H), 8.72 (d, J = 5.1 Hz, 1H), 8.11 (dd, J = 4.4, 1.4 Hz, 2H), 8.07 (s, 1H), 7.95 (d, J = 9.0 Hz, 2H), 7.93 (d, J = 9.0 Hz, 2H), 7.60 (d, J = 5.1 Hz, 1H), 7.35 (d, J = 8.2 Hz, 2H), 7.07 (d, J = 8.2 Hz, 2H), 2.23 (s, 3H); 13C NMR (150 MHz, DMSO-d6): δ 166.7, 162.0, 160.5, 156.3, 151.1 (2C), 144.2, 144.1 (2C), 137.5, 131.2, 129.5 (2C), 128.9 (2C), 125.6, 121.4 (2C), 119.1(2C), 118.3 (2C), 109.8, 20.8; HRMS (ESI, m/z): calcd for C24H22N7O2+ [M + H]+ 440.1829, found 440.1824, calcd for C24H21N7O2Na+ [M + Na]+ 462.1649, found 462.1642.
4.1.5.6. N-(2,4-difluorophenyl)-2–(4-((4-(pyridin-4-yl)pyrimidin-2-yl)amino)benzoyl) hydrazine-1-carboxamide (6f)
Yellow solid (93.67%), 0.065 g, yield: 71%, mp: 252–254 °C. 1H NMR (600 MHz, DMSO-d6): δ 10.23 (br s, 1H), 10.17 (s, 1H), 8.79 (dd, J = 4.4, 1.4 Hz, 2H), 8.72 (d, J = 5.1 Hz, 1H), 8.65 (br s, 1H), 8.42 (s, 1H), 8.11 (dd, J = 4.4, 1.4 Hz, 2H), 7.95 (d, J = 9.0 Hz, 2H), 7.90 (d, J = 9.0 Hz, 2H), 7.61 (d, J = 5.1 Hz, 1H), 7.26–7.31 (m, 1H), 6.99–7.08 (m, 1H); 13C NMR (150 MHz, DMSO-d6): δ 166.6, 162.0, 160.5, 160.4, 156.1, 151.1(2C), 144.2, 144.1(2C), 128.9 (2C), 125.4, 124.2, 121.4, 118.4 (2C), 111.6, 111.4, 109.8, 104.5, 104.3, 104.3, 104.1; HRMS (ESI, m/z): calcd for C23H18F2N7O2+ [M + H]+ 462.1485, found 462.1476, calcd for C23H17F2N7O2Na+ [M + Na]+ 484.1304, found 484.1296.
4.1.5.7. N-(3-chloro-4-methylphenyl)-2-(4-((4-(pyridin-4-yl)pyrimidin-2-yl)amino) benzoyl)hydrazine-1-carboxamide (6g)
Yellow solid (99.12%), 0.063 g, yield: 64%, mp: 209–210 °C. 1H NMR (600 MHz, DMSO-d6): δ 10.18 (s, 1H), 10.15 (s, 1H), 8.97 (br s, 1H), 8.81 (dd, J = 4.4, 1.4 Hz, 2H), 8.73 (d, J = 5.1 Hz, 1H), 8.23 (br s, 1H), 8.12 (dd, J = 4.4, 1.4 Hz, 2H), 7.96 (d, J = 9.0 Hz, 2H), 7.93 (d, J = 9.0 Hz, 2H), 7.68 (d, J = 1.4 Hz, 1H), 7.62 (d, J = 5.1 Hz, 1H), 7.29 (br s, 1H), 7.22 (d, J = 8.2 Hz, 1H), 2.25 (s, 3H); 13C NMR (150 MHz, DMSO-d6): δ 166.7, 162.0, 160.5, 160.4, 151.0(2C), 144.3, 144.1 (2C), 139.4, 133.4, 131.5, 128.9 (2C), 128.7, 125.6 (2C), 121.5 (2C), 118.3 (2C), 109.8, 19.2; HRMS (ESI, m/z): calcd for C24H21ClN7O2+ [M + H]+ 474.1440, found 474.1440, calcd for C24H20ClN7O2Na+ [M + Na]+ 496.1259, found 496.1259.
4.1.5.8. N-phenethyl-2-(4-((4-(pyridin-4-yl)pyrimidin-2-yl)amino)benzoyl)hydrazine-1-carboxamide (6h)
Yellow solid (96.95%), 0.072 g, yield: 75%, mp: 240–241 °C. 1H NMR (600 MHz, DMSO-d6): δ 10.17 (s, 1H), 9.99 (s, 1H), 8.81 (dd, J = 4.4, 1.4 Hz, 2H), 8.74 (d, J = 5.1 Hz, 1H), 8.12 (dd, J = 4.4, 1.4 Hz, 2H), 7.94 (d, J = 9.0 Hz, 2H), 7.91 (d, J = 9.0 Hz, 2H), 7.86 (s, 1H), 7.62 (d, J = 5.1 Hz, 1H), 7.29 (t, J = 7.6 Hz, 3H), 7.20–7.24 (m, 3H), 7.17–7.20 (m, 1H), 6.51 (br s, 1H), 3.27 (d, J = 7.3 Hz, 2H), 2.72 (t, J = 7.3 Hz, 2H); 13C NMR (150 MHz, DMSO-d6): δ 166.5, 162.0, 160.5, 158.9, 151.1(2C), 144.2, 144.0 (2C), 140.1, 129.1 (2C), 128.9 (2C), 128.8, 128.8, 126.5, 125.8, 121.4 (2C), 118.3 (2C), 109.8, 41.4, 36.5; HRMS (ESI, m/z): calcd for C25H24N7O2+ [M + H]+ 454.1986, found 454.1979, calcd for C25H23N7NaO2+ [M + Na]+ 476.1805, found 476.1799.
4.1.5.9. N-butyl-2-(4-((4-(pyridin-3-yl)pyrimidin-2-yl)amino)benzoyl)hydrazine-1-carboxamide (6i)
Yellow solid (99.63%), 0.076 g, yield: 81%, mp: 179–181 °C. 1H NMR (600 MHz, DMSO-d6): δ 10.10 (s, 1H), 9.96 (br s, 1H), 9.36 (d, J = 2.0 Hz, 1H), 8.75 (dd, J = 4.7, 2.0, Hz, 1H), 8.67 (d, J = 5.1 Hz, 1H), 8.53 (dt, J = 8.0, 2.0 Hz, 1H), 7.93 (d, J = 9.0 Hz, 2H), 7.89 (d, J = 9.0 Hz, 2H), 7.73 (s, 1H), 7.61 (dd, J = 8.0, 4.7, Hz, 1H), 7.59 (d, J = 5.1 Hz, 1H), 6.45 (br s, 1H), 3.03–3.04 (m, 2H), 1.38 (quin, J = 7.3 Hz, 2H), 1.28 (sxt, J = 7.3 Hz, 2H), 0.87 (t, J = 7.3 Hz, 3H); 13C NMR (150 MHz, DMSO- d6): δ 166.5, 162.2, 160.3, 160.0, 159.0, 152.1, 148.6, 144.0, 134.9, 132.5, 128.9 (2C), 125.7, 124.5, 118.2 (2C), 109.5, 39.3, 32.5, 19.9, 14.2; HRMS (ESI, m/z): calcd for C21H24N7O2+ [M + H]+ 406.1986, found 406.1980.
4.1.5.10. N-cyclopentyl-2-(4-((4-(pyridin-3-yl)pyrimidin-2-yl)amino)benzoyl) hydrazine-1-carboxamide (6j)
Yellow solid (95.47%), 0.071 g, yield: 75%, mp: 159–161 °C. 1H NMR (600 MHz, DMSO-d6): δ 10.11 (s, 1H), 9.94 (s, 1H), 9.37 (d, J = 1.6 Hz, 1H), 8.75 (dd, J = 4.7, 1.6, Hz, 1H), 8.68 (d, J = 5.1 Hz, 1H), 8.53 (d, J = 7.5 Hz, 1H), 7.92–7.95 (d, J = 9.0 Hz, 2H), 7.88 (d, J = 9.0 Hz, 2H), 7.66 (s, 1H), 7.61–7.63 (m, 1H), 7.59 (d, J = 5.1 Hz, 1H), 6.35 (d, J = 7.5 Hz, 1H), 3.91 (m, 1H), 1.76–1.83 (m, 2H), 1.58–1.65 (m, 2H), 1.45–1.53 (m, 2H), 1.33−1.41 (m, 2H); 13C NMR (150 MHz, DMSO-d6): δ 166.4, 162.2, 160.4, 159.9, 158.5, 152.1, 148.7, 144.0, 134.9, 132.5, 128.8 (2C), 125.7, 124.5, 118.2 (2C), 109.5, 51.5, 33.1 (2C), 23.7 (2C); HRMS (ESI, m/z): calcd for C22H24N7O2+ [M + H]+ 418.1986, found 418.1980, calcd for C22H23N7NaO2+ [M + Na]+ 440.1805, found 440.1799.
