Abstract
The carbonic anhydrases (CAs, EC 4.2.1.1) catalyse a simple but physiologically crucial reversible reaction, the carbon dioxide hydration with the production of bicarbonate and protons. In the last years, and especially, to the rapid emergence of the bacterial antibiotic resistance that is occurring worldwide, the understanding of the function of bacterial CAs has increased significantly. Recently, a new CA-class (ι-CA) was discovered in the marine diatom T. pseudonana. It has been reported that bacterial genomes may contain genes with relevant homology to the diatom ι-class CA. Still, the catalytic activity of the enzyme encoded by the gene was not investigated. Thus, herein, for the first time, we cloned, expressed, and purified the recombinant bacterial ι-CA (acronym BteCAι) identified in the genome of Burkholderia territorii. The recombinant BteCAι resulted in a good catalyst for the hydration of CO2 to bicarbonate and protons, with a kcat of 3.0 × 105 s −1 and kcat/KM of 3.9 × 107 M −1 s −1, and is also sensitive to inhibition by the sulphonamide acetazolamide. Furthermore, with the aid of the protonography, it has been demonstrated that BteCAι can be present as a dimer. This result is corroborated by the construction of a molecular model of BteCAι, which showed that the enzyme is formed by two equivalent monomers having a structure similar to a butterfly.
Introduction
The metalloenzyme carbonic anhydrases (CAs, EC 4.2.1.1) catalyse a simple but physiologically crucial reversible reaction, the carbon dioxide hydration with the production of bicarbonate and protons (CO2 + H2O ⇄ HCO3– + H+)Citation1. The CO2 hydration reaction is catalysed at very high rates, with a pseudo-first-order kinetic constant (kcat) ranging from 104 to 106 s−1 Citation2,Citation3. In Eukaryotes, CAs are involved in the transport and supply of CO2 or HCO3–, pH homeostasis, secretion of electrolytes, biosynthetic processes, and photosynthesisCitation4,Citation5. Since the CA hydratase/dehydratase activity contributes to numerous physiological functions involving dissolved inorganic carbon, CAs are crucial biomolecules in many physiological and pathological conditions in all types of organismsCitation6. In the last years, and especially, to the rapid emergence of the bacterial antibiotic resistance that is occurring worldwide, the understanding of the function of bacterial CAs has increased significantlyCitation7,Citation8. CAs are essential for the survival of the microbes and their pathogenicity and virulenceCitation9,Citation10. It has been demonstrated in vivo that the bacterial growth at ambient CO2 concentration is dependent on CA activity for several species, such as in the organisms discussed belowCitation11,Citation12. Thus, CAs encoded in the genome of Helicobacter pylori, are essential for the acid acclimatisation of the pathogen within the stomachCitation13–15; Vibrio cholerae uses CAs as a system to colonise the host since CAs are involved in the production of sodium bicarbonate, which induces cholera toxin expressionCitation16; Brucella suis needed functional CAs for growingCitation17–20; β-CA from Salmonella enterica is highly expressed during the bacterial infectionCitation21. Finally, the deletion of the gene encoding for the β-CA in Pseudomonas aeruginosa (psCA1) impaired virulence of the pathogen by reducing calcium salt depositionsCitation22. Starting from 1920, when was observed for the first time the CA activity in hemolyzed bloodCitation23, the knowledge of the enzymes responsible for that activity, i.e., the CA, has extensively been improved. Briefly, in all the living organisms, eight genetically distinct classes, named with the Greek letters, represent the CA superfamily: α-, β-, γ-, δ-, ζ-, η, θ, and ιCitation7,Citation9. The last three classes were only recently discoveredCitation24,Citation25. All the catalytically active CAs contain, independently of the genetic groups, a metal ion cofactor, which is necessary for enzyme catalysisCitation7,Citation9,Citation26,Citation27. The α-, β-, δ-, η- and, perhaps θ-CAs use as catalytic metal the Zn2+ ion; γ-CAs are Fe2+ enzymes, but they are active with bound Zn(II) or Co2+ ions tooCitation28–35. ζ-CAs are cambialistic enzymes, which are active both with Cd2+ or Zn2+ Citation2,Citation36,Citation37. Unexpectedly, the last identified ι-CA, which is encoded in the genome of the marine diatom Thalassiosira pseudonana, prefers Mn2+ to Zn2+ as a cofactorCitation24. In the CA active site, the metal is coordinated by three amino acid residues and, the fourth ligand is a water molecule/hydroxide ion acting as the nucleophile in the catalytic enzyme cycleCitation3,Citation7,Citation26,Citation27,Citation38,Citation39. The metal coordination is rather variegated among the CA-classes since in the α-, γ-, δ- and, probably, θ-classes the ion cofactor is coordinated by three His residues; by one His, and two Cys residues in β- and ζ-CAs; by two His and one Gln residues in the η-classCitation40. In the diatom ι-CAs the putative residues able to coordinate the Mn(II) are probably two His, one Asp and one Glu, but this has not yet been clearly proved by any biophysical techniqueCitation24. The CA classes are assembled with different folding and structures. α-CAs are usually active as monomers or dimers; β-CAs are active only as dimers, tetramers, or octamers, whereas γ-CAs must be trimers for accomplishing their physiological functionCitation29,Citation30,Citation33,Citation41. θ-CAs seem to have an X-ray crystal structure very similar to the β-CAsCitation42. The crystal structure of ζ-CA showed three slightly different active sites on the same polypeptide chainCitation37. X-ray crystal structures of δ-, η-, and ι-CAs are not yet available. Intriguing, α-, η-, θ- and ι-CAs were reported to catalyse the hydrolysis of esters/thioesters, while no esterase activity was detected for the other CA familiesCitation3,Citation24,Citation43. Finally, the CA-classes are differently distributed among the living organisms. For example, CAs present in mammals belong to α-classCitation44,Citation45, plants and algae have α-, β-, γ-, δ- and θ-classes; fungi encode for α- and β-CAs; protozoa for α-, β- and/or η-CAsCitation9. In metazoans, the α-CAs are the predominant enzymes showing CO2 hydratase activityCitation46,Citation47. α-, β-, and γ-CAs are the typical classes present in BacteriaCitation7,Citation26,Citation27,Citation48–51. In 2019, Gontero et al. discovered the ι-CA class in the diatom T. pseudonana, as mentioned aboveCitation24. They also reported that bacterial genomes may contain genes with relevant homology to the diatom ι-class CA, but such proteins were never characterised so far. However, most of these new bacterial sequences were annotated in the data bank as oxidoreductases. In the only work on ι-CAs published so far, the authors did not investigate if bacterial such enzymes possess a catalytic activity similar to the one reported for the diatom enzymeCitation24.
