?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
The β-carbonic anhydrase (CA, EC 4.2.1.1) from the genome of the opportunistic pathogen Malassezia restricta (MreCA), which was recently cloned and characterised, herein has been investigated for enzymatic activation by a panel of amines and amino acids. Of the 24 compounds tested in this study, the most effective MreCA activators were L-adrenaline (KA of 15 nM), 2-aminoethyl-piperazine/morpholine (KAs of 0.25–0.33 µM), histamine, L-4-amino-phenylalanine, D-Phe, L-/D-DOPA, and L-/D-Trp (KAs of 0.32 − 0.90 µM). The least effective activators were L-/D-Tyr, L-Asp, L-/D-Glu, and L-His, with activation constants ranging between 4.04 and 12.8 µM. As MreCA is involved in dandruff and seborrhoeic dermatitis, these results are of interest to identify modulators of the activity of enzymes involved in the metabolic processes of such fungi.
1. Introduction
Carbonic anhydrases (CAs; EC 4.2.1.1) are present in most organisms investigated to dateCitation1–5, with eight genetically distinct classes of such enzymes, the α-, β-, γ-, δ-, ζ-, η-, θ-, and ι-CA classes being encoded in the genome of various organismsCitation6–8. They all catalyse the simple but fundamental interconversion reaction between carbon dioxide and bicarbonate, with the comcomitant generation of hydronium ions:
α-CAs are Zn2+ metalloproteins expressed in vertebrates, fungi, protozoa, algae, plants and prokaryotesCitation4–9. The β-CAs are also Zn2+ enzymes and they are present in bacteria, fungi, protozoa and chloroplasts of mono-/dicotyledon plantsCitation4,Citation5. γ-CAs are probably Zn2+ or Fe2+ enzymes, although it has been shown they are also active with Co2+ within their active site, and are present in archaea, bacteria and plantsCitation3,Citation4. Limited information is known about δ-CAs, which are zinc- or cobalt-containing enzymes present in marine diatomsCitation7,Citation10. The ζ-CAs are active with Cd2+ or Zn2+ at their active site, and were also identified in marine diatomsCitation11. η-CA are Zn2+ metalloproteins identified in Plasmodium spp. and other protozoansCitation12. The recently identified θ- and ι-CAs are also present in marine diatomsCitation11,Citation13, and the latter class is also expressed in bacteria and are likely Mn(II) metalloenzymes, as recently reportedCitation13.
Various classes of inhibitors of these enzymes, mainly targeting mammalian CAs, are in clinical use as diuretics, antiglaucoma, antiepileptic or antiobesity agents for decades, whereas their use as anticancer agents started to be contemplated only in the last decadeCitation1,Citation2,Citation6,Citation14–18. There has also been recent interest in inhibiting CAs in various pathogenic bacteria to develop anti-infective applicationsCitation6–8. These diverse applications are due to the fact that at least 15 different α-CA isoforms are present in humans, being involved in critical physiological and pathological processesCitation14–18.
Activation studies of various classes of CAs, among which the β-, γ-, δ-, ζ-, and η-CA classes were explored only recently, and only with two classes of modulators of activity, the amines and the amino acidsCitation3,Citation19. The catalytic mechanism of these enzymes is also well understood and explains also their activation mechanismCitation3. A metal hydroxide species present in the active site of these enzymes acts as a strong nucleophile (at physiologic pH) for converting the CO2 to bicarbonate, which is thereafter coordinated to the catalytic metal ion. This adduct is not very stable and its reaction with an incoming water molecule leads to the liberation of bicarbonate in solution and generation of an acidic form of the enzyme incorporating an M2+(OH2) species at the metal centre, which is catalytically ineffective for the hydration of CO2Citation1–3. To generate the nucleophilic M2+(OH–) species, a proton transfer reaction occurs, which determines the rate for the catalytic cycle in many of these types of very efficient enzymes. For many α-CAs this step is assisted by a proton shuttle residue, which is His64 in most mammalian isoformsCitation20. This is one of the few residues in α-CAs possessing a flexible conformation, with an inward (in conformation) and outward (out) conformation. For this reason, the imidazole moiety of this histidine, with a pKa of 6.0–7.5 (depending on the isoformCitation3) is an appropriate proton shuttling residue which transfers the proton from the metal coordinated water to the reaction medium, in a crucially important and rate-determining step of the catalytic cycleCitation1–3. The process can also be assisted by endogenous molecules, which bind within the enzyme active site, as proven by X-ray crystallography and other techniques, which have been termed CA activators (CAAs)Citation19. Such activators facilitate the proton transfer reactions between the metal ion centre and the external medium by an alternative pathway to the proton shuttle residue. The two reactions of the CA catalytic cycle are shown by EquationEquations (1)(1)
(1) and Equation(2)
(2)
(2) , with the deprotonation of zinc-bound water being the rate-determining step (Equation (2)Citation19,Citation21. This leads to the generation of the active form of the enzymeCitation3,Citation19,Citation22:
(1)
(1)
(2)
(2)
In the presence of an activator molecule “A”, EquationEquation (2)(2)
(2) becomes EquationEquation (3)
(3)
(3) ; that is, in the enzyme-activator complex the proton transfer reaction is no longer intermolecular but intramolecular, and thus favouredCitation3,Citation19:
(3)
(3)
Enzyme–activator complexes
CAAs were recently demonstrated to have potential pharmacologic applicationsCitation23, as the activation of mammalian enzymes was shown to enhance cognition and memory in experimental animalsCitation23a,b, whereas its inhibition had the opposite effectCitation14.