4.1.5.11. 2-(4-((4-(pyridin-3-yl)pyrimidin-2-yl)amino)benzoyl)-N-(p-tolyl)hydrazine-1-carboxamide (6k)
Yellow solid (96.10%), 0.056 g, yield:59%, mp: 209–210 °C. 1H NMR (600 MHz, DMSO-d6): δ 10.13 (s, 2H), 9.37 (d, J = 1.6 Hz, 1H), 8.76 (dd, J = 4.7, 1.6 Hz, 1H), 8.74 (br s, 1H), 8.68 (d, J = 5.1 Hz, 1H), 8.53 (dt, J = 8.0, 1.6 Hz, 1H), 8.07 (s, 1H), 7.95 (d, J = 9.0 Hz, 2H), 7.92 (d, J = 9.0 Hz, 2H), 7.61–7.63 (m, 1H), 7.60 (d, J = 5.1 Hz, 1H), 7.36 (d, J = 8.2 Hz, 2H), 7.07 (d, J = 8.2 Hz, 2H), 2.24 (s, 3H); 13C NMR (150 MHz, DMSO-d6): δ 166.6, 162.2, 160.4, 160.0, 152.1, 148.7, 144.2, 137.6, 135.0, 132.5, 131.1, 129.6, 129.5 (2C), 128.9 (2C), 125.5, 124.5, 121.5 (2C), 118.3 (2C), 109.5, 20.8; HRMS (ESI, m/z): calcd for C24H22N7O2+ [M + H]+ 440.1829, found 440.1823, calcd for C24H21N7NaO2+ [M + Na]+ 462.1649, found 462.1643.
4.1.5.12. N-phenethyl-2-(4-((4-(pyridin-3-yl)pyrimidin-2-yl)amino)benzoyl) hydrazine-1-carboxamide (6l)
Yellow solid (95.57%), 0.054 g, yield: 56%, mp: 177–178 °C. 1H NMR (600 MHz, DMSO-d6): δ 10.09 (s, 1H), 9.99 (s, 1H), 9.35 (d, J = 2.0 Hz, 1H), 8.74 (dd, J = 4.7, 1.4 Hz, 1H), 8.66 (d, J = 5.1 Hz, 1H), 8.52 (dt, J = 8.0, 1.4 Hz, 1H), 7.93 (d, J = 9.0 Hz, 2H), 7.88–7.90 (d, J = 9.0 Hz, 2H), 7.86 (s, 1H), 7.61 (dd, J = 8.0, 4.7 Hz, 1H), 7.58 (d, J = 5.1 Hz, 1H), 7.26−7.30 (m, 2H), 7.20–7.23 (m, 2H), 7.17–7.19 (m, 1H), 6.53 (br s, 1H), 3.22–3.29 (m, 2H), 2.71 (t, J = 7.3 Hz, 2H); 13C NMR (150 MHz, DMSO-d6): δ 166.6, 162.2, 160.3, 160.0, 159.0, 152.1, 148.6, 144.1, 140.0, 135.0, 132.5, 129.1 (2C), 128.9 (2C), 128.8 (2C), 126.5, 125.6, 124.5, 118.2 (2C), 109.5, 41.4, 36.4;HRMS (ESI, m/z): calcd for C25H24N7O2+ [M + H]+ 454.1986, found 454.1981.
4.1.5.13. N-(tert-butyl)-2-(4-((4-(pyridin-2-yl)pyrimidin-2-yl)amino)benzoyl) hydrazine-1-carboxamide (6m)
Yellow solid (98.73%), 0.076 g, yield: 79%, mp: 213–214 °C. 1H NMR (600 MHz, DMSO-d6): δ 10.10 (s, 1H), 9.95 (s, 1H), 8.76 (s, 1H), 8.71 (d, J = 4.9 Hz, 1H), 8.43 (d, J = 7.8 Hz, 1H), 8.07 (td, J = 7.8, 1.6 Hz, 1H), 7.94 (d, J = 8.6 Hz, 2H), 7.88 (d, J = 8.6 Hz, 2H), 7.80 (d, J = 4.9 Hz, 1H), 7.58–7.61 (m, 2H), 6.08 (br s, 1H), 1.27 (s, 9H); 13C NMR (150 MHz, DMSO-d6): δ 166.3, 163.4, 160.2, 160.1, 157.8, 153.9, 150.2, 144.1, 138.1, 128.7 (2C), 126.3, 125.6, 121.6, 118.2 (2C), 109.2, 49.9, 29.6 (3 C); HRMS (ESI, m/z): calcd for C21H24N7O2+ [M + H]+ 406.1986, found 406.1982.
4.1.5.14. N-butyl-2-(4-((4-(pyridin-2-yl)pyrimidin-2-yl)amino)benzoyl)hydrazine-1-carboxamide (6n)
White solid (96.55%), 0.072 g, yield: 78%, mp: 228–229 °C. 1H NMR (600 MHz, DMSO-d6): δ 10.10 (s, 1H), 9.96 (s, 1H), 8.77 (s, 1H), 8.71 (d, J = 4.9 Hz, 1H), 8.43 (d, J = 7.8 Hz, 1H), 8.07 (td, J = 7.8, 1.6 Hz, 1H), 7.95 (d, J = 8.6 Hz, 2H), 7.90 (d, J = 8.6 Hz, 2H), 7.80 (d, J = 4.9 Hz, 1H), 7.74 (s, 1H), 7.57–7.62 (m, 1H), 6.44 (br s, 1H), 3.03 (m, 2H), 1.39 (quin, J = 7.3 Hz, 2H), 1.28 (sxt, J = 7.3 Hz, 2H), 0.88 (t, J = 7.3 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 166.6, 163.4, 160.2, 160.1, 159.1, 153.9, 150.1, 144.1, 138.1, 128.9 (2C), 126.3, 125.6, 121.6, 118.2 (2C), 109.2, 32.4, 19.9, 14.2; HRMS (ESI, m/z): calcd for C21H24N7O2+ [M + H]+ 406.1986, found 406.1985, calcd for C21H23N7O2Na+ [M + Na]+ 428.1805, found 428.1806.
4.1.5.15. N-cyclopentyl-2-(4-((4-(pyridin-2-yl)pyrimidin-2-yl)amino)benzoyl) hydrazine-1-carboxamide (6o)
White solid (95.44%), 0.063 g, yield: 73%, mp: 156–158 °C. 1H NMR (600 MHz, DMSO-d6): δ 10.10 (s, 1H), 9.94 (s, 1H), 8.77 (d, J = 4.0 Hz, 1H), 8.71 (d, J = 4.9 Hz, 1H), 8.43 (d, J = 7.8 Hz, 1H), 8.07 (td, J = 7.8, 1.6 Hz, 1H), 7.94–7.97 (d, J = 8.6 Hz, 2H), 7.90 (d, J = 8.6 Hz, 2H), 7.80 (d, J = 4.9 Hz, 1H), 7.66 (s, 1H), 7.59 (m, 1H), 6.35 (d, J = 7.5 Hz, 1H), 3.91 (m, 1H), 1.76–1.84 (m, 2H), 1.58–1.66 (m, 2H), 1.46–1.54 (m, 2H), 1.34–1.41 (m, 2H); 13C NMR (150 MHz, DMSO-d6): δ 166.4, 163.4, 160.2, 160.1, 158.5, 153.9, 150.2, 144.1, 138.1, 128.8 (2C), 126.3, 125.6, 121.6, 118.2 (2C), 109.2, 51.5, 33.1 (2C), 23.7 (2C); HRMS (ESI, m/z): calcd for C22H24N7O2+ [M + H]+ 418.1986, found 418.1980.