Thus, herein, for the first time, we cloned, expressed, and purified the recombinant bacterial ι-CA (acronym BteCAι) identified in the genome of Burkholderia territorii, a Gram-negative bacterium found in soil and water, which is often resistant to common antibioticsCitation52.
Materials and methods
Chemicals and instruments
IPTG and antibiotic were purchased from Sigma, Affinity column (His-Trap FF), molecular weight markers from GE Healthcare. All other chemicals used in this study were of reagent grade. AKTA-Prime purification system was purchased by GE Healthcare. The SX20 Stopped-Flow was obtained by the AppliedPhotophysics, while SDS–PAGE apparatus was procured by BioRAD.
Enzyme cloning, expression and purification
The synthetic B. territorii gene encoding for the BteCAι was cloned into the expression vector pET100D-Topo/BteCAι and used to transform the Competent Escherichia coli BL21 (DE3) codon plus cells (Agilent) as reported by Del Prete et al.Citation53. The cellular culture was induced with Isopropyl β-D-1-thiogalactopyranoside (IPTG) to overexpress the recombinant BteCAι. After the growth, the cells were harvested and disrupted by sonication. Cellular extract was purified using a nickel affinity column (His-Trap FF). HisTrap column (1.0 mL) was equilibrated with 20 mL equilibration buffer (50 mM Tris, 20 mM imidazole and 150 mM sodium chloride, pH 7.5) at 1 mL/min. The supernatant from the cellular lysate was loaded onto the column at 1.0 mL/min, connected with AKTA Prime. The recombinant BteCAι was eluted from the column with a flow of 0.5 mL/min and the elution buffer composed of 50 mM Tris, 500 mM imidazole and 300 mM sodium chloride, pH 7.5. The recovered BteCAι was 90% pure. The protein quantification was carried out by Bradford method (BioRAD)Citation54.
Sds-Page, protonography and Western blot
A 12% Sodium Dodecyl Sulfate-polyacrylamide gel electrophoresis (SDS-Page) prepared as described by described by LaemmliCitation55 was running loading on the gel the recovered BteCAι from the affinity column. The gel was stained with Coomassie Brilliant Blue-R. To perform the protonography, wells of 12% SDS-PAGE gel were loaded with BteCAι and bCA mixed with loading buffer without 2-mercaptoethanol and without boiling the samples, in order to avoid protein denaturation. Moreover, aliquots of the purified BteCAι were mixed with a loading solution buffer (LSB) containing different concentrations of SDS, ranging from 1.0 to 0.1%. The gel was run at 150 V until the dye front ran off the gel. Following the electrophoresis, the 12% SDS-PAGE gel was subject to protonography to detect the yellows bands due to the hydratase activity on the gel as described by Capasso and co-workersCitation56–59. Besides, the BteCAι subjected to a 12% (w/v) SDS-PAGE and followed by electrophoretic was transferred to a PVDF membrane with transfer buffer (25 mM Tris, 192 mM glycine, 20% methanol) using Trans-Plot SD Cell (Bio-Rad, Hercules, CA, USA). His-Tag Western blot was carried out using the Pierce Fast Western Blot Kit (Thermo Scientific,Waltham, MA, USA). Blotted membrane had been placed in the wash blot solution Fast Western 1 Wash Buffer to remove transfer buffer. Primary Antibody Working Dilution was added to the blot and incubated for 30 min at room temperature (RT) with shaking. Afterwards, the blot was removed from the primary antibody solution and incubated for 10 min with the FastWestern Optimised HRP ReagentWorking Dilution. Subsequently, the membrane was washed two times in about 20 mL of FastWestern 1 Wash Buffer. Finally, the membrane was incubated with the detection reagent working solution and incubated for 1.0 min at room temperature and then developed with X-ray film.
Assay for carbonic anhydrase using CO2 as substrate
CA activity assay was performed as described by Capasso et al.Citation60. Briefly, the assay was based on the monitoring of pH variation due to the catalysed conversion of CO2 to bicarbonate. Bromothymol blue was used as the indicator of pH variation and the assay was performed at 0 °C. The CO2-satured solution was used as substrate. To test the activity of carbonic anhydrase, 1.0 mL of 25 mM Tris, pH 8.3, containing bromothymol blue as a dye (to give a distinct and visible blue colour) was added to two test tubes chilled in an ice bath. An appropriate amount of the enzyme solution was added to one tube, and an equivalent amount of buffer was added to the second tube as control. One millilitre of CO2 solution was added, and the time required for the solution to change from blue to yellow was recorded (transition point of bromothymol blue is pH 6.0–7.6). The time required for the colour change is inversely related to the quantity of enzyme present in the sample. Wilbur-Anderson units were calculated according to the following definition: One Wilbur-Anderson unit (WAU) of activity is defined as (T0 − T)/T, where T0 (uncatalyzed reaction) and T (catalysed reaction) are recorded as the time (in seconds) required for the pH to drop from 8.3 to the transition point of the dye in a control buffer and in the presence of enzyme, respectively. The enzyme restoring activity with the metal ion was determined on samples aliquots kept overnight in a solution containing 150 mM NaCl and 20 mM Tris-HCl, pH 8.0, with or without 5 mM EDTA. EDTA-treated samples were mixed with 10 mM of CaCl2, MnCl2, and ZnCl2 and incubated at room temperature for 45 min before to performing the CO2 hydratase assay.