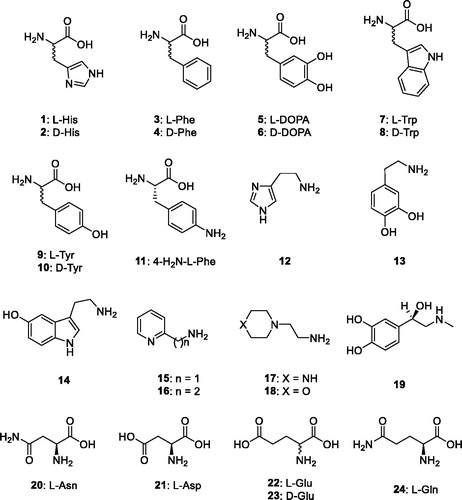
The activation of CAs from pathogenic bacteria may also be relevant for understanding the factors governing virulence and colonisation of the host, because pH in the tissues surrounding the pathogens likely plays a key role in such processesCitation3,Citation5 and many compounds that are CAAs (biogenic amines and amino acid derivatives) are abundant in such tissues. Considering such evidence, in this study we report an activation study with amines and amino acids (compounds 1–24, ) of the β-CA recently reported and characterised biochemically from the dandruff producing organism Malassezia restrictaCitation24.
2. Materials and methods
2.1. Enzymes production and purification
The protocol described in the previous worksCitation24 has been used to obtain purified recombinant MreCA.
2.2. Ca activity/activation measurements
An Sx.18Mv-R Applied Photophysics (Oxford, UK) stopped-flow instrument has been used to assay the catalytic activity of various CA isozymes for CO2 hydration reactionCitation25. Phenol red (at a concentration of 0.2 mM) was used as indicator, working at the absorbance maximum of 557 nm, with 10 mM Hepes (pH 7.5, for α-CAs)Citation26–29 or TRIS (pH 8.3, for β-CAs)Citation30–33 as buffers, 0.1 M NaClO4 (for maintaining constant ionic strength), following the CA-catalyzed CO2 hydration reaction for a period of 10 s at 25 °C. The CO2 concentrations ranged from 1.7 to 17 mM for the determination of the kinetic parameters and inhibition constants. For each activator at least six traces of the initial 5–10% of the reaction have been used for determining the initial velocity. The uncatalyzed rates were determined in the same manner and subtracted from the total observed rates. Stock solutions of activators (at 0.1 mM) were prepared in distilled-deionized water and dilutions up to 1 nM were made thereafter with the assay buffer. Enzyme and activator solutions were pre-incubated together for 15 min prior to assay, in order to allow for the formation of the enzyme–activator complexes. The activation constant (KA), defined similarly with the inhibition constant KI, can be obtained by considering the classical Michaelis–Menten equation (EquationEquation (4)(4)
(4) , which has been fitted by non-linear least squares by using PRISM 3:
(4)
(4)
where [A]f is the free concentration of activator.
Working at substrate concentrations considerably lower than KM ([S] ≪KM), and considering that [A]f can be represented in the form of the total concentration of the enzyme ([E]t) and activator ([A]t), the obtained competitive steady-state equation for determining the activation constant is given by EquationEquation (5)(5)
(5) :
(5)
(5)
where v0 represents the initial velocity of the enzyme-catalyzed reaction in the absence of activatorCitation3,Citation30–35. This type of approach to measuring enzyme-ligand interactions is in excellent agreement with recent results from native mass spectrometry measurementsCitation36.
2.3. Reagents
Amines and amino acid derivatives 1–24 were obtained in the highest purity that was available commercially from Sigma-Aldrich (Milan, Italy).
3. Results and discussion
We measured the kinetics constants (kcat and KM) of the recently described β-CA from M. restricta, MreCA, for comparison to those of the thoroughly studied human (h) CA isoforms hCA I and II, belonging to the α-CA class (). The experiments were also performed in the presence of 10 μM L-Trp as activator.