4.1.5.16. 2-(4-((4-(pyridin-2-yl)pyrimidin-2-yl)amino)benzoyl)-N-(m-tolyl) hydrazine-1-carboxamide (6p)
Yellow solid (97.54%), 0.054 g, yield: 67%, mp: 229–230 °C. 1H NMR (600 MHz, DMSO-d6): δ 10.13 (s, 1H), 8.77 (d, J = 4.0 Hz, 2H), 8.72 (d, J = 5.1 Hz, 1H), 8.44 (d, J = 7.7 Hz, 1H), 8.11 (s, 1H), 8.07 (dt, J = 7.7, 1.6 Hz, 1H), 7.98 (d, J = 8.6 Hz, 2H), 7.93 (d, J = 8.6 Hz, 2H), 7.81 (d, J = 5.1 Hz, 1H), 7.58–7.61 (m, 1H), 7.31 (s, 1H), 7.29 (d, J = 7.7 Hz, 1H), 7.14 (t, J = 7.7 Hz, 1H), 6.78 (d, J = 7.7 Hz, 1H), 2.26 (s, 3H); 13C NMR (150 MHz, DMSO-d6): δ 166.6, 163.4, 160.3, 160.1, 156.3, 153.9, 150.2, 144.2, 140.1, 138.2, 138.1, 128.9 (2C), 128.9, 126.3, 125.5, 123.1, 121.6, 119.5, 118.2 (2C), 116.1, 109.2, 21.7; HRMS (ESI, m/z): calcd for C24H22N7O2+ [M + H]+ 440.1829, found 440.1826, calcd for C24H21N7NaO2+ [M + Na]+ 462.1649, found 462.1646.
4.1.5.17. N-(3,5-dimethylphenyl)-2-(4-((4-(pyridin-2-yl)pyrimidin-2-yl)amino) benzoyl)hydrazine-1-carboxamide (6q)
Yellow solid (98.69%), 0.073 g, yield: 79%, mp: 245–246 °C. 1H NMR (600 MHz, DMSO-d6): δ 10.13 (s, 1H), 10.12 (s, 1H), 8.77 (d, J = 4.0 Hz, 1H), 8.72 (d, J = 5.1 Hz, 1H), 8.68 (br s, 1H), 8.44 (d, J = 7.8 Hz, 1H), 8.06–8.10 (m, 2H), 7.98 (d, J = 8.6 Hz, 2H), 7.93 (d, J = 8.6 Hz, 2H), 7.81 (d, J = 5.1 Hz, 1H), 7.58–7.61 (m, 1H), 7.11 (s, 2H), 6.60 (s, 1H), 2.22 (s, 6H); 13C NMR (150 MHz, DMSO-d6): δ 166.6, 163.4, 160.2, 160.1, 156.2, 153.9, 150.2, 144.2, 140.0, 138.1, 138.0 (2C), 128.9 (2C), 126.3, 125.5, 123.9 (2C), 121.6, 118.2 (2C), 116.7, 109.2, 21.6 (2C); HRMS (ESI, m/z): calcd for C25H24N7O2+ [M + H]+ 454.1986, found 454.1983.
4.1.5.18. N-(tert-butyl)-2-(4-((4–(4-chlorophenyl)pyrimidin-2-yl)amino)benzoyl) hydrazine-1-carboxamide (6r)
White solid (92.63%), 0.065 g, yield:76%, mp: 170–172 °C. 1H NMR (600 MHz, DMSO-d6): δ 10.05 (s, 1H), 9.93 (s, 1H), 8.63 (d, J = 5.1 Hz, 1H), 8.21 (d, J = 8.6 Hz, 2H), 7.92 (d, J = 8.8 Hz, 2H), 7.86 (d, J = 8.8 Hz, 2H), 7.64 (d, J = 8.6 Hz, 2H), 7.59 (s, 1H), 7.51 (d, J = 5.1 Hz, 1H), 6.07 (br s, 1H), 1.26 (s, 9H); 13C NMR (150 MHz, DMSO-d6): δ 166.3, 163.0, 160.3, 159.8, 157.8, 144.1, 136.3, 135.8, 129.5 (2C), 129. 2(2C), 128.7 (2C), 125.5, 118.2 (2C), 109.1, 49.9, 29.6 (3 C); HRMS (ESI, m/z): calcd for C22H24ClN6O2+ [M + H]+ 439.1644, found 439.1641, calcd for C22H23ClN6NaO2+ [M + Na]+ 461.1463, found 461.1459.
4.1.5.19. 2-(4-((4-(4-chlorophenyl)pyrimidin-2-yl)amino)benzoyl)-N-ethylhydrazine-1-carboxamide (6s)
White solid (97.87%), 0.077 g, yield: 82%, mp: 241–242 °C. 1H NMR (600 MHz, DMSO-d6): δ 10.06 (s, 1H), 9.95 (s, 1H), 8.64 (d, J = 5.1 Hz, 1H), 8.22 (d, J = 8.6 Hz, 2H), 7.93 (d, J = 8.8 Hz, 2H), 7.88 (d, J = 8.8 Hz, 2H), 7.76 (s, 1H), 7.62–7.68 (m, 2H), 7.52 (d, J = 5.1 Hz, 1H), 6.45 (br s, 1H), 3.01–3.11 (m, 2H), 1.02 (t, J = 7.0 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 166.6, 163.0, 160.3, 159.8, 158.9, 144.1, 136.3, 135.7, 129.5 (2C), 129.2 (2C), 128.9 (2C), 125.5, 118.2 (2C), 109.1, 34.5, 16.0; HRMS (ESI, m/z): calcd for C20H20ClN6O2+ [M + H]+ 411.1331, found 411.1330, calcd for C20H19ClN6NaO2+ [M + Na]+ 433.1150, found 433.1149.
4.1.5.20. 2-(4-((4-(4-chlorophenyl)pyrimidin-2-yl)amino)benzoyl)-N-cyclohexyl-hydrazine-1-carboxamide (6t)
White solid (97.94%), 0.046 g, yield: 48%, mp: 165–167 °C. 1H NMR (600 MHz, DMSO-d6): δ 10.06 (s, 1H), 9.94 (s, 1H), 8.64 (d, J = 5.1 Hz, 1H), 8.20–8.23 (m, 2H), 7.92 (d, J = 8.8 Hz, 2H), 7.88 (d, J = 8.8 Hz, 2H), 7.69 (s, 1H), 7.63–7.66 (m, 2H), 7.52 (d, J = 5.1 Hz, 1H), 6.24 (d, J = 8.0 Hz, 1H), 3.38–3.47 (m, 1H), 1.73–1.78 (m, 2H), 1.66 (m, 2H), 1.51–1.56 (m, 1H), 1.22–1.31 (m, 2H), 1.10–1.21 (m, 3H); 13C NMR (150 MHz, DMSO-d6): δ 166.4, 163.0, 160.3, 159.8, 158.1, 144.1, 136.3, 135.8, 129.5 (2C), 129.2 (2C), 128.8 (2C), 125.6, 118.1 (2C), 109.1, 48.6, 33.5 (2C), 25.7, 25.1 (2C); HRMS (ESI, m/z): calcd for C24H26ClN6O2+ [M + H]+ 465.1800, found 465.1799, calcd for C24H25ClN6NaO2+ [M + Na]+ 487.1620, found 487.1619.
4.1.5.21. N-phenyl-2-(4-((4-(pyridin-4-yl)pyrimidin-2-yl)amino)benzoyl)hydrazine-1-carbothioamide (6A)
Yellow solid (97.57%), 0.054 g, yield: 66%, mp: 267–269 °C. 1H NMR (600 MHz, DMSO-d6): δ 10.40 (s, 1H), 10.19 (s, 1H), 9.79 (br s, 1H), 9.68 (br s, 1H), 8.80 (dd, J = 4.4, 1.4 Hz, 2H), 8.74 (d, J = 4.9 Hz, 1H), 8.11 (dd, J = 4.4, 1.4 Hz, 2H), 7.96 (br s, 4H), 7.63 (d, J = 4.9 Hz, 1H), 7.46 (br s, 2H), 7.33 (t, J = 7.7 Hz, 2H), 7.16 (t, J = 7.7 Hz, 1H); 13C NMR (150 MHz, DMSO-d6): δ 166.1, 162.0, 160.5, 160.4, 151.1(2C), 144.2, 139.8 (2C), 129.3 (2C), 128.4 (2C), 128.2, 126.4 (2C), 125.5(2C), 121.4, 118.4, 118.1 (2C), 109.8; HRMS (ESI, m/z): calcd for C23H20N7OS+ [M + H]+ 442.1445, found 442.1440, calcd for C23H19N7NaOS+ [M + Na]+ 464.1264, found 464.1261.