Determination of the kinetic parameters and inhibition constants
The CO2 hydration activity performed by the BteCAι was monitored using an Applied Photophysics stopped-flow instrumentCitation61. Phenol red (at a concentration of 0.2 mM) was used as indicator, working at the absorbance maximum of 557 nm, with 20 mM TRIS (pH 7.5) as buffer, and 20 mM NaClO4 (for maintaining constant the ionic strength), following the initial rates of the CA-catalysed CO2 hydration reaction for a period of 10–100 s. To determine the kinetic parameters by Lineweaver-Burk plots and the inhibition constants, a concentration of CO2 between 1.7 to 17 mM was used. At least six measurements of the original 5–10% reaction were used to assess the initial velocity for each inhibitor. The uncatalyzed rates were identically determined and detracted from the total observed rates. Stock inhibitor solutions (10–100 mM) were prepared in distilled-deionized water and dilutions up to 0.01 mM were done with the buffer test. Inhibitor and enzyme solutions were preincubated together for 15 min at room temperature prior to assay, in order to allow for the formation of the E-I complex or for the eventual active site mediated hydrolysis of the inhibitor. The inhibition constants were obtained by non-linear least-squares methods using PRISM 6 and the Cheng-Prusoff equation, as reported earlierCitation62–64, and represent the mean from at least three different determinations. All CA isoforms were recombinant ones obtained in-house. All salts/small molecules were of the highest purity available, from Sigma-Aldrich (Milan, Italy).
Results and discussion
The genome of diatoms encodes for a diversity of CA-classes, which are probably involved in the algal metabolism, pH regulation and balance of CO2/HCO3– concentrations. In the marine diatom Phaeodactylum tricornutum five α-CAs have been identified, which are confined in the matrices of the four-layered plastid membranesCitation25,Citation65, together with two β-CAs (PtCA1 and PtCA2) located in the pyrenoid, two mitochondrial γ-CAsCitation25, and one θ-CA in the lumen of the pyrenoid-penetrating thylakoidCitation25. The genome of Thalassiosira weissflogii encodes for two CAs (TWCA1 and CDCA1) classified as the first members of the δ- and ζ-class, respectivelyCitation66. Recently, as mentioned above, it has been demonstrated that the low-CO2-inducible protein (LCIP63) with the molecular weight of 63.0 kDa is a new class of CA (i.e., the ι-CA) and is present in the marine diatom Thalassiosira pseudonana. This CA showed peculiar characteristics: presence of an endoplasmic reticulum signal peptide (22 amino acid residues), and a chloroplast signal peptide (34 amino acid residues) at the N-terminal part. It is a multidomain protein with four repeated domains (multidomain protein), each of them homologous to the calcium/calmodulin dependent protein kinase II Association Domain. Furthermore, Mn(II) is reported to be the preferred catalytic ion cofactorCitation24. LCIP63 homologues were found in other diatoms, algae, bacteria, and archaeaCitation24 and this prompted us to investigate the biochemical properties of the bacterial ι-CAs.
Primary sequence and phylogenetic analysis
Using LCIP63 as query sequence and the similarity searching programme BLASTCitation67, a ι-CA homologous to the LCIP63 was identified in the genome of the bacterium Burkholderia territorii. The bacterial ι-CA nucleotide sequence shows an open reading frame encoding for a polypeptide chain (BteCAι) of 162 amino acid residues (), having a theoretical molecular mass of 17.8 kDa.
Figure 1. Graphical representation of the nucleotide sequence and translated amino acid sequence of BteCAι. The open reading frame (ORF) is indicated by bold capital letters, while the two nucleotide strands (5′ and 3′) by lower-case letters. The dark box (top of the figure) contains the amino acid residues identified as the protein signal peptide. The lightbox (bottom of the picture) contains the three histidines motifs, which is a unique characteristic of the ι-CA, and thus, not present in the other CA-classes reported up to now.

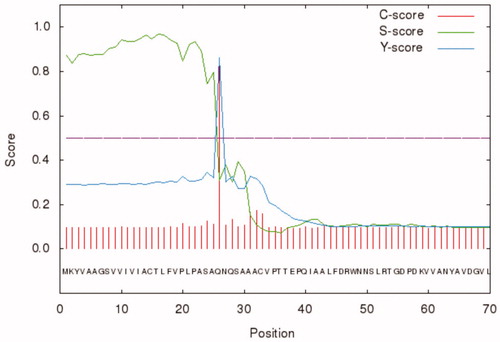
Besides, BteCAι was investigated for the presence of the secretion signal peptide at the N-terminal using the bioinformatics tool “SignalP 4.1” (http://www.cbs.dtu.dk/services/SignalP/). As shown in , the programme identified a putative signal peptide with a cleavage site between amino acid residues 25 and 26 (ASA-QN) of the BteCAι N-terminal amino acid sequence ().
Figure 2. Graphical output generated by the software SignalP 4.1. The graph shows three different scores for the first 70 amino acid residues of BteCAι. Legend: X-axis, amino acid position; Y-axis, the following three scores: C-score (red line), raw cleavage site score; S-score (green line), signal peptide score; Y-score (blue line), combined cleavage site score.
We should mention that a common feature of the bacterial α-CAs identified in the Gram-negative bacteria is the presence of a secretory signal peptide at the N-terminal of the amino acid sequence. The secretory signal peptide allows the periplasmic localisation of the α-CAs. In our investigation on the bacterial CAs, it has also been noted that β- or γ-CA primary structures belonging to some pathogenic Gram-negative bacteria are typified by a secretory signal peptide of 18 or more amino acid residuesCitation7,Citation8. These findings coupled with the evidence that also the bacterial ι-CAs have a putative secretory signal peptide suggest that the β-, γ- and/or ι-CAs characterised by the presence of a signal peptide might have in the Gram-negative bacteria a periplasmic localisation supporting or complementing the same role of the α-CAsCitation7,Citation8.