Table 1. Activation of hCA I, II and MreCA with L-Trp, at 25 °C, for the CO2 hydration reactionCitation25.
The data in indicates that the presence of L-Trp does not change the KM both for the two enzymes belonging to the α-class (hCA I/II) as well as for MreCA, a situation also observed for all CA classes for which CA activators have been investigated so farCitation3,Citation29–33. In fact, as proven by kinetic and crystallographic dataCitation3,Citation20, the activator binds in a diverse binding region within the active site of the substrate binding site. Thus, the activator does not influence KM but has an effect only on the kcat. Indeed, a 10 µM concentration of L-Trp leads to a 9-fold enhancement of the kinetic constant of MreCA compared to the same parameter in the absence of the activator (). For hCA I and II, the enhancement of the kinetic constant in the presence of L-Trp was rather modest, as these enzymes show a weaker affinity for this activator (). On the other hand, L-Trp has a submicromolar affinity for MreCa which explains its potent activating effect (see discussion later in the text).
Thus, an entire range of amines and amino acids, of types 1–24, were tested for their efficacy as MreCA activators (). These compounds were also investigated earlierCitation3 for their activating properties against hCAs and many enzymes from pathogenic organisms, as reported previouslyCitation26–33. The following structure–activity relationship (SAR) for the activation of MreCA with compounds 1–24 has been documented considering the data in :
Table 2. Activation constants of hCA I, hCA II and MreCA with amino acids and amines 1–24 by a stopped-flow CO2 hydrase assayCitation25.
The compounds which showed the least effective for activating MreCA were L-His, L-/D-Tyr, L-Asp, and L-/D-Glu, with activation constants ranging between 4.04 and 12.8 µM. These compounds belong to a rather heterogeneous group of amino acids, with both deprotonated (Asp, Glu), neutral (Tyr) and protonated (His) side chains at pH 7.4. On the other hand, it seems that in some cases the enantiomer is relevant for this activity, if one compares the differences in KA between L and D-His, with the last compound being 6.95 times a better activator compared to its diastereoisomer ().
Compounds possessing a medium activating effect were D-His, L-Phe, dopamine and 2-(2aminoethyl)pyridine (derivative 16), which showed activation constants in the range of 1.84–2.71 μM (). Again, small structural changes, as in the pair of compounds 15/16, leads to drastic changes of activity. The two compounds only differ by a CH2 group, but 15 is 6.26 times a more effective activator compared to 16.
The effective, submicromolar CAAs against MreCA detected here were D-Phe, L-/D-DOPA, and L-/D-Trp, 4-amino-L-phenylalanine, 2-aminoethyl-piperazine/morpholine, histamine, serotonin, some pyridyl-alkylamines, L-Gln, and L-Asn, with KAs of 0.25–0.93 µM. L-adrenaline, with a KA of 15 nM, was the most effective among all compounds investigated here for the activation of MreCA ().
The activation profile of this fungal enzyme with amino acids and amines is very different from that of the human isoforms hCA I and II, with only L-Asn and L-Gln showing some selectivity for the activation of the fungal versus the human enzymes.
4. Conclusions
CAs were shown to be involved in metabolic and signalling pathways in fungi, including pathogenic ones, and this mechanism has been proposed to be exploited for the development of antifungals with different mechanisms of action compared to the clinically used agents, for which extensive drug resistance has been documentedCitation7,Citation24,Citation34. Indeed, in an animal model of dandruff provoked by M. globosa, a related species to M. restricta, it has been shown that β-CA inhibition with sulphonamides has a potent antifungal effectCitation35. However, there are no studies to date on the role of CAAs on the life cycle of fungal pathogens. Considering the fact that amines and amino acids as those investigated here are found in high concentrations in many tissues, our present finding may be relevant for a better understanding of processes connected with infectivity and growth of fungal pathogens. L-adrenaline was observed to be the best MreCA activator. Is it a coincidence that stress, i.e. higher circulating amounts of catecholamines such as L-adrenaline, is associated with a worsening of seborrhoeic dermatitis and dandruff?
Acknowledgements
WAD and CTS thank the Australian Research Council for funding. This work was financed in part by l’Oréal.
Disclosure statement
The authors have no relevant affiliations of financial involvement with any organisation or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript.
References
- (a) Supuran CT. Carbonic anhydrases: novel therapeutic applications for inhibitors and activators. Nat Rev Drug Discov 2008;7:168–81. (b) Nocentini A, Supuran CT. Advances in the structural annotation of human carbonic anhydrases and impact on future drug discovery. Expert Opin Drug Discov 2019;14:1175–97.
