?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
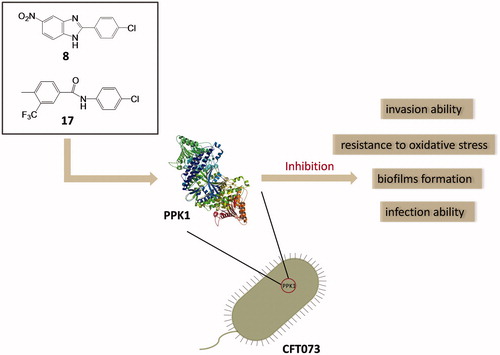
Novel antibacterial agents are urgently needed to address the infections caused by multi-drug resistant bacteria. Urinary tract infections are common infectious diseases in clinical. Most of these infections are caused by drug-resistant uropathogenic Escherichia coli. PPK1 is an essential kinase for bacterial motility, biofilm formation, quorum sensing, and virulence factors in the expression of uropathogenic E. coli. In the present study, two small molecules potentially targeting PPK1 were discovered through virtual screening and biological assays. The in vitro and in vivo results suggested that the interaction of these compounds with PPK1 can disrupt biofilm formation of uropathogenic E. coli and reduce invasive ability and resistance to oxidative stress of this strain. Moreover, the compounds exhibit good antibacterial bacterial activity in the mice with urinary tract infection. Taken together, our findings could provide a new chemotype for the development of antibacterials targeting PPK1.
Graphical Abstract
Keywords:
1. Introduction
The use of clinical antibiotics to treat bacterial infections becomes more and more difficult because bacteria have developed resistance to antibiotics at an alarming speed. Urinary tract infections (UTIs) are very common infectious diseases in clinical, especially among women, elderly, and childrenCitation1–3. With respect to the worldwide dissemination of drug-resistant bacterial strains, clinical treatment of UTI becomes a serious challengeCitation4. Although either Gram-positive or -negative bacteria can cause UTIs, more than 80% of community-acquired UTIs are caused by uropathogenic Escherichia coli (UPEC)Citation4,Citation5. Antimicrobial susceptibility tests indicate that UPEC is resistant to conventional antibiotics, such as furadantin, and even the last-line of defenders like colistinCitation6,Citation7. Therefore, the development of new types of antibacterial agents to combat UPEC is urgently needed.
Most of the conventional antibiotics target essential bacterial cell functions. For example, methicillin and vancomycin target bacterial cell wall synthesis, polymyxins target cell membrane, and quinolones inhibit the critical enzymes of the bacterial growth process. However, the resistance to most of these drugs has been observed to dateCitation8. To reduce bacterial virulence instead of killing the pathogen seems an alternative strategy to treat bacterial infectionsCitation9–11. Polyphosphate kinase 1 (PPK1) is essential in bacterial pathogens and is responsible for the synthesis of polyphosphate (poly P) and metabolism of ATPCitation11,Citation12. Previous studies revealed that PPK1 is essential for bacterial motility, biofilm formation, quorum sensing, and virulence factors expression and it is also an important target for the investigation of antibiotics resistance and pathogenicityCitation13,Citation14. Our previous study showed that, in the PPK1 knockout mutant of E. coli, the amount of poly P and the abilities of adhesion/invasion were significantly decreased compared with the wild type E. coli. In addition, poor survival rates during oxidative stress, osmotic shock, and acidic challenge also were found in this Δppk1 strainCitation15. Since E. coli PPK1 has been fully investigated and characterisedCitation16,Citation17. It is absent in the human body and becomes a potential drug target for the development of novel antibacterial agentsCitation18.
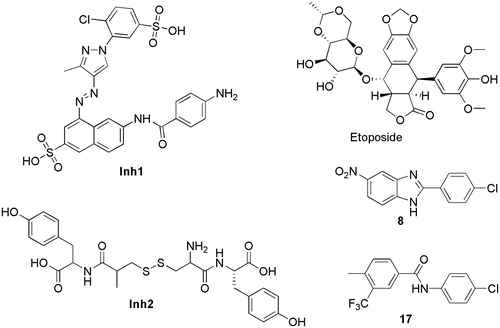
Over the past decade, only a few compounds were reported to interact with PPK1 of E. coli. For example, through in silico and in vitro methods, Inh1 and Inh2 () were predicted to disrupt the function of PPK1 in E. coli and to inhibit biofilm formationCitation19. Etoposide () was predicted to bind into the PPK1 through virtual screeningCitation20. However, no further in-depth study has been reported thus far. In this study, we identified two PPK1 inhibitors (8 and 17) through the integration of virtual screening and biological assays. These two small molecules were investigated in a mice model and the results suggested that the compounds are able to prevent UTI effectively through the disruption of the PPK1 function.