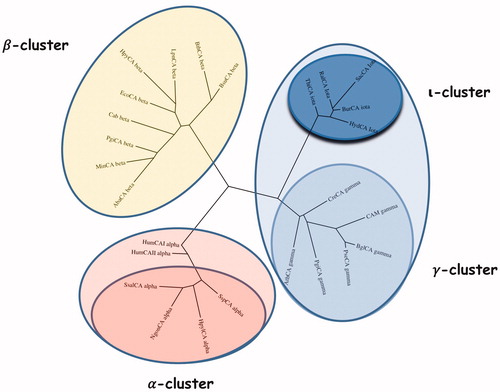
shows the alignment of the ι-CA amino acid sequences identified in B. territorii and T. pseudonana. BteCAι displayed a 35% identity with LCIP63 domain, and, as expected, showed the typical ι-CA consensus sequence “HHHSS” at the C-terminus of the polypeptide chain, which is a distinctive character of this new CA-class. Generally, the analysis of the sequence hallmarks is far from being exhaustive, as it does not take into account all the amino acid substitutions that differentiate this new class from the other bacterial CA classes. Hence, a phylogenetic tree has been constructed to better evidence the relationship of the bacterial ι-CA sequences with respect to the other bacterial CA-classes (α, β, and γ) (). The two human isoforms hCA I and hCA II were added in this analysis, too. From the dendrogram shown in , the bacterial ι-CAs clustered in a group closely associated with the bacterial γ-CAs. Although closer to the γ-CA, the ι-CA cluster appears well separated from them as well as distinct from all other CA-clusters. Thus, this finding reinforces the fact that these proteins were classified in a new CA class. We even speculate that the bacterial ι-CAs may be the result of a modification of an “ancestor gene” commune with that of the γ-CAs, which we demonstrated earlier to represent the ancestral class among all CA-classesCitation7,Citation9.
Figure 3. Alignment of BteCAι and LCIP63. The peptide signal (in red) are enclosed for both proteins. In the case of LCIP63, only the first domain of the four repeated domains was considered. The grey box represents the common consensus of the ι-CAs.

Figure 4. Phylogenetic analysis was carried out using the PhyML programme. The bootstrap consensus tree (100 replicates) was obtained using all four classes of CAs identified in the genome of different bacteria. The two human α-CA isoforms hCA I and II were included in the phylogenetic analysis, too. Legend: α-cluster (pink), β-cluster (yellow), γ-cluster (light blue), and ι-cluster (dark blue).
Recombinant enzyme preparation and protonography
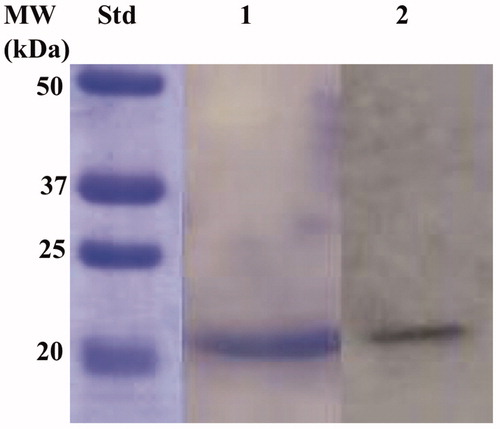
The recombinant BteCAι was produced as a fusion protein with a tail containing six histidines (His-Tag) at the N-terminal end, without its secretory signal peptide. The heterologous overexpression was induced by the addition of IPTG to the E. coli BL21 DE3 Codon plus cells. A supplement of 0.5 mM ZnCl2 was added to the host cells to allow the correct protein folding. Most of the ι-CA activity was recovered in the soluble bacterial cellular extract prepared by sonication and centrifugation. Using the affinity column (His-select HF Nickel affinity gel), the BteCAι fusion protein was purified to the homogeneity as a subunit with an apparent molecular weight of about 19,0 kDa as indicated by SDS-PAGE and Western Blot ().
Figure 5. Combined lanes of SDS-Page and Western blot of BteCAι. The affinity purified recombinant BteCAι was subjected to SDS-PAGE (Lane 1) and then electro-blotted and incubated with the anti-HisTag (Lane 2, Panel A). Lane Std, molecular markers.
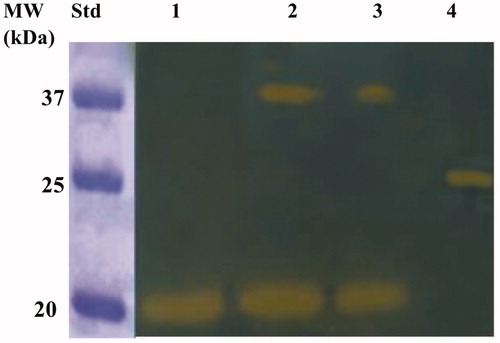
The purified BteCAι was investigated for its catalytic activity on the polyacrylamide gel through the use of a technique developed in our laboratories, the so-called protonographyCitation56. The method allows the monitoring of the pH variation in the polyacrylamide gel due to the CA-catalysed conversion of CO2 to bicarbonate and protons. Besides, to investigate the oligomeric protein state, aliquots of the purified BteCAι were mixed with a loading solution buffer (LSB) containing different concentrations of SDS, ranging from 1.0 to 0.1%. The resulting protonogram shown in evidenced the yellow bands due to the production of ions (H+) during the CO2 hydration reaction. The protonogram shows that the ι-CA is present only in the monomeric state when BteCAι was treated with LSB containing 1% of SDS. When the concentration of SDS in the LSB is less than 1% (0.5 or 0.1%), it was possible to see the two oligomeric states of the enzyme with an apparent molecular weight of 19.0 and 40 kDa, which correspond to the monomer and dimer form, respectively (). Of course, with the aid of protonography is not possible to establish if the ι-CA monomer can perform the hydration reaction since the protonography technique requires the SDS removal from the gel.Citation56 This procedure potentially led to the rearrangement of ι-CA monomers in the polyacrylamide gel, reconstituting the dimeric form of the enzyme. Thus, if the monomer is inactive, we always will see the presence of a yellow band due to the reconstituted dimer at the position of the ι-CA monomers. Thus, the activity of the ι-CA dimer can be explain by the two following conditions: (1) the ι-CA dimer is subject to a structural arrangement of the ι-CA monomers similar to that observed in the β- and γ-CAs, whose monomers assemble into dimer (β-CA) or trimer (γ-CA) to produce the catalytic active form of the enzymeCitation29,Citation68; (2) the ι-CA monomer is active as reported for the bacterial α-CAs, which crystallise as a dimerCitation33.