- (a) Supuran CT. How many carbonic anhydrase inhibition mechanisms exist?. J Enzyme Inhib Med Chem 2016;31:345–60. (b) Supuran CT. Advances in structure-based drug discovery of carbonic anhydrase inhibitors. Expert Opin Drug Discov 2017;12:61–88. (c) Supuran CT. Structure and function of carbonic anhydrases. Biochem J 2016;473:2023–32.
- (a) Supuran CT. Carbonic anhydrase activators. Future Med Chem 2018;10:561–73. (b) Temperini C, Scozzafava A, Supuran CT. Carbonic anhydrase activation and the drug design. Curr Pharm Des 2008;14:708–15. (c) Akocak S, Supuran CT. Activation of α-, β-, γ- δ-, ζ- and η- class of carbonic anhydrases with amines and amino acids: a review. J Enzyme Inhib Med Chem 2019;34:1652–9.
- (a) Zoccola D, Innocenti A, Bertucci A, et al. Coral carbonic anhydrases: regulation by ocean acidification. Mar Drugs 2016;14:109. (b) Bertucci A, Moya A, Tambutté S, et al. Carbonic anhydrases in anthozoan corals – a review. Bioorg Med Chem 2013;21:1437–50. (c) Del Prete S, Vullo D, Zoccola D, et al. Activation profile analysis of CruCA4, an α-carbonic anhydrase involved in skeleton formation of the mediterranean red coral, Corallium rubrum. Molecules 2017;23:66.(d) Dathan NA, Alterio V, Troiano E, et al. Biochemical characterization of the chloroplastic β-carbonic anhydrase from Flaveria bidentis (L.) "Kuntze". J Enzyme Inhib Med Chem 2014;29:500–4.
- (a) Capasso C, Supuran CT. An overview of the alpha-, beta- and gamma-carbonic anhydrases from Bacteria: can bacterial carbonic anhydrases shed new light on evolution of bacteria?. J Enzyme Inhib Med Chem 2015;30:325–32. (b) Smith KS, Jakubzick C, Whittam TS, Ferry JG. Carbonic anhydrase is an ancient enzyme widespread in prokaryotes. Proc Natl Acad Sci USA 1999;96:15184–9.
- (a) Supuran CT. Applications of carbonic anhydrases inhibitors in renal and central nervous system diseases. Expert Opin Ther Pat 2018;28:713–21. (b) Supuran CT. Carbonic anhydrase inhibitors and their potential in a range of therapeutic areas. Expert Opin Ther Pat 2018;28:709–12. (c) Supuran CT. Carbonic anhydrase inhibitors as emerging agents for the treatment and imaging of hypoxic tumors. Expert Opin Investig Drugs 2018;27:963–70. (d) Nocentini A, Supuran CT. Carbonic anhydrase inhibitors as antitumor/antimetastatic agents: a patent review (2008–2018). Expert Opin Ther Pat 201828:729–40.
- (a) Supuran CT, Capasso C. The eta-class carbonic anhydrases as drug targets for antimalarial agents. Expert Opin Ther Targets 2015;19:551–63. (b) Capasso C, Supuran CT. Bacterial, fungal and protozoan carbonic anhydrases as drug targets. Expert Opin Ther Targets 2015;19:1689–704. (c) Supuran CT, Capasso C. Biomedical applications of prokaryotic carbonic anhydrases. Expert Opin Ther Pat 2018;28:745–54.
- (a) Ozensoy Guler O, Capasso C, Supuran CT. A magnificent enzyme superfamily: carbonic anhydrases, their purification and characterization. J Enzym Inhib Med Chem 2016;31:689–94. (b) De Simone G, Supuran CT. (In)organic anions as carbonic anhydrase inhibitors. J Inorg Biochem 2012;111:117–29. (c) Supuran CT. Carbonic anhydrase inhibition and the management of hypoxic tumors. Metabolites 2017;7:E48.
- (a) Alterio V, Esposito D, Monti SM, et al. Crystal structure of the human carbonic anhydrase II adduct with 1-(4-sulfamoylphenyl-ethyl)-2,4,6-triphenylpyridinium perchlorate, a membrane-impermeant, isoform selective inhibitor. J Enzyme Inhib Med Chem 2018;33:151–7. (b) Innocenti A, Vullo D, Scozzafava A, et al. Carbonic anhydrase inhibitors. Inhibition of mammalian isoforms I–XIV with a series of substituted phenols including paracetamol and salicylic acid. Bioorg Med Chem 2008;16:7424–8. (c) Nocentini A, Bonardi A, Gratteri P, et al. Steroids interfere with human carbonic anhydrase activity by using alternative binding mechanisms. J Enzyme Inhib Med Chem 2018;33:1453–9. (d) Tars K, Vullo D, Kazaks A, et al. Sulfocoumarins (1,2-benzoxathiine-2,2-dioxides): a class of potent and isoform-selective inhibitors of tumor-associated carbonic anhydrases. J Med Chem 2013;56:293–300.