2. Results and discussions
2.1. Screening of potential compound targeting PPK1
Virtual screening was carried out to identify novel inhibitors of PPK1. We performed virtual screening on PKU-CNCL database and SPECS chemical library by molecular docking method. Finally, 44 compounds were selected for the following bioactivity evaluations. The structures of these 44 compounds are listed in the Supporting Information (Figure S1). A surface plasmon resonance (SPR) assay was then performed to rapidly screen candidates by evaluating the binding responses between PPK1 and the selected compounds. Etoposide was used as a positive control in the screening assay with SPR. At the concentration of 100 μM, etoposide showed a relative binding response of 3.06 100RU/Da. On the other hand, two small molecules (8 and 17) showed a relative response of 6.555 and 4.119, respectively. The binding responses were significantly higher than etoposide (Figure S2 and Table S1). Moreover, the sensorgrams (Figure S3) suggested that these two compounds interacted with PPK1 in vitro. Therefore, 8 and 17 were selected to further investigation.
2.2. 8. And 17 were able to decrease the invasion ability of CFT073l without any effect on bacterial growth
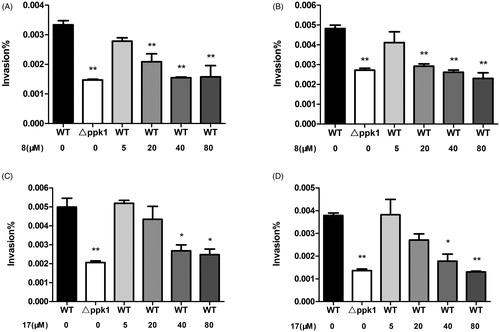
Since PPK1 is essential for the invasion ability of E. coli, 8 and 17 were subjected to a cell-based assay to evaluate their effects against the wild-type (WT) of uropathogenic E. coli strain (CFT0731) and its ppk1 knockout (△ppk1) mutant was used as a control strain in the assay. To study whether these compounds can affect the growth of CFT073, the bacterial cultures of CFT0731 were incubated with 8 and 17 at the concentration of 50, 100, or 200 μM. As shown in Figure S4, even at the concentration of 200 μM, neither 8 nor 17 showed an impact on the growth of CFT073. And then, we explored their effects on the bacterial invasion in two human cell lines (5637 and SV-HUC-1). The results of the invasion of the WT or △ppk1 strain in 5637 and SV-HUC-1 cells obtained upon colony counts are shown in . As expected, the invasion ability of △ppk1 strain on both cells was very low when compared with the WT strain. On the other hand, 8 and 17 decreased, dose-dependently, the invasion of the WT CFT073 in 5637 and SV-HUC-1 cells. These results suggest that 8 and 17 may inhibit the invasion ability of CFT073 through interacting PPK1.
2.3. 8. And 17 were able to reduce the resistance to oxidative stress of CFT073
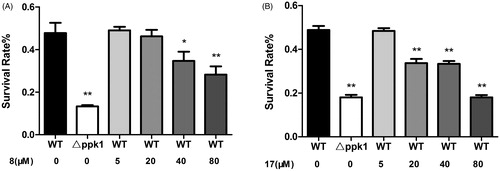
Oxidative stress resistance assay was further used to determine whether these compounds can alter the sensitivity of CFT073 to hydrogen peroxide. The survival rates of CFT073 in the presence and absence of the compounds were investigated. In the assay, △ppk1 strain was used as a control. The results showed that △ppk1 strain was very sensitive to hydrogen peroxide. On the other hand, the WT CFT073 after treated with 8 or 17 caused the strain exhibiting high susceptibility to hydrogen peroxide. As shown in , 8 and 17 enhanced the susceptibility of the WT CFT073 to hydrogen peroxide in a dose-dependent manner. These results suggest that 8 and 17 may cause E. coli cells more sensitive to oxidative stress by interfering with the biological function of PPK1.