Figure 6. Developed protonogram showing the CO2 hydratase activity of BteCAι. The purified bacterial ι-CA was mixed with the Loading Solution Buffer (LSB) containing SDS at different concentrations (1.0, 0.5 and 0.1%) and loaded on the gel at 10 µg/well. The yellow bands correspond to the enzyme activity responsible for the drop of pH from 8.2 to the transition point of the dye in the control buffer. Legend: Lane 1, BteCAι with 1% SDS (protein in a monomeric state, MW: 19.0 kDa); Lane Std, Molecular markers. Lane 2 and 3 purified BteCAι mixed with 0.5 and 0.1% SDS, respectively (monomer and dimer); Lane 4, commercial bovine CA, used as positive controls.
Enzyme activity and kinetic parameters
The CO2 hydratase activity and the kinetic constants of the purified and soluble BteCAι were determined using the stopped-flow technique. The enzyme had a high catalytic activity for the physiological reaction of CO2 hydration to bicarbonate and protons, with a kcat 3.0 × 105 s−1 and a catalytic efficiency (kcat/KM) of 3.9 × 107 M−1 s−1, and was also sensitive to inhibition by the sulphonamide acetazolamide (KI = 64.9 nM). The BteCAι kinetic parameters are shown in and compared with those of the α-CAs from the Homo sapiens (isoforms hCAI and hCAII) as well as with a representative belonging to the bacterial α, β, and γ-class.
Table 1. BteCAι kinetic parameters compared with those calculated for the two human isoforms hCA I and II (α-class), and the α-, β-, γ- and ι-CAs from different bacterial species (all of them with Zn(II) at the active site).
BteCAι kcat value is similar to that obtained for the other bacterial CAs, as well as for the hCA I. These results are of extreme importance in the field of the inhibition of the bacterial CAs, which are crucial molecules for supporting the microbial functions involving dissolved inorganic carbon. Thus, among the new antibacterial should also be considered those new inhibitors able to inhibit the activity of the bacterial i-CA for contrasting the bacterial growth and their virulence.
Catalytic ion cofactor
As described above, the recombinant BteCAι was overexpressed to supplement the bacterial culture medium with 0,5 mM of ZnCl2. Following the strategy proposed by Jensen et al.Citation24, the amino acid sequence of BteCAι was submitted to a bioinformatics programme able to predict the binding of nine metal ions (Zn2+, Cu2+, Fe2+, Fe3+, Ca2+, Mg2+, Mn2+, Na+, K+) with the query sequence protein (BteCAι) (https://zhanglab.ccmb.med.umich.edu/IonCom/)Citation69. The output generated by the programme is shown in .
Figure 7. Output generated by the bioinformatics programme IonCom using as query sequence the BteCAι amino acid polypeptide chain. The output evidenced the ions not detected for the binding (point 1); and the possible ions, such as Zn2+ and Ca2+ (points 2 and 3). Besides, it reports the BteCAι amino acid sequence with the binding residues indicated in red.

The programme predicted that the recombinant BteCAι could bind only Zn2+ and Ca2+. To verify this, an aliquot of BteCAι was treated with EDTA overnight. The treatment with EDTA inactivated the enzyme as reported for the other CAs having Zn2+ as a catalytic cofactor. Intriguing, the addition of Zn2+ or Ca2+ fully restored the enzyme activity (). We also try to restore the enzyme activity using a metal, which was not predicted by the programme IonCom, such as Mn2+. From , it is readily apparent that this ion did not restore the enzyme activity, diversely as described for the ι-CA identified in the genome of the marine diatom T. pseudonanaCitation24.
Table 2. Effect of the addition of the Zn2+, Ca2+, and Mg2+ on the activity of the EDTA-treated BteCAι.