4.1.5.22. N-benzyl-2-(4-((4-(pyridin-4-yl)pyrimidin-2-yl)amino)benzoyl)hydrazine-1-carbothioamide (6B)
Yellow solid (98.33%), 0.065 g, yield: 73%, mp: 240–241 °C. 1H NMR (600 MHz, DMSO-d6): δ 10.27 (s, 1H), 10.18 (s, 1H), 9.42 (s, 1H), 8.80 (dd, J = 4.4, 1.4 Hz, 2H), 8.73 (d, J = 5.1 Hz, 1H), 8.65 (br s, 1H), 8.11 (dd, J = 4.4, 1.4 Hz, 2H), 7.91–7.97 (m, 4H), 7.62 (d, J = 5.1 Hz, 1H), 7.29– 7.33 (m, 4H), 7.20–7.24 (m, 1H), 4.75 (d, J = 6.0 Hz, 2H); 13C NMR (150 MHz, DMSO-d6): δ 166.1, 162.0, 160.5, 160.4, 151.1(2C), 144.1, 140.0 (2C), 129.2 (2C), 128.5 (2C), 127.5, 127.0 (2C), 125.5, 121.4(2C), 118.1(2C), 109.8, 47.2; HRMS (ESI, m/z): calcd for C24H22N7OS+ [M + H]+ 456.1601, found 456.1595, calcd for C24H21N7OSNa+ [M + Na]+ 478.1421, found 478.1415.
4.1.5.23. N-benzyl-2-(4-((4-(pyridin-3-yl)pyrimidin-2-yl)amino)benzoyl)hydrazine-1-carbothioamide (6C)
Yellow solid (98.60%), 0.072 g, yield: 75%, mp: 159–160 °C. 1H NMR (600 MHz, DMSO-d6): δ 10.27 (s, 1H), 10.12 (s, 1H), 9.42 (s, 1H), 9.35–9.38 (s, 1H), 8.75 (dd, J = 4.9, 1.8 Hz, 1H), 8.68 (d, J = 5.1 Hz, 1H), 8.66 (d, J = 4.9 Hz, 1H), 8.52 (td, J = 8.0, 1.8 Hz, 1H), 7.92–7.96 (br s, 4H), 7.61–7.63 (m, 1H), 7.60 (d, J = 5.1 Hz, 1H), 7.28–7.34 (m, 4H), 7.20–7.24 (m, 1H), 4.75 (d, J = 6.0 Hz, 2H); 13C NMR (150 MHz, DMSO- d6): δ 166.2, 162.2, 160.3, 160.0, 152.1, 148.7, 144.2, 140.0, 134.9, 132.5, 129.2 (2C), 128.5 (2C), 127.5, 127.0 (2C), 125.4, 124.5, 118.1 (2C), 109.5, 47.2; HRMS (ESI, m/z): calcd for C24H22N7OS+[M + H]+ 456.1601, found 456.1599, calcd for C24H21N7NaOS+ [M + Na]+ 478.1421, found 478.1419.
4.1.5.24. N-phenyl-2-(4-((4-(pyridin-2-yl)pyrimidin-2-yl)amino)benzoyl)hydrazine-1-carbothioamide (6D)
Yellow solid (99.24%), 0.046 g, yield: 48%, mp: 200–202 °C. 1H NMR (600 MHz, DMSO-d6): δ 10.39 (s, 1H), 10.13 (s, 1H), 9.80 (br s, 1H), 9.68 (s, 1H), 8.76–8.78 (d, J = 4.0 Hz, 1H), 8.72 (d, J = 5.1 Hz, 1H), 8.43 (d, J = 7.7 Hz, 1H), 8.07 (td, J = 7.7, 1.8 Hz, 1H), 7.98 (s, 4H), 7.81 (d, J = 5.1 Hz, 1H), 7.59 (m, 1H), 7.46 (d, J = 1.8 Hz, 2H), 7.34 (t, J = 7.7 Hz, 2H), 7.13–7.19 (m, 1H); 13C NMR (150 MHz, DMSO-d6): δ 166.2, 163.4, 160.2, 160.1, 153.9, 150.2, 144.3, 139.8, 138.1, 129.3 (2C), 128.4 (2C), 126.5, 126.3 (2C), 125.4, 121.6, 118.0 (2C), 109.3, 40.5; HRMS (ESI, m/z): calcd for C23H20N7OS+ [M + H]+ 442.1445, found 442.1444, calcd for C23H19N7OSNa+ [M + Na]+ 464.1264, found 464.1265.
4.1.6.25. N-benzyl-2-(4-((4-(pyridin-2-yl)pyrimidin-2-yl)amino)benzoyl)hydrazine-1-carbothioamide (6E)
White solid (99.14%), 0.045 g, yield: 67%, mp: 222–223 °C. 1H NMR (600 MHz, DMSO-d6): δ 10.27 (s, 1H), 10.12 (s, 1H), 9.42 (s, 1H), 8.76–8.78 (m, 1H), 8.71 (d, J = 4.9 Hz, 1H), 8.65 (br s, 1H), 8.42 (d, J = 7.7 Hz, 1H), 8.07 (td, J = 7.7, 1.8 Hz, 1H), 7.92–7.98 (br s, 4H), 7.80 (d, J = 4.9 Hz, 1H), 7.59 (m, 1H), 7.28–7.34 (m, 4H), 7.20–7.24 (m, 1H), 4.75 (m, 2H); 13C NMR (150 MHz, DMSO-d6): δ 166.2, 163.4, 160.2, 160.2, 153.9, 150.2, 144.3, 140.0, 138.1, 129.3 (2C), 128.5 (2C), 127.5, 127.0 (2C), 126.3, 125.3, 121.6, 118.0 (2C), 109.3, 70.2, 47.2; HRMS (ESI, m/z): calcd for C24H22N7OS+ [M + H]+ 456.1601, found 456.1599, calcd for C24H21N7NaOS+ [M + Na]+ 478.1421, found 478.1419.
4.1.6.26. N-cyclohexyl-2-(4-((4-phenylpyrimidin-2-yl)amino)benzoyl)hydrazine-1-carbothioamide (6F)
White solid (98.99%), 0.032 g, yield:59%, mp: 230–232 °C. 1H NMR (600 MHz, DMSO-d6): δ 10.10 (br s, 1H), 10.05 (s, 1H), 9.16 (br s, 1H), 8.63 (d, J = 5.1 Hz, 1H), 8.20–8.21 (m, 1H), 8.18–8.20 (m, 1H), 7.96 (d, J = 8.8 Hz, 2H), 7.92 (d, J = 8.8 Hz, 2H), 7.68 (d, J = 3.8 Hz, 1H), 7.56–7.60 (m, 3H), 7.51 (d, J = 5.1 Hz, 1H), 4.15 (br s, 1H), 1.79 (br s, 2H), 1.64–1.72 (m, 2H), 1.57 (d, J = 13.0 Hz, 1H), 1.28–1.33 (m, 2H), 1.22–1.27 (m, 2H), 1.01–1.12 (m, 1H); 13C NMR (150 MHz, DMSO-d6): δ 166.0, 164.2, 160.3, 159.6, 144.4, 137.0, 131.5 (2C), 129.5 (2C), 129.2, 127.4 (2C), 125.2, 118.0 (2C), 109.3, 53.4, 32.3 (2C), 25.6, 25.4 (2C); HRMS (ESI, m/z): calcd for C24H27N6OS+ [M + H]+ 447.1962, found 447.1959, calcd for C24H26N6NaOS+ [M + Na]+ 469.1781, found 469.1780.
4.1.5.27. N-phenyl-2-(4-((4-phenylpyrimidin-2-yl)amino)benzoyl)hydrazine-1-carbothioamide (6G)
White solid (97.80%), 0.034 g, yield: 50%, mp: 210–212 °C. 1H NMR (600 MHz, DMSO-d6): δ 10.38 (br s, 1H), 10.06 (s, 1H), 9.79 (br s, 1H), 9.67 (br s, 1H), 8.63 (d, J = 5.3 Hz, 1H), 8.19−8.21 (m, 2H), 7.98 (d, J = 8.8 Hz, 2H), 7.95 (d, J = 8.8 Hz, 2H), 7.57–7.59 (m, 3H), 7.51 (d, J = 5.3 Hz, 1H), 7.43–7.49 (m, 2H), 7.33 (t, J = 7.8 Hz, 2H), 7.13–7.19 (m, 1H); 13C NMR (150 MHz, DMSO-d6): δ 166.2, 164.2, 160.3, 159.7, 144.5, 139.8, 137.0, 131.5 (2C), 129.5 (2C), 129.3 (2C), 128.4, 127.4 (2C), 126.5 (2C), 125.5, 125.3, 118.0 (2C), 109.3; HRMS (ESI, m/z): calcd for C24H21N6OS+ [M + H]+ 441.1492, found 441.1490, calcd for C24H20N6NaOS+ [M + Na]+ 463.1312, found 463.1309.