- (a) Del Prete S, Vullo D, Scozzafava A, et al. Cloning, characterization and anion inhibition study of the δ-class carbonic anhydrase (TweCA) from the marine diatom Thalassiosira weissflogii. Bioorg Med Chem 2014;22:531–7. (b) Vullo D, Del Prete S, Osman SM, et al. Sulfonamide inhibition studies of the δ-carbonic anhydrase from the diatom Thalassiosira weissflogii. Bioorg Med Chem Lett 2014;24:275–9. (c) Del Prete S, Vullo D, De Luca V, et al. Biochemical characterization of the δ-carbonic anhydrase from the marine diatom Thalassiosira weissflogii. J Enzyme Inhib Med Chem 2014;29:906–11.
- (a) Supuran CT, Capasso C. An overview of the bacterial carbonic anhydrases. Metabolites 2017; 7:56. (b) Clare BW, Supuran CT. Carbonic anhydrase activators. 3: structure–activity correlations for a series of isozyme II activators. J Pharm Sci 1994;83:768–73. (c) Supuran CT. Carbonic anhydrases and metabolism. Metabolites 2018;8:25. (d) Supuran CT. Carbon- versus sulphur-based zinc binding groups for carbonic anhydrase inhibitors? J Enzyme Inhib Med Chem 2018;33:485–95.
- (a) Del Prete S, Vullo D, Fisher GM, et al. Discovery of new family of carbonic anhydrases in the malaria pathogen Plasmodium falciparum- the η-carbonic anhydrases. Bioorg Med Chem 2014;24:4389–96. (b) De Simone G, Di Fiore A, Capasso C, Supuran CT. The zinc coordination pattern in the η-carbonic anhydrase from Plasmodium falciparum is different from all other carbonic anhydrase genetic families. Bioorg Med Chem Lett 2015;25:1385–9.
- Jensen EL, Clement R, Kosta A, et al. A new widespread subclass of carbonic anhydrase in marine phytoplankton. ISME J 2019;13:2094–106.
- (a) Alterio V, Di Fiore A, D'Ambrosio K, et al. Multiple binding modes of inhibitors to carbonic anhydrases: how to design specific drugs targeting 15 different isoforms? Chem Rev 2012;112:4421–68. (b) Briganti F, Pierattelli R, Scozzafava A, Supuran CT. Carbonic anhydrase inhibitors. Part 37. Novel classes of carbonic anhydrase inhibitors and their interaction with the native and cobalt-substituted enzyme: kinetic and spectroscopic investigations. Eur J Med Chem 1996;31:1001–10. (c) Neri D, Supuran CT. Interfering with pH regulation in tumours as a therapeutic strategy. Nat Rev Drug Discov 2011;10:767–77. (e) Supuran CT, Vullo D, Manole G, et al. Designing of novel carbonic anhydrase inhibitors and activators. Curr Med Chem Cardiovasc Hematol Agents 2004;2:49–68.
- (a) Pacchiano F, Carta F, McDonald PC, et al. Ureido-substituted benzenesulfonamides potently inhibit carbonic anhydrase IX and show antimetastatic activity in a model of breast cancer metastasis. J Med Chem 2011;54:1896–902. (b) Lou Y, McDonald PC, Oloumi A, et al. Targeting tumor hypoxia: suppression of breast tumor growth and metastasis by novel carbonic anhydrase IX inhibitors. Cancer Res 2011;71:3364–76. (c) Pacchiano F, Aggarwal M, Avvaru BS, et al. Selective hydrophobic pocket binding observed within the carbonic anhydrase II active site accommodate different 4-substituted-ureido-benzenesulfonamides and correlate to inhibitor potency. Chem Commun (Camb) 2010;46:8371–3. (d) Köhler K, Hillebrecht A, Schulze Wischeler J, et al. Saccharin inhibits carbonic anhydrases: possible explanation for its unpleasant metallic aftertaste. Angew Chem Int Ed Engl 2007;46:7697–9.
- (a) Krall N, Pretto F, Decurtins W, et al. A small‐molecule drug conjugate for the treatment of carbonic anhydrase IX expressing tumors. Angew Chem Int Ed Engl 2014;53:4231–5. (b) El-Gazzar MG, Nafie NH, Nocentini A, et al. Carbonic anhydrase inhibition with a series of novel benzenesulfonamide-triazole conjugates. J Enzyme Inhib Med Chem 2005;20:333–40. (c) Eldehna WM, Abo-Ashour MF, Berrino E, et al. SLC-0111 enaminone analogs, 3/4-(3-aryl-3-oxopropenyl) aminobenzenesulfonamides, as novel selective subnanomolar inhibitors of the tumor-associated carbonic anhydrase isoform IX. Bioorg Chem 2019;83:549–58. (d) Abo-Ashour MF, Eldehna WM, Nocentini A, et al. Novel synthesized SLC-0111 thiazole and thiadiazole analogues: Determination of their carbonic anhydrase inhibitory activity and molecular modeling studies. Bioorg Chem 2019;87:794–802.