Homology modelling
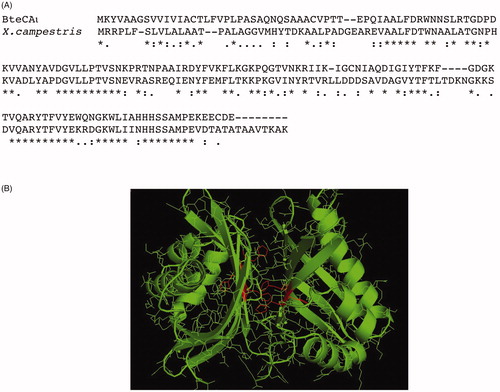
To date, no X-ray crystallographic or NMR structures are available for ι-CA and, as obtaining them is an intricate and time consuming process. However, the computational approach to predict protein three-dimensional structures starting from the amino acid sequence is an appealing alternative. Thus, using a fully automated protein homology modelling server SWISS-MODEL (https://swissmodel.expasy.org), we generated a raw first model of the bacterial BteCAι. The automated mode selects the structural templates that maximise the expected quality of the model. Two main templates were identified with a 50% identity (the other templates showed an identity ranging from 16 to 27%). Surprisingly, all the templates were identified as putative calcium/calmodulin-dependent protein kinase II Association Domain. We focalised our attention on the first template homologous to BteCAι and coming from Xanthomonas campestris, a bacterial species that causes a variety of plant diseasesCitation70. Its PDB code is 3h51, while NP_636218.1 is the protein accession number. Astonishingly, the code NP_6362218.1 (now WP_011036063.1 in the Pubmed (https://www.ncbi.nlm.nih.gov/protein/WP_011036063.1)) corresponded to a sequence classified as SgcJ/EcaC family oxidoreductase, which is the annotation of most of the sequences found during the search of the bacterial ι-CAs in the data bank. This prompt us to align the amino acid sequence of BteCAι with that identified in X. campestris (). The ι-CA amino acid sequences identified in B. territorii displays a 42% identity with the X. campestris amino acid sequence (38%identity with LCIP63 domain), and, as expected, the typical ι-CA consensus “HHHSS” at the C-terminus of the polypeptide chain is present (). In , we report the generated model of BteCAι with the consensus histidines of each monomer highlighted in red. We want stress that the resulting model predicted a dimeric organisation of the protein. This confirms the results obtained from the protonography performed at different concentration of SDS, which evidenced the presence of an hydratase activity at a molecular weight of 40.0 kDa (enzyme dimeric state). Of course, work is in progress for obtaining the crystal structure of BteCAι especially for understanding the coordination of the catalytic ion cofactor, if Zn2+, Mn2+ or another metal ion, as well as the metal coordination pattern.
Figure 8. Panel A: Alignment of the amino acid sequences from B. territory and X. campestris. Panel B: Model of the bacterial BteCAι generated using the automated procedure on the server SWISS-MODEL (https://swissmodel.expasy.org).
Conclusions
The analysis of BteCAι primary structure evidenced a secretory signal peptide consisting of 25 amino acid residues at the N-terminal part of the protein. The phylogenetic analysis carried out using bacterial α-, β-, γ-, and ι-CAs evidenced that the new ι-CAs were closer to the γ-classes than the other bacterial CAs (α and β). The determination of the kinetic constants showed that the bacterial ι-CA is a good catalyst for the hydration of CO2 to bicarbonate and protons, with a kcat of 3.0 × 105 s−1 and kcat/KM of 3.9 × 107 M−1 s−1, and is also sensitive to inhibition by the sulphonamide acetazolamide (KI= 64.9 nM). With the aid of the protonography, a technique developed by our groups, which allows the detection of the hydratase activity onto a polyacrylamide gel, it has been demonstrated that BteCAι can be present as a dimer. The structure prediction model showed that BteCAι is formed by two equivalent monomers, which appears as a “butterfly” structure where the Zn2+ ion cofactor of the active site might be coordinated by two histidines of a monomer and one histidine of the other monomer. Of course, the metal coordination will be thoroughly investigated with the resolution of the three-dimensional structure of this new class of CA, which is in progress in our laboratories.
Acknowledgements
We are grateful to Giovanni Del Monaco for technical assistance.
Disclosure statement
The authors state no conflict of interests.
References
- Alterio V, Di Fiore A, D’Ambrosio K, et al. Multiple binding modes of inhibitors to carbonic anhydrases: how to design specific drugs targeting 15 different isoforms? Chem Rev 2012;112:4421–68.
- Supuran CT. Advances in structure-based drug discovery of carbonic anhydrase inhibitors. Expert Opin Drug Discov 2017;12:61–88.
- Supuran CT. Structure and function of carbonic anhydrases. Biochem J 2016;473:2023–32.
- Nishimori I, Onishi S, Takeuchi H, Supuran CT. The alpha and beta classes carbonic anhydrases from Helicobacter pylori as novel drug targets. Curr Pharm Des 2008;14:622–30.
- Morishita S, Nishimori I, Minakuchi T, et al. Cloning, polymorphism, and inhibition of beta-carbonic anhydrase of Helicobacter pylori. J Gastroenterol 2008;43:849–57.
- Neri D, Supuran CT. Interfering with pH regulation in tumours as a therapeutic strategy. Nat Rev Drug Discov 2011;10:767–77.
- Capasso C, Supuran CT. An overview of the alpha-, beta- and gamma-carbonic anhydrases from Bacteria: can bacterial carbonic anhydrases shed new light on evolution of bacteria? J Enzyme Inhib Med Chem 2015;30:325–32.
- Supuran CT, Capasso C. New light on bacterial carbonic anhydrases phylogeny based on the analysis of signal peptide sequences. J Enzyme Inhib Med Chem 2016;31:1254–60.
- Supuran CT, Capasso C. An overview of the bacterial carbonic anhydrases. Metabolites 2017;7:56.
- Vullo D, Del Prete S, Di Fonzo P, et al. Comparison of the sulfonamide inhibition profiles of the beta- and gamma-carbonic anhydrases from the pathogenic bacterium burkholderia pseudomallei. Molecules 2017;22:421.
- Kusian B, Sültemeyer D, Bowien B. Carbonic anhydrase is essential for growth of Ralstonia eutropha at ambient CO(2) concentrations. J Bacteriol 2002;184:5018–26.
- Merlin C, Masters M, McAteer S, Coulson A. Why is carbonic anhydrase essential to Escherichia coli? J Bacteriol 2003;185:6415–24.
- Modak JK, Tikhomirova A, Gorrell RJ, et al. Anti-helicobacter pylori activity of ethoxzolamide. J Enzyme Inhib Med Chem 2019;34:1660–7.
- Ronci M, Del Prete S, Puca V, et al. Identification and characterization of the alpha-CA in the outer membrane vesicles produced by Helicobacter pylori. J Enzyme Inhib Med Chem 2019;34:189–95.
- Buzas GM. [Helicobacter pylori – 2010]. Orv Hetil 2010;151:2003–10.
- Abuaita BH, Withey JH. Bicarbonate Induces Vibrio cholerae virulence gene expression by enhancing ToxT activity. Infect Immun 2009;77:4111–20.