4.1.5.28. N-benzyl-2-(4-((4-phenylpyrimidin-2-yl)amino)benzoyl)hydrazine-1-carbothioamide (6H)
White solid (99.08%), 0.054 g, yield: 62%, mp: 289–291 °C. 1H NMR (600 MHz, DMSO-d6): δ 10.26 (s, 1H), 10.05 (s, 1H), 9.41 (s, 1H), 8.65 (br s, 1H), 8.62 (d, J = 5.1 Hz, 1H), 8.17–8.22 (m, 2H), 7.97 (d, J = 8.8 Hz, 2H), 7.93 (d, J = 8.8 Hz, 2H), 7.57 (m, 3H), 7.51 (dd, J = 5.1, 1.6 Hz, 1H), 7.28–7.34 (m, 4H), 7.22 (d, J = 5.5 Hz, 1H), 4.75 (d, J = 5.5 Hz, 2H); 13C NMR (150 MHz, DMSO-d6): δ 166.2, 164.2, 160.3, 159.7, 144.4, 140.0, 136.9, 131.6 (2C), 129.5 (2C), 129.2, 128.5 (2C), 127.5, 127.4 (2C), 127.0 (3 C), 125.2, 117.9 (2C), 109.3, 47.2; HRMS (ESI, m/z): calcd for C25H23N6OS+ [M + H]+ 455.1649, found 455.1644, calcd for C25H22N6NaOS+ [M + Na]+ 477.1468, found 477.1465.
4.1.5.29. 2-(4-((4–(4-chlorophenyl)pyrimidin-2-yl)amino)benzoyl)-N-phenylhydrazine-1-carbothioamide (6I)
White solid (98.95%), 0.045 g, yield: 48%, mp: 275–277 °C. 1H NMR (600 MHz, DMSO-d6): δ 10.38 (s, 1H), 10.09 (s, 1H), 9.79 (br s, 1H), 9.68 (s, 1H), 8.65 ((d, J = 5.1, 1H), 8.22 (d, J = 8.6 Hz, 2H), 7.95 (br s, 4H), 7.65 (d, J = 8.6 Hz, 2H), 7.52 (d, J = 5.1 Hz, 1H), 7.46 (br s, 2H), 7.33 (m, 2H), 7.13–7.19 (m, 1H). 13C NMR (150 MHz, DMSO-d6): δ 163.0, 160.3, 159.9, 144.3, 139.8, 136.3, 135.8, 129.5 (2C), 129.3 (2C), 129.2 (2C), 128.4 (2C), 126.5 (2C), 125.5, 125.4, 118.0 (2C), 109.2; HRMS (ESI, m/z): calcd for C24H20ClN6OS+ [M + H]+ 475.1102, found 475.1100, calcd for C24H19ClN6NaOS+ [M + Na]+ 497.0922, found 497.0920.
4.1.5.30. N-benzyl-2-(4-((4–(4-chlorophenyl)pyrimidin-2-yl)amino)benzoyl) hydrazine-1-carbothioamide (6J)
White solid (95.91%), 0.035 g, yield: 56%, mp: 235–237 °C. 1H NMR (600 MHz, DMSO-d6): δ 10.26 (s, 1H), 10.08 (s, 1H), 9.42 (s, 1H), 8.63 (d, J = 5.1 Hz, 2H), 8.20 (d, J = 8.6 Hz, 2H), 7.91 − 7.95 (br s, 4H), 7.65 (d, J = 8.6 Hz, 2H), 7.52 (d, J = 5.1 Hz, 1H), 7.29–7.33 (m, 4H), 7.20– 7.24 (m, 1H), 4.75 (d, J = 6.0 Hz, 2H); 13C NMR (150 MHz, DMSO-d6): δ 166.2, 163.0, 160.3, 159.9, 144.3, 140.0, 136.3, 135.8, 129.5 (2C), 129.2 (2C), 128.5 (2C), 127.5 (2C), 127.0 (2C), 125.3, 118.0 (2C), 109.2, 47.2; HRMS (ESI, m/z): calcd for C25H22ClN6OS+ [M + H]+ 489.1259, found 489.1255, calcd for C25H21ClN6NaOS+ [M + Na]+ 511.1078, found 511.1075.
4.1.6. General procedure for the synthesis of compounds 7a–7f and 7A–7D (Series B)
Compounds 5A–5C (1.00 eq) were separately used to react with an appropriate isocyanate or isothiocyanate (1.00 eq) in ethanol at 70 °C for 4 h, then cooling to room temperature, filtered to get crude products of compounds 7a–7f and 7A–7D, which were further purified by silica gel column chromatography (DCM:MeOH).
4.1.6.1. N-(tert-butyl)-2-(3-((4-(pyridin-4-yl)pyrimidin-2-yl)amino)benzoyl) hydrazine-1-carboxamide (7a)
Yellow solid (98.13%), 0.076 g, yield: 83%, mp: 161–162 °C. 1H NMR (600 MHz, DMSO-d6): δ 10.09 (br s, 1H), 10.06 (s, 1H), 8.84 (d, J = 6.0 Hz, 2H), 8.75 (d, J = 5.1 Hz, 1H), 8.56 (s, 1H), 8.20 (d, J = 6.0 Hz, 2H), 7.94 (dd, J = 8.0, 1.4 Hz, 1H), 7.72 (s, 1H), 7.64 (d, J = 5.1 Hz, 1H), 7.54 (d, J = 7.7 Hz, 1H), 7.48 (t, J = 7.7 Hz, 1H), 6.04–6.20 (m, 1H), 1.33 (s, 9H); 13C NMR (150 MHz, DMSO-d6): δ 167.0, 161.8, 160.7, 160.5, 157.7, 151.1, 144.2 (2C), 140.9, 133.9, 129.0, 122.5, 121.5 (2C), 120.7, 119.0, 109.3, 50.0, 29.6 (3 C); HRMS (ESI, m/z): calcd for C21H24N7O2+ [M + H]+ 406.1986, found 406.1982, calcd for C21H23N7NaO2+ [M + Na]+ 428.1805, found 428.1801.
4.1.6.2. N-cyclopentyl-2-(3-((4-(pyridin-4-yl)pyrimidin-2-yl)amino)benzoyl) hydrazine-1-carboxamide (7b)
Yellow solid (99.30%), 0.076 g, Yield: 79%, mp: 221–223 °C. 1H NMR (600 MHz, DMSO-d6): δ 10.05 (s, 1H), 10.01 (s, 1H), 8.79 (d, J = 6.0 Hz, 2H), 8.70 (d, J = 5.1 Hz, 1H), 8.50 (s, 1H), 8.15 (d, J = 6.0 Hz, 2H), 7.91 (dd, J = 7.7, 1.5 Hz, 1H), 7.75 (s, 1H), 7.58 (d, J = 5.1 Hz, 1H), 7.51 (d, J = 7.7 Hz, 1H), 7.40–7.45 (J = 7.7 Hz, 1H), 6.36 (m, 1H), 3.93 (m, 1H), 1.78–1.85 (m, 2H), 1.58–1.67 (m, 2H), 1.47–1.55 (m, 2H), 1.35–1.43 (m, 2H); 13C NMR (150 MHz, DMSO-d6): δ 167.2, 161.8, 160.6, 160.5, 158.4, 151.0, 144.2 (2C), 140.9, 133.9, 129.0, 122.5, 121.4 (2C), 120.8, 119.0, 109.3, 51.6, 33.1 (2C), 23.7 (2C); HRMS (ESI, m/z): calcd for C22H24N7O2+ [M + H]+ 418.1986, found 418.1983, calcd for C22H13N7NaO+ [M + Na]+ 440.1805, found 440.1803.