- (a) Akocak S, Lolak N, Nocentini A, et al. Synthesis and biological evaluation of novel aromatic and heterocyclic bis-sulfonamide Schiff bases as carbonic anhydrase I, II, VII and IX inhibitors. Bioorg Med Chem 2017;25:3093–7. (b) Akocak S, Lolak N, Bua S, et al. Synthesis and biological evaluation of novel N,N`-diaryl cyanoguanidines acting as potent and selective carbonic anhydrase II inhibitors. Bioorg Chem 2018;77:245–51. (c) Lolak N, Akocak S, Bua S, et al. Design and synthesis of novel 1,3-diaryltriazene-substituted sulfonamides as potent and selective carbonic anhydrase II inhibitors. Bioorg Chem 2018; 77542–7. (d) Akocak S, Lolak N, Bua S, et al. Discovery of novel 1,3-diaryltriazene sulfonamides as carbonic anhydrase I, II, VII, and IX inhibitors. J Enzyme Inhib Med Chem 2018; 33:1575–80.
- (a) Lolak N, Akocak S, Bua S, Supuran CT. Design, synthesis and biological evaluation of novel ureido benzenesulfonamides incorporating 1,3,5-triazine moieties as potent carbonic anhydrase IX inhibitors. Bioorg Chem 2019;82:117–22. (b) Lolak N, Akocak S, Bua S, et al. Discovery of new ureido benzenesulfonamides incorporating 1,3,5-triazine moieties as carbonic anhydrase I, II, IX and inhibitors. Bioorg Med Chem 2019;27:1588–94. (c) Shabana AM, Mondal UK, Alam R, et al. pH-sensitive multiligand gold nanoplatform targeting carbonic anhydrase IX enhances the delivery of Doxorubicin to hypoxic tumor spheroids and overcomes the hypoxia-induced chemoresistance. ACS Appl Mater Interfaces 2018;10:17792–808.
- (a) Briganti F, Mangani S, Orioli P, et al. Carbonic anhydrase activators: X-ray crystallographic and spectroscopic investigations for the interaction of isozymes I and II with histamine. Biochemistry 1997;36:10384–92. (b) Temperini C, Scozzafava A, Vullo D, Supuran CT. Carbonic anhydrase activators. Activation of isozymes I, II, IV, VA, VII, and XIV with l- and d-histidine and crystallographic analysis of their adducts with isoform II: engineering proton-transfer processes within the active site of an enzyme. Chemistry 2006;12:7057–66. (c) Temperini C, Scozzafava A, Vullo D, Supuran CT. Carbonic anhydrase activators. Activation of isoforms I, II, IV, VA, VII, and XIV with L- and D-phenylalanine and crystallographic analysis of their adducts with isozyme II: stereospecific recognition within the active site of an enzyme and its consequences for the drug design. J Med Chem 2006;49:3019–27. (d) Temperini C, Innocenti A, Scozzafava A, Supuran CT. Carbonic anhydrase activators: kinetic and X-ray crystallographic study for the interaction of D- and L-tryptophan with the mammalian isoforms I-XIV. Bioorg Med Chem 2008;16:8373–8. (e) Temperini C, Innocenti A, Scozzafava A, et al. Carbonic anhydrase activators: L-Adrenaline plugs the active site entrance of isozyme II, activating better isoforms I, IV, VA, VII, and XIV. Bioorg Med Chem Lett 2007;17:628–35.
- (a) De Simone G, Di Fiore A, Truppo E, et al. Exploration of the residues modulating the catalytic features of human carbonic anhydrase XIII by a site-specific mutagenesis approach. J Enzyme Inhib Med Chem 2019;34:1506–10. (b) Buonanno M, Di Fiore A, Langella E, et al. The crystal structure of a hCA VII variant provides insights into the molecular determinants responsible for its catalytic behavior. Int J Mol Sci 2018;19:1571. (c) Mikulski R, West D, Sippel KH, et al. Water networks in fast proton transfer during catalysis by human carbonic anhydrase II. Biochemistry 2013;52:125–31.