- Kohler S, Ouahrani-Bettache S, Winum JY. Brucella suis carbonic anhydrases and their inhibitors: towards alternative antibiotics? J Enzyme Inhib Med Chem 2017;32:683–7.
- Singh S, Supuran CT. 3D-QSAR CoMFA studies on sulfonamide inhibitors of the Rv3588c beta-carbonic anhydrase from Mycobacterium tuberculosis and design of not yet synthesized new molecules. J Enzyme Inhib Med Chem 2014;29:449–55.
- Ceruso M, Vullo D, Scozzafava A, Supuran CT. Sulfonamides incorporating fluorine and 1,3,5-triazine moieties are effective inhibitors of three beta-class carbonic anhydrases from Mycobacterium tuberculosis. J Enzyme Inhib Med Chem 2014;29:686–9.
- Carta F, Maresca A, Covarrubias AS, et al. Carbonic anhydrase inhibitors. Characterization and inhibition studies of the most active beta-carbonic anhydrase from Mycobacterium tuberculosis, Rv3588c. Bioorg Med Chem Lett 2009;19:6649–54.
- Rollenhagen C, Bumann D. Salmonella enterica highly expressed genes are disease specific. Infect Immun 2006;74:1649–60.
- Lotlikar SR, Kayastha BB, Vullo D, Khanam SS, et al. Pseudomonas aeruginosa β-carbonic anhydrase, psCA1, is required for calcium deposition and contributes to virulence. Cell Calcium 2019;84:102080.
- Ozensoy Guler O, Capasso C, Supuran CT. A magnificent enzyme superfamily: carbonic anhydrases, their purification and characterization. J Enzyme Inhib Med Chem 2015;31:1–694.
- Jensen EL, Clement R, Kosta A, et al. A new widespread subclass of carbonic anhydrase in marine phytoplankton. Isme J 2019;13:2094–106.
- Kikutani S, Nakajima K, Nagasato C, et al. Thylakoid luminal theta-carbonic anhydrase critical for growth and photosynthesis in the marine diatom Phaeodactylum tricornutum. Proc Natl Acad Sci USA 2016;113:9828–33.
- Capasso C, Supuran CT. An overview of the carbonic anhydrases from two pathogens of the oral cavity: streptococcus mutans and porphyromonas gingivalis. Curr Topics Med Chem 2016;16:2359–68.
- Capasso C, Supuran CT. An overview of the selectivity and efficiency of the bacterial carbonic anhydrase inhibitors. Curr Med Chem 2015;22:2130–9.
- Pinard MA, Lotlikar SR, Boone CD, et al. Structure and inhibition studies of a type II beta-carbonic anhydrase psCA3 from Pseudomonas aeruginosa. Bioorg Med Chem 2015;23:4831–8.
- Ferraroni M, Del Prete S, Vullo D, et al. Crystal structure and kinetic studies of a tetrameric type II beta-carbonic anhydrase from the pathogenic bacterium Vibrio cholerae. Acta Crystallogr D Biol Crystallogr 2015;71:2449–56.
- De Simone G, Monti SM, Alterio V, et al. Crystal structure of the most catalytically effective carbonic anhydrase enzyme known, SazCA from the thermophilic bacterium Sulfurihydrogenibium azorense. Bioorg Med Chem Lett 2015;25:2002–6.
- Żołnowska B, Sławiński J, Pogorzelska A, et al. Carbonic anhydrase inhibitors. Synthesis, and molecular structure of novel series N-substituted N′-(2-arylmethylthio-4-chloro-5-methylbenzenesulfonyl)guanidines and their inhibition of human cytosolic isozymes I and II and the transmembrane tumor-associated isozymes IX and XII. Euro J Med Chem 2014;71:135–47.
- De Luca L, Ferro S, Damiano FM, et al. Structure-based screening for the discovery of new carbonic anhydrase VII inhibitors. Europ J Med Chem 2014;71:105–11.
- Di Fiore A, Capasso C, De Luca V, et al. ray structure of the first ‘extremo-alpha-carbonic anhydrase’, a dimeric enzyme from the thermophilic bacterium Sulfurihydrogenibium yellowstonense YO3AOP1. Acta Crystallogr D Biol Crystallogr 2013;69:1150–9.
- Supuran CT. Structure-based drug discovery of carbonic anhydrase inhibitors. J Enzyme Inhib Med Chem 2012;27:759–72.
- Supuran CT. Carbonic anhydrases–an overview. Curr Pharmaceut Design 2008;14:603–14.
- Bhatt A, Mahon BP, Cruzeiro VW, et al. Structure-activity relationships of benzenesulfonamide-based inhibitors towards carbonic anhydrase isoform specificity. Chembiochem: Euro J Chem Biol 2017;18:213–22.
- Alterio V, Langella E, Viparelli F, et al. Structural and inhibition insights into carbonic anhydrase CDCA1 from the marine diatom Thalassiosira weissflogii. Biochimie 2012;94:1232–41.
- Buzas GM, Supuran CT. The history and rationale of using carbonic anhydrase inhibitors in the treatment of peptic ulcers. In memoriam Ioan Puscas (1932–2015). J Enzyme Inhib Med Chem 2016;31:527–33.
- Carta F, Supuran CT, Scozzafava A. Sulfonamides and their isosters as carbonic anhydrase inhibitors. Future Med Chem 2014;6:1149–65.
- De Simone G, Di Fiore A, Capasso C, Supuran CT. The zinc coordination pattern in the eta-carbonic anhydrase from Plasmodium falciparum is different from all other carbonic anhydrase genetic families. Bioorg Med Chem Lett 2015;25:1385–9.
- Lomelino CL, Mahon BP, McKenna R, et al. Kinetic and X-ray crystallographic investigations on carbonic anhydrase isoforms I, II, IX and XII of a thioureido analog of SLC-0111. Bioorg Med Chem 2016;24:976–81.