4.1.6.3. N-(tert-butyl)-2-(3-((4-(pyridin-3-yl)pyrimidin-2-yl)amino)benzoyl) hydrazine-1-carboxamide (7c)
White solid (97.12%), 0.079 g, yield: 81%, mp: 206–207 °C. 1H NMR (600 MHz, DMSO-d6): δ 10.04 (br s 1H), 9.94 (s, 1H), 9.38 (d, J = 1.6 Hz, 1H), 8.74 (dd, J = 4.8, 1.8 Hz, 1H), 8.64 (d, J = 5.1 Hz, 1H), 8.57 (dt, J = 7.9, 1.8 Hz, 1H), 8.45 (s, 1H), 7.93 (dd, J = 7.9, 1.8 Hz, 1H), 7.66 (s, 1H), 7.59 (dd, J = 7.9, 4.8 Hz, 1H), 7.56 (d, J = 5.1 Hz, 1H), 7.49 (d, J = 7.7 Hz, 1H), 7.40–7.43 (t, J = 7.7 Hz 1H), 6.08 (br s, 1H), 1.28 (s, 9H); 13C NMR (150 MHz, DMSO-d6): δ 166.9, 162.1, 160.6, 159.9, 157.7, 152.0, 148.7, 141.0, 135.0, 133.8, 132.6, 128.9, 124.4, 122.4, 120.6, 119.0, 108.9, 49.9, 29.6 (3 C); HRMS (ESI, m/z): calcd for C21H24N7O2+ [M + H]+ 406.1986, found 406.1982, calcd for C21H23N7NaO2+ [M + Na]+ 428.1805, found 428.1801.
4.1.6.4. N-butyl-2-(3-((4-(pyridin-3-yl)pyrimidin-2-yl)amino)benzoyl)hydrazine-1-carboxamide (7d)
Yellow solid (99.36%), 0.066 g, Yield: 79%, mp: 196–198 °C. 1H NMR (600 MHz, DMSO-d6): δ 10.05 (s, 1H), 9.94 (s, 1H), 9.38 (d, J = 1.9 Hz, 1H), 8.74 (dd, J = 4.7, 1.4 Hz, 1H), 8.64 (d, J = 5.1 Hz, 1H), 8.57 (dt, J = 8.0, 1.9 Hz, 1H), 8.45 (s, 1H), 7.94 (dd, J = 8.0, 1.4 Hz, 1H), 7.82 (s, 1H), 7.59 (dd, J = 8.0, 4.7 Hz, 1H), 7.56 (d, J = 5.1 Hz, 1H), 7.52 (d, J = 7.7 Hz, 1H), 7.42 (t, J = 7.7 Hz, 1H), 6.42 (t, J = 7.7 Hz, 1H), 3.04 (q, J = 6.7 Hz, 2H), 1.39 (quin, J = 7.3 Hz, 2H), 1.28 (sxt, J = 7.3 Hz, 2H), 0.88 (t, J = 7.3 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 167.1, 162.1, 160.6, 159.9, 158.8, 152.0, 148.7, 141.0, 135.0, 133.9, 132.6, 128.8, 124.4, 122.5, 120.8, 119.1, 108.9, 39.4, 32.5, 19.9, 14.2; HRMS (ESI, m/z): calcd for C21H24N7O2+ [M + H]+ 406.1986, found 406.1980, calcd for C21H23N7NaO2+ [M + Na]+ 428.1805, found 428.1802.
4.1.6.5. N-(2,4-dimethoxyphenyl)-2-(3-((4-(pyridin-3-yl)pyrimidin-2-yl)amino) benzoyl)hydrazine-1-carboxamide (7e)
White solid (94.02%), 0.032 g, yield: 55%, mp: 231–232 °C. 1H NMR (600 MHz, DMSO- d6): δ 10.28 (br s, 1H), 9.97 (s, 1H), 9.38 (d, J = 1.8 Hz, 1H), 8.73 (dd, J = 4.7, 1.4 Hz, 1H), 8.64 (d, J = 5.1 Hz, 1H), 8.61 (s, 1H), 8.57 (dt, J = 8.0, 1.8 Hz, 1H), 8.48 (s, 1H), 8.03 (br s, 1H), 7.95 (dd, J = 8.0, 1.4 Hz, 1H), 7.88 (d, J = 8.8 Hz, 1H), 7.57–7.59 (m, 1H), 7.55–7.57 (m, 1H), 7.49–7.52 (m, 1H), 7.43–7.47 (m, 1H), 6.62 (d, J = 2.5 Hz, 1H), 6.48 (dd, J = 8.8, 2.5 Hz, 1H), 3.84 (s, 3H), 3.73 (s, 3H); 13C NMR (150 MHz, DMSO- d6): δ 167.3, 162.1, 160.6, 160.0, 156.0, 155.4, 152.0, 149.6, 148.7, 141.1, 135.0, 133.7, 132.6, 129.0, 124.4, 122.6, 122.2, 120.6, 120.0, 118.9, 109.0, 104.6, 99.2, 56.3, 55.7; HRMS (ESI, m/z): calcd for C25H24N7O4+ [M + H]+ 486.1884, found 486.1883, calcd for C25H23N7NaO4+ [M + Na]+ 508.1704, found 508.1703.
4.1.6.6. N-butyl-2-(3-((4-(pyridin-4-yl)pyrimidin-2-yl)amino)benzoyl)hydrazine-1-carboxamide (7f)
Yellow solid (98.48%), 0.043 g, yield: 80%, mp: 183–184 °C. 1H NMR (600 MHz, DMSO-d6): δ 10.05 (s, 1H), 9.94 (s, 1H), 8.74–8.77 (m, 1H), 8.68 (d, J = 4.9 Hz, 1H), 8.53 (s, 1H), 8.51 (d, J = 7.9 Hz, 1H), 8.04 (td, J = 7.9, 1.7 Hz, 1H), 7.92 (dd, J = 7.9, 1.7 Hz, 1H), 7.83 (s, 1H), 7.77 (d, J = 5.1 Hz, 1H), 7.58 (m, 1H), 7.52 (d, J = 7.9 Hz, 1H), 7.42 (t, J = 7.9 Hz, 1H), 6.43 (t, J = 5.1 Hz, 1H), 3.05 (q, J = 6.8 Hz, 2H), 1.39 (quin, J = 7.3 Hz, 2H), 1.29 (sxt, J = 7.3 Hz, 2H), 0.88 (t, J = 7.3 Hz, 3H); 13C NMR (150 MHz, DMSO-d6): δ 167.2, 163.3, 160.5, 160.1, 158.8, 154.0, 150.1, 141.1, 138.1, 133.9, 128.9, 126.2, 122.4, 121.8, 120.7, 119.0, 108.7, 39.4, 32.5, 19.9, 14.2;HRMS (ESI, m/z): calcd for C21H24N7O2+ [M + H]+ 406.1986, found 406.1980, calcd for C21H23N7NaO2+ [M + Na]+ 428.1805, found 428.1802.
4.1.6.7. N-benzyl-2-(3-((4-(pyridin-4-yl)pyrimidin-2-yl)amino)benzoyl)hydrazine-1-carbothioamide (7A)
Yellow solid (98.66%), 0.042 g, yield: 53%, mp: 218–220 °C. 1H NMR (600 MHz, DMSO- d6): δ 10.40 (s, 1H), 10.01 (s, 1H), 9.51 (s, 1H), 8.79 (d, J = 6.0 Hz, 2H), 8.69 (d, J = 5.1 Hz, 2H), 8.54 (br s, 1H), 8.15 (d, J = 6.0 Hz, 2H), 7.94 (dd, J = 1.2, 8.0 Hz, 1H), 7.57–7.61 (m, 2H), 7.44 (t, J = 7.8 Hz, 1H), 7.31–7.34 (m, 2H), 7.27– 7.31 (m, 2H), 7.19–7.23 (m, 1H), 4.76 (d, J = 5.8 Hz, 2H); 13C NMR (150 MHz, DMSO- d6): δ 166.8, 161.8, 160.6, 160.5, 151.0, 144.2 (2C), 140.9, 139.9, 133.5, 128.8 (2C), 128.5, 127.5, 127.0 (2C), 122.8, 121.4, 121.3 (2C), 119.4, 109.3, 47.2; HRMS (ESI, m/z): calcd for C24H22N7OS+ [M + H]+ 456.1601, found 456.1599, calcd for C24H21N7NaOS+ [M + Na]+ 478.1421, found 478.1418.