- (a) Akocak S, Lolak N, Vullo D, et al. Synthesis and biological evaluation of histamine Schiff bases as carbonic anhydrase I, II, IV, VII, and IX activators. J Enzyme Inhib Med Chem 2017;32:1305–12. (b) Akocak S, Lolak N, Bua S, et al. α-Carbonic anhydrases are strongly activated by spinaceamine derivatives. Bioorg Med Chem 2019;27:800–4. (c) Akocak S, Lolak N, Bua S, et al. Activation of human α-carbonic anhydrase isoforms I, II, IV and VII with bis-histamine Schiff bases and bis-spinaceamine substituted derivatives. J Enzyme Inhib Med Chem 2019;34:1193–8. (d) Dave K, Scozzafava A, Vullo D, et al. Pyridinium derivatives of histamine are potent activators of cytosolic carbonic anhydrase isoforms, I, II and VII. Org Biomol Chem 2011;9:2790–800. (e) Dave K, Ilies MA, Scozzafava A, et al. An inhibitor-like binding mode of a carbonic anhydrase activator within the active site of isoform II. Bioorg Med Chem Lett 2011;21:2764–8.
- (a) Bhatt A, Mondal UK, Supuran CT, et al. Crystal structure of carbonic anhydrase ii in complex with an activating ligand: implications in neuronal function. Mol Neurobiol 2018;55:7431–7. (b) Temperini C, Scozzafava A, Supuran CT. Carbonic anhydrase activators: the first X-ray crystallographic study of an adduct of isoform I. Bioorg Med Chem Lett 2006;16:5152–6. (c) Temperini C, Scozzafava A, Puccetti L, Supuran CT. Carbonic anhydrase activators: X-ray crystal structure of the adduct of human isozyme II with L-histidine as a platform for the design of stronger activators. Bioorg Med Chem Lett 2005;15:5136–41. (d) Draghici B, Vullo D, Akocak S, et al. Ethylene bis-imidazoles are highly potent and selective activators for isozymes VA and VII of carbonic anhydrase, with a potential nootropic effect. Chem Commun 2014;50:5980–3.
- (a) Canto de Souza L, Provensi G, Vullo D, et al. Carbonic anhydrase activation enhances object recognition memory in mice through phosphorylation of the extracellular signal-regulated kinase in the cortex and the hippocampus. Neuropharmacology 2017;118:148–56. (b) Sanku RKK, John JS, Salkovitz M, et al. Potential learning and memory disruptors and enhancers in a simple, 1-day operant task in mice. Behavioural Pharmacol 2018;29:482–92. (c) Wang X, Schröder HC, Schlossmacher U, et al. Modulation of the initial mineralization process of SaOS-2 cells by carbonic anhydrase activators and polyphosphate. Calcif Tissue Int 2014;94:495–509.
- (a) Del Prete S, Vullo D, Ghobril C, et al. Cloning, purification, and characterization of a β-carbonic anhydrase from Malassezia restricta, an opportunistic pathogen involved in dandruff and seborrheic dermatitis. Int J Mol Sci 2019;20:2447. (b) Prete SD, Angeli A, Ghobril C, et al. Anion inhibition profile of the β-carbonic anhydrase from the opportunist pathogenic fungus Malassezia restricta involved in dandruff and seborrheic dermatitis. Metabolites 2019;9:E147. (c) Del Prete S, Angeli A, Ghobril C, et al. Sulfonamide inhibition profile of the β-carbonic anhydrase from Malassezia restricta, an opportunistic pathogen triggering scalp conditions. Metabolites 2020;10:39.
- Khalifah RG. The carbon dioxide hydration activity of carbonic anhydrase. I. Stop-flow kinetic studies on the native human isoenzymes B and C. J Biol Chem 1971;246:2561–73.
- (a) Vullo D, Innocenti A, Nishimori I, et al. Carbonic anhydrase activators: Activation of the human isoforms VII (cytosolic) and XIV (transmembrane) with amino acids and amines. Bioorg Med Chem Lett 2007;17:4107–12. (b) Pastorekova S, Vullo D, Nishimori I, et al. Carbonic anhydrase activators: Activation of the human tumor-associated isozymes IX and XII with amino acids and amines. Bioorg Med Chem 2008;16:3530–6. (c) Innocenti A, Hilvo M, Parkkila S, et al. Carbonic anhydrase activators: activation of the membrane-associated isoform XV with amino acids and amines. Bioorg Med Chem Lett 2009;19:3430–3.
- (a) Innocenti A, Zimmerman SA, Scozzafava A, et al. Carbonic anhydrase activators: Activation of the archaeal β-class (Cab) and γ-class (Cam) carbonic anhydrases with amino acids and amines. Bioorg Med Chem Lett 2008;18:6194–8. (b) Vullo D, Del Prete S, Capasso C, Supuran CT. Carbonic anhydrase activators: activation of the β-carbonic anhydrase from Malassezia globosa with amino acids and amines. Bioorg Med Chem Lett 2016;26:1381–5. (c) Isik S, Kockar F, Aydin M, et al. Carbonic anhydrase activators: activation of the β-carbonic anhydrase Nce103 from the yeast Saccharomyces cerevisiae with amino acids and amines. Bioorg Med Chem Lett 2009;19:1662–5.