- D’Ambrosio K, Di Fiore A, Buonanno M, et al. Eta and Teta-carbonic anhydrases. Supuran CT, Nocentini A, ed. Journal of the American Chemical Society. London (UK): Elsevier; 2019.
- Supuran CT. How many carbonic anhydrase inhibition mechanisms exist? J Enzyme Inhib Med Chem 2016;31:345–60.
- Aspatwar A, Tolvanen ME, Ortutay C, Parkkila S. Carbonic anhydrase related proteins: molecular biology and evolution. Subcell Biochem 2014;75:135–56.
- Supuran CT. Carbonic anhydrases as drug targets–an overview. Curr Topics Med Chem 2007;7:825–33.
- Perfetto R, Del Prete S, Vullo D, et al. Biochemical characterization of the native alpha-carbonic anhydrase purified from the mantle of the Mediterranean mussel, Mytilus galloprovincialis. J Enzyme Inhib Med Chem 2017;32:632–9.
- Del Prete S, Vullo D, Zoccola D, et al. Kinetic properties and affinities for sulfonamide inhibitors of an alpha-carbonic anhydrase (CruCA4) involved in coral biomineralization in the Mediterranean red coral Corallium rubrum. Bioorg Med Chem 2017;25:3525–30.
- Capasso C, Supuran CT. Bacterial, fungal and protozoan carbonic anhydrases as drug targets. Expert Opin Ther Tar 2015;19:1689–704.
- Supuran CT, Capasso C. The eta-class carbonic anhydrases as drug targets for antimalarial agents. Expert Opin Ther Targets 2015;19:551–63.
- Capasso C, Supuran CT. Sulfa and trimethoprim-like drugs – antimetabolites acting as carbonic anhydrase, dihydropteroate synthase and dihydrofolate reductase inhibitors. J Enzyme Inhib Med Chem 2014;29:379–87.
- Capasso C, Supuran CT. Anti-infective carbonic anhydrase inhibitors: a patent and literature review. Expert Opin Ther Patents 2013;23:693–704.
- De Smet B, Mayo M, Peeters C, et al. Burkholderia stagnalis sp. nov. and Burkholderia territorii sp. nov., two novel Burkholderia cepacia complex species from environmental and human sources. Int J Syst Evol Microbiol 2015;65:2265–71.
- Del Prete S, Vullo D, Ghobril C, et al. Cloning, purification, and characterization of a beta-carbonic anhydrase from Malassezia restricta, an opportunistic pathogen involved in dandruff and seborrheic dermatitis. Int J Mol Sci 2019;20:pii:E2447.
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 1976;72:248–54.
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970;227:680–5.
- De Luca V, Del Prete S, Supuran CT, Capasso C. Protonography, a new technique for the analysis of carbonic anhydrase activity. J Enzyme Inhib Med Chem 2015;30:277–82.
- Del Prete S, De Luca V, Iandolo E, et al. Protonography, a powerful tool for analyzing the activity and the oligomeric state of the gamma-carbonic anhydrase identified in the genome of Porphyromonas gingivalis. Bioorg Med Chem 2015;23:3747–50.
- Del Prete S, De Luca V, Supuran CT, Capasso C. Protonography, a technique applicable for the analysis of eta-carbonic anhydrase activity. J Enzyme Inhib Med Chem 2015;30:920–4.
- Del Prete S, Vullo D, Caminiti-Segonds N, et al. Protonography and anion inhibition profile of the alpha-carbonic anhydrase (CruCA4) identified in the Mediterranean red coral Corallium rubrum. Bioorg Chem 2018;76:281–7.
- Capasso C, De Luca V, Carginale V, et al. Biochemical properties of a novel and highly thermostable bacterial alpha-carbonic anhydrase from Sulfurihydrogenibium yellowstonense YO3AOP1. J Enzyme Inhib Med Chem 2012;27:892–7.
- Khalifah RG. The carbon dioxide hydration activity of carbonic anhydrase. I. Stop-flow kinetic studies on the native human isoenzymes B and C. J Biolog Chem 1971;246:2561–73.
- Del Prete S, Vullo D, De Luca V, et al. Anion inhibition profiles of alpha-, beta- and gamma-carbonic anhydrases from the pathogenic bacterium Vibrio cholerae. Bioorg Med Chem 2016;24:3413–7.
- Del Prete S, Vullo D, De Luca V, et al. Anion inhibition profiles of the complete domain of the eta-carbonic anhydrase from Plasmodium falciparum. Bioorg Med Chem 2016;24:4410–4.
- De Luca V, Vullo D, Del Prete S, et al. Cloning, characterization and anion inhibition studies of a gamma-carbonic anhydrase from the Antarctic bacterium Colwellia psychrerythraea. Bioorg Med Chem 2016;24:835–40.
- Harada H, Nakatsuma D, Ishida M, Matsuda Y. Regulation of the expression of intracellular beta-carbonic anhydrase in response to CO2 and light in the marine diatom Phaeodactylum tricornutum. Plant Physiol 2005;139:1041–50.
- Park H, Song B, Morel FM. Diversity of the cadmium-containing carbonic anhydrase in marine diatoms and natural waters. Environ Microbiol 2007;9:403–13.
- Pearson WR. An introduction to sequence similarity (“homology”) searching. Current Protocols in Bioinformatics. Hoboken: John Wiley and Sons, Inc; 2013. Chapter Unit3:3 1.
- Tripp BC, Ferry JG. A structure-function study of a proton transport pathway in the gamma-class carbonic anhydrase from Methanosarcina thermophila. Biochemistry 2000;39:9232–40.
- Hu X, Dong Q, Yang J, Zhang Y. Recognizing metal and acid radical ion-binding sites by integrating ab initio modeling with template-based transferals. Bioinformatics 2016;32:3694.
- Schatschneider S, Schneider J, Blom J, et al. Systems and synthetic biology perspective of the versatile plant-pathogenic and polysaccharide-producing bacterium Xanthomonas campestris. Microbiology 2017;163:1117–1144.