4.1.6.8. N-benzyl-2-(3-((4-(pyridin-3-yl)pyrimidin-2-yl)amino)benzoyl)hydrazine-1-carbothioamide (7B)
White solid (96.84%), 0.055 g, yield: 58%; mp: 212–213 °C . 1H NMR (600 MHz, DMSO- d6): δ 10.38 (s, 1H), 9.95 (s, 1H), 9.49 (s, 1H), 9.38 (d, J = 1.8 Hz, 1H), 8.74 (dd, J = 4.7, 1.6 Hz, 1H), 8.66 (br s, 1H), 8.63 (d, J = 5.1 Hz, 1H), 8.57 (dt, J = 8.0, 1.8 Hz, 1H), 8.50 (s, 1H), 7.95 (dd, J = 8.0, 1.6 Hz, 1H), 7.57–7.60 (m, 2H), 7.56 (d, J = 5.1 Hz, 1H), 7.43 (t, J = 8.0 Hz, 1H), 7.27–7.34 (m, 4H), 7.19–7.23 (m, 1H), 4.75 (d, J = 5.8 Hz, 2H); 13C NMR (150 MHz, DMSO- d6): δ 166.7, 162.1, 160.6, 160.0, 152.1, 148.7, 141.0, 139.9, 135.0, 133.5, 132.5, 128.7, 128.5, 127.5 (2C), 127.0 (2C), 124.4, 122.7, 121.1, 119.3, 108.9, 47.2; HRMS (ESI, m/z): calcd for C24H22N7OS+ [M + H] + 456.1601, found 456.1599, calcd for C24H21N7NaOS+[M + Na]+ 478.1421, found 478.1418.
4.1.6.9. N-phenyl-2-(3-((4-(pyridin-2-yl)pyrimidin-2-yl)amino)benzoyl)hydrazine-1-carbothioamide (7C)
Yellow solid (97.88%), 0.043 g, Yield: 59%, mp: 202–203 °C . 1H NMR (600 MHz, DMSO-d6): δ 10.51 (s, 1H), 9.93–10.00 (s, 1H), 9.83 (br s, 1H), 9.75 (br s, 1H), 8.75 (m, 1H), 8.68 (d, J = 5.0 Hz, 1H), 8.60 (br s, 1H), 8.51 (d, J = 7.8 Hz, 1H), 8.00 (t, J = 7.8 Hz, 1H), 7.94 (d, J = 8.0 Hz, 1H), 7.77 (d, J = 5.0 Hz, 1H), 7.62 (d, J = 6.9 Hz, 1H), 7.56 (dd, J = 6.9, 5.0 Hz, 1H), 7.42–7.50 (m, 3H), 7.34 (t, J = 7.3 Hz, 2H), 7.14–7.19 (t, J = 7.3 Hz, 1H); 13C NMR (150 MHz, DMSO-d6): δ 166.7, 163.3, 160.5, 160.2, 154.0, 150.1, 141.1, 139.8, 138.1, 133.6, 128.8, 128.4 (2C), 126.2, 125.5 (2C), 122.6, 121.8, 121.2, 119.2, 108.7; HRMS (ESI, m/z): calcd for C23H20N7OS+ [M + H]+ 442.1445, found 442.1445, calcd for C23H19N7NaOS+ [M + Na]+ 464.1264, found 464.1261.
4.1.6.10. N-benzyl-2-(3-((4-(pyridin-2-yl)pyrimidin-2-yl)amino)benzoyl)hydrazine-1-carbothioamide (7D)
Yellow solid (98.95%), 0.052 g, yield: 56%, mp: 203–205 °C . 1H NMR (600 MHz, DMSO-d6): δ 10.40 (s, 1H), 9.95 (s, 1H), 9.50 (s, 1H), 8.75 (d, J = 4.5 Hz, 1H), 8.68 (t, J = 5.1 Hz, 1H), 8.67 (d, J = 5.1 Hz, 1H), 8.59 (br s, 1H), 8.52 (d, J = 7.8 Hz, 1H), 8.00 (dt, J = 8.0, 1.6 Hz, 1H), 7.93 (dd, J = 8.0, 1.6 Hz, 1H), 7.77 (d, J = 5.1 Hz, 1H), 7.56–7.60 (m, 2H), 7.44 (t, J = 7.8 Hz, 1H), 7.32 (t, J = 7.3 Hz, 2H), 7.29 (t, J = 7.3 Hz, 2H), 7.22 (t, J = 7.3 Hz, 1H), 4.76 (d, J = 5.8 Hz, 2H); 13C NMR (150 MHz, DMSO-d6): δ 166.8, 163.3, 160.4, 160.2, 154.0, 150.1, 141.1, 139.9, 138.1, 133.5, 128.8, 128.5 (2C), 127.5, 127.0 (3 C), 126.3, 122.6, 121.8, 121.1, 119.2, 108.7, 47.2; HRMS (ESI, m/z): calcd for C24H22N7OS+ [M + H]+ 456.1601, found 456.1599, calcd for C24H21N7NaOS+ [M + Na]+ 478.1421, found 478.1418.
4.2. Bioassay
All the human cancer cell lines were purchased from the American type culture collection (ATCC, Shanghai, China). Goat antirabbit and anti-mouse secondary antibody conjugated to horseradish peroxidase (HRP) was obtained from Thermo Fisher Scientific (Waltham, MA), anti-rabbit IgG conjugated with Cy3 from Chemicon International (Temecula, CA), anti-RXRα (D-20, sc-553) from Santa Cruz Biotechnology (Santa Cruz, CA), anti-PARP (46D11, 9542) and anti-Clevage Caspase 3 (Asp175, 9661S) from Cell Signalling Technology (Boston, MA), anti-α-tublin antibody from Proteintech (Rosemont, IL), and JC-1 Probe (Cat. T3168) from Thermo Fisher Scientific.
4.2.1. Cell culture and transfection
SW620, LS174T, HepG2, and A549 were maintained in Dulbecco’s Modified Eagles Medium supplemented with 10% foetal bovine serum (FBS), HeLa was maintained in Modified Eagles Medium supplemented with 10% FBS and were cultured at 37 °C, under 90% humidity with 5% carbon-di-oxide (CO2).
4.2.2. Antiproliferative activity assay
The logarithmic phase of HepG2 and A549 cells were seeded in 96-well plates at about 1 × 104 cells per well. treated with various concentrations of compounds for 24 h. then 15 μL MTT was added and incubated for 4 h at 37°C and 5% CO2. the light intensity was measured by microplate reader (PerkinElmer, Waltham, MA) at a wavelength of 490 nm. Experiments were repeated three times. The results were analysed by GraphPad Prism 5.01 software, La Jolla, CA.
4.2.3. Luciferase reporter gene assay
HEK293T cells were cotransfected with pG5-luc reporter (50.0 ng) and Pbind-RXRα-LBD (50.0 ng). One day after transfection, cells were treated with DMSO, compounds (1 μM) or 9-cis-RA (0.1 μM). After 12 h, cells were lysed and quantitated using the Dual-Luciferase Reporter Assay System (Thermo Fisher Scientific). Transfection and expression efficiency were normalised to Renilla luciferase activity.
4.2.4. RXRE-mediated transcription assay
HEK293T cells were seeded into 48-well plates at 40% confluence and cotransfected with RXRE luciferase reporter gene (100.0 ng), Myc-RXRα (50.0 ng), and Renilla control plasmid (1.0 ng). After 24 h post transfection, the compounds or vehicle were added. Cells were harvested after 12 h of incubation with ligands and then quantitated using the Dual-Luciferase Reporter Assay System (Promega). Luminescence of firefly luciferase values was normalised to the Renilla luciferase activity.
4.2.5. EC50 determination
The EC50 values were determined from full dose-response curves for compounds tested ranging from 0.10 to 20.0 μM. HEK293T cells were plated, transfected, and treated as above using Luciferase Reporter Gene Assay. Then cells were treated with DMSO vehicle (0.1%) or compounds tested (final concentration of 0.1, 0.3125, 0.625, 1.25, 2.5, 5.0, 10.0, and 20.0 μM) for 24 h. The amount of rexinoid activity at each concentration was measured using the same luciferase assay described above and EC50 values were derived from dose-response curves of ligand concentration versus normalised luciferase activity using Graph Pad Prism 5 software.
4.2.6. Western blot analysis
Equal amounts of cell lysates were boiled in sodium dodecyl sulphate (SDS) sample loading buffer, then separated through 10% SDS-polyacrylamide gel electrophoresis and electro transferred to nitrocellulose membranes. The membranes were blocked with 5% non-fat milk in 10 mM TriseHCl (pH 8.0), 150 mM NaCl, 0.05% Tween 20 (TBST) buffer for 1 h at room temperature. The membranes were incubated overnight at 4 °C, with specific primary antibodies, after washing, followed by incubated with HRP-conjugated anti-mouse or anti-rabbit secondary antibodies, immunoreactive products were visualised with a chemiluminescence solution.