- Innocenti A, Leewattanapasuk W, Manole G, et al. Carbonic anhydrase activators: activation of the β-carbonic anhydrase from the pathogenic yeast Candida glabrata with amino acids and amines. Bioorg Med Chem Lett 2010;20:1701–4.
- (a) Angeli A, Del Prete S, Osman SM, et al. Activation studies of the α- and β-carbonic anhydrases from the pathogenic bacterium Vibrio cholerae with amines and amino acids. J Enzyme Inhib Med Chem 2018;33:227–33. (b) Angeli A, Del Prete S, Donald WA, et al. The γ-carbonic anhydrase from the pathogenic bacterium Vibrio cholerae is potently activated by amines and amino acids. Bioorg Chem 2018;77:1–5. (c) Angeli A, Del Prete S, Osman SM, et al. Activation studies with amines and amino acids of the β-carbonic anhydrase encoded by the Rv3273 gene from the pathogenic bacterium Mycobacterium tuberculosis. J Enzyme Inhib Med Chem 2018;33:364–9.
- (a) Angeli A, Del Prete S, Pinteala M, et al. The first activation study of the β-carbonic anhydrases from the pathogenic bacteria Brucella suis and Francisella tularensis with amines and amino acids. J Enzyme Inhib Med Chem 2019;34:1178–85. (b) Stefanucci A, Angeli A, Dimmito MP, et al. Activation of β- and γ-carbonic anhydrases from pathogenic bacteria with tripeptides. J Enzyme Inhib Med Chem 2018;33:945–50. (c) Bua S, Haapanen S, Kuuslahti M, et al. Activation studies of the β-carbonic anhydrase from the pathogenic protozoan Entamoeba histolytica with amino acids and amines. Metabolites 2019;9:26–33.
- (a) Vullo D, Del Prete S, Osman SM, et al. Burkholderia pseudomallei γ-carbonic anhydrase is strongly activated by amino acids and amines. Bioorg Med Chem Lett 2017;27:77–80. (b) Vullo D, Del Prete S, Osman SM, et al. Comparison of the amine/amino acid activation profiles of the β- and γ-carbonic anhydrases from the pathogenic bacterium Burkholderia pseudomallei. J Enzyme Inhib Med Chem 2018;33:25–30.
- (a) Angeli A, Del Prete S, Osman SM, et al. Activation studies of the γ-carbonic anhydrases from the Antarctic marine bacteria Pseudoalteromonas haloplanktis and Colwellia psychrerythraea with amino acids and amines. Marine Drugs 2019;17:238–46. (b) Angeli A, Alasmary FAS, Del Prete S, et al. The first study of a δ-carbonic anhydrase: TweCAδ from the diatom Thalassiosira weissflogii is effectively activated by amines and amino acids. J Enzyme Inhib Med Chem 2018;33:680–5. (c) Angeli A, Buonanno M, Donald WA, et al. The zinc – but not cadmium – containing ζ-carbonic from the diatom Thalassiosira weissflogii is potently activated by amines and amino acids. Bioorg Chem 2018;80:261–5.
- (a) Angeli A, Del Prete S, Alasmary FAS, et al. The first activation studies of the η-carbonic anhydrase from the malaria parasite Plasmodium falciparum with amines and amino acids. Bioorg Chem 2018; 80:94–8. (b) Angeli A, Donald WA, Parkkila S, Supuran CT. Activation studies with amines and amino acids of the β-carbonic anhydrase from the pathogenic protozoan Leishmania donovani chagasi. Bioorg Chem 2018;78:406–10.
- Capasso C, Supuran CT, Protozoan, fungal and bacterial carbonic anhydrases targeting for obtaining antiinfectives. In: Supuran CT, Capasso, C. ed. Targeting carbonic anhydrases. London: Future Science Ltd.; 2014:133–141. (b) Schlicker C, Hall RA, Vullo D, et al. Structure and inhibition of the CO2-sensing carbonic anhydrase Can2 from the pathogenic fungus Cryptococcus neoformans. J Mol Biol 2009;385:1207–20.
- Hewitson KS, Vullo D, Scozzafava A, et al. Molecular cloning, characterization and inhibition studies of a β-carbonic anhydrase from Malassezia globosa, a potential antidandruff target. J Med Chem 2012;55:3513–20.
- Nguyen GTH, Tran TN, Podgorski MN, et al. Nanoscale ion emitters in native mass spectrometry for measuring ligand-protein binding affinities. ACS Cent Sci 2019;5:308–18.