4.2.7. Generation of RXRα knock-out cell by CRISPR/Cas9 system
For RXRα genes, sgRNAs were designed based on the original Cas9 approach. Then RXRα sgRNA were colonised into gRNA cloning vector PX330 (add gene 71707) and confirmed by sequencing. The cells were infected with control vector and gRNA expression vectors for 24 h and selected single colonies using G418 (0.5 mg/mL). The knockout cell was determined using western blot analysis
4.2.8. SPR
BIAcore-T200 machine (GE Healthcare) was used to measure the binding kinetics of compound 6A with RXRα-LBD, ERα-LBD, PPARγ-LBD, and TR3-LBD. Purified RXRα-LBD, ERα-LBD, PPARγ-LBD, and TR3-LBD were respectively immobilised on the CM5 chips according to the manufacturer’s instructions. Then various concentrations of 6A was injected into the flow cells in running buffer (phosphate buffer saline (PBS), 0.4% DMSO) at a flow rate of 30.0 μL/min, when all cycle were collected, The results were analysed using Biacore T200 Evaluation Software.
4.2.9. Analysis of mitochondrial membrane potential
For detection of apoptosis by FACS, A549 cells were incubated with 6A at 3.0 μM for 12 h. After treatment, the cells were harvested and washed with PBS followed by stained with JC-1 for 30 min at room temperature in the dark, Flow-cytometric analysis was performed using FACS.
4.2.10. Immunofluorescence microscopy
A549 cells were seeded on glass cover slips, incubated for 12 h in the presence of compound 6A at a final concentration of 3.0 μM. Cells mounted on glass slides were fixed in 4% paraformaldehyde and permeabilised with PBS containing 0.1% Triton X-100 and 0.10 mol/l glycine for 10 min, and then blocked with 1% bovine serum albumin in PBS for 30 min at room temperature, followed with incubation with cleaved-caspase 3 antibodies (1:100) at room temperature for 2 h, and detected by anti-rabbit IgG conjugated with Cy3 (1:200) at room temperature for 2 h. Cells were contained with 4’6′-diamidino-2-phenylindole (DAPI) (1:10,000) to visualise nuclei. The images were analysed by using MRC-1024 MP laser-scanning confocal microscope (Leica).
4.3. Molecular docking studies
The molecular docking studies were performed with the Schrodinger 2015.1 GlideCitation26. The crystal structure of RXRα (PDB code: 3A9E) were downloaded from the RCSB protein data bank and prepared with Protein Preparation Wizard panel as implemented in Maestro 10.5: water molecules were removed, missing hydrogens and residues were added. The glide grid centre was setting according the geometrical centre of original inhibitor (LG100754) and the grid size was 20 × 20 × 20 Å3. Compound 6A were prepared with LigPrep panel with OPLS2005 force field, generating the protonation and tautomeric states at 7 ± 2 pH unitsCitation27. Compound 6A was docked into the RXRα binding site using Glide in XP mode. All of docking results were processed maestro 10.1 and PyMOL software.
Supplemental Material
Download Zip (7 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Huang P, Chandra V, Rastinejad F. Retinoic acid actions through mammalian nuclear receptors. Chem Rev 2014;114:233–54.
- Evans RM, Mangelsdorf DJ. Nuclear receptors, RXR, and the big bang. Cell 2014;157:255–66.
- Chambon P. The nuclear receptor superfamily: a personal retrospect on the first two decades. Mol Endocrinol 2005;19:1418–28.
- Bourguet W, Vivat V, Wurtz JM, et al. Crystal structure of a heterodimeric complex of RAR and RXR ligand-binding domains. Mol Cell 2000;5:289–98.
- Shulman AI, Larson C, Mangelsdorf DJ, Ranganathan R. Structural determinants of allosteric ligand activation in RXR heterodimers. Cell 2004;116:417–29.
- Chandra V, Huang PX, Hamuro Y, et al. Structure of the intact PPAR-gamma-RXR-alpha nuclear receptor complex on DNA. Nature 2008;456:350–U333.
- Cesario RM, Stone J, Yen WC, et al. Differentiation and growth inhibition mediated via the rxr: PPAR-gamma heterodimer in colon cancer. Cancer Lett 2006;240:225–33.
- Szanto A, Narkar V, Shen Q, et al. Retinoid x receptors: X-ploring their (patho)physiological functions. Cell Death Differ 2004;11: S126–S143.
- Germain P, Iyer J, Zechel C, Gronemeyer H. Co-regulator recruitment and the mechanism of retinoic acid receptor synergy. Nature 2002;415:187–92.
- Altucci L, Leibowitz MD, Ogilvie KM, et al. RAR and RXR modulation in cancer and metabolic disease. Nat Rev Drug Discov 2007;6:793–810.
- Gianni M, Ponzanelli I, Mologni L, et al. Retinoid-dependent growth inhibition, differentiation and apoptosis in acute promyelocytic leukemia cells. Expression and activation of caspases. Cell Death Differ 2000;7:447–60.
- Alvarez R, Vaz B, Gronemeyer H, de Lera AR. Functions, therapeutic applications, and synthesis of retinoids and carotenoids. Chem Rev 2014;114:1–125.
- Tanaka T, Suh KS, Lo AM, De Luca LM. P21waf1/cip1 is a common transcriptional target of retinoid receptors: Pleiotropic regulatory mechanism through retinoic acid receptor (rar)/retinoid x receptor (RXR) heterodimer and RXR/RXR homodimer. J Biol Chem 2007;282:29987–97.
- Park EJ, Kondratyuk TP, Morrell A, et al. Induction of retinoid X receptor activity and consequent upregulation of p21WAF1/CIP1 by indenoisoquinolines in MCF7 cells. Cancer Prev Res 2011;4:592–607.
- Cao X, Liu W, Lin F, et al. Retinoid X receptor regulates NUR77/TR3-dependent apoptosis [corrected] by modulating its nuclear export and mitochondrial targeting. Mol Cell Biol 2004;24:9705–25.
- Tunctan B, Kucukkavruk SP, Temiz-Resitoglu M, et al. Bexarotene, a selective RXR alpha agonist, reverses hypotension associated with inflammation and tissue injury in a rat model of septic shock. Inflammation 2018;41:337–55.
- Zhou H, Liu W, Su Y, et al. NSAID sulindac and its analog bind RXR alpha and inhibit RXR alpha-dependent AKT signaling. Cancer Cell 2010;17:560–73.
- Wang GH, Jiang FQ, Duan YH, et al. Targeting truncated retinoid X receptor-alpha by CF31 induces TNF-alpha-dependent apoptosis. Cancer Res 2013;73:307–18.
- Heck MC, Wagner CE, Shahani PH, et al. Modeling, synthesis, and biological evaluation of potential retinoid X receptor (RXR)-selective agonists: analogues of 4-[1-(3,5,5,8,8-pentamethyl-5,6,7,8-tetrahydro-2-naphthyl)ethynyl]benzoic acid (bexarotene) and 6-(ethyl(5,5,8,8-tetrahydronaphthalen-2-yl)amino)nicotinic acid (net-TMN). J Med Chem 2016;59:8924–40.
- Xu D, Guo S, Chen Z, et al. Binding characterization, synthesis and biological evaluation of RXRalpha antagonists targeting the coactivator binding site. Bioorg Med Chem Lett 2016;26:3846–9.
- de Lera AR, Bourguet W, Altucci L, Gronemeyer H. Design of selective nuclear receptor modulators: RAR and RXR as a case study. Nat Rev Drug Discov 2007;6:811–20.
- Boehm MF, Zhang L, Zhi L, et al. Design and synthesis of potent retinoid X receptor selective ligands that induce apoptosis in leukemia cells. J Med Chem 1995;38:3146–55.
- Gottardis MM, Bischoff ED, Shirley MA, et al. Chemoprevention of mammary carcinoma by lgd1069 (targretin): An RXR-selective ligand. Cancer Res 1996;56:5566–70.
- de Vries-van der Weij J, de Haan W, Hu L, et al. Bexarotene induces dyslipidemia by increased very low-density lipoprotein production and cholesteryl ester transfer protein-mediated reduction of high-density lipoprotein. Endocrinology 2009;150:2368–75.
- Sato Y, Ramalanjaona N, Huet T, et al. The “phantom effect” of the rexinoid lg100754: structural and functional insights. PLoS One 2010;5:e15119.
- Schrödinger Release 2015-1: LigPrep. New York (NY): Schrödinger, LLC; 2015.
- Schrödinger Release 2015-1: Glide. New York (NY): Schrödinger, LLC; 2015.