2.4. 8. And 17 inhibit biofilm formation of CFT073
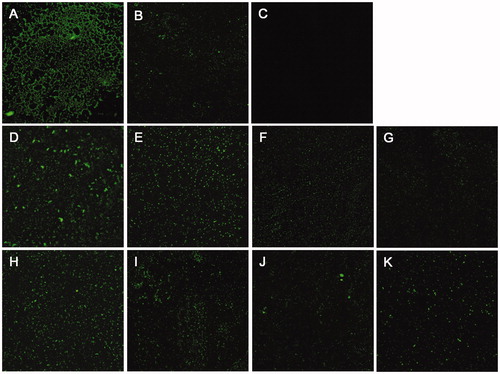
Since biofilm formation is related to the biological function of PPK1, colonisation, and morphology of biofilms on the glass slides in the presence and absence of 8 or 17 were evaluated. As expected, the biofilm formation can be observed obviously on the WT sample (). On the other hand, only a few biofilms can be detected on the △ppk1 strain (). and showed the results with the presence of 8 or 17 on the biofilm formation of CFT073 at different concentrations. These images obviously revealed that 8 and 17 can reduce dose-dependently the biofilm formation compared to the control (). The results suggested that 8 and 17 inhibit biofilm formation of CFT073 through disrupting the function of PPK1.
Figure 4. Effects of 8 and 17 on biofilms formation of CFT073. CLSM images (400×) of biofilms formed by WT CFT073 (A) and △ppk1 strain (B). (C) Fluorescent images of blank group. (D–G) Biofilms formed by WT CFT073 in the presence of 8 (5, 20, 40, and 80 μM, respectively). (H–K) Biofilms formed by WT CFT073 in the presence of 17 (5, 20, 40, and 80 μM, respectively).
2.5. 8. And 17 may prevent CFT073 infection in mice
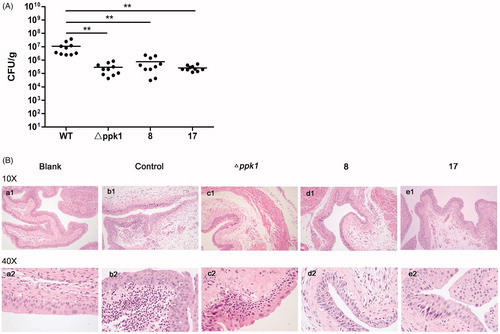
Since PPK1 is essential for virulence of bacteriaCitation21–23 and CFT073 is a uropathogenic strain, we further investigated the efficacy of 8 and 17 in the treatment and/or prevention of UTIs. To examine the therapeutic potential, 8 and 17 were determined for their ability to reduce bacterial burdens in the urinary tracts of C57BL/6J mice during acute UTIs. The results showed that high levels of bladder colonisation formed by CFT073 can be found in the control group. On the other hand, 8 and 17 can significantly reduce bacterial burdens in the bladder (p < 0.05) to the same level of △ppk1 group (). Furthermore, from the results of the morphological examination, a lot of neutrophil infiltration, edoema, and urinary bladder epithelium can be found in the control group (CFT073 treated, ) when compared with the blank group (). In contrast, inflammatory cells and the tissue edoema and epithelial damage were decreased in the group with compound treated (). These phenomena were observed in △ppk1 group (). These results reveal that the in vivo anti-UTI activity of 8 and 17 may be due to their inhibitory effect on PPK1.
Figure 5. Effects of 8 and 17 on the pathogenicity of CFT073. (A) The evaluation of the bacterial burdens in the bladder at 48 h after infection. All values represent the means of determinations. Each experiment was performed three times. Significant differences are marked by asterisks (*p < 0.05 vs. wild-type group; **p < 0.01 vs. wild-type group). (B) The histologic analysis of bladder tissues stained with HE staining (100× and 400×). (a, Blank: no infection; b, Control: infection by CFT073; c, △ppk1: infection by △ppk1 strain; d, 8: infection by CFT073 with 8; e: infection by CFT073 with 17).
2.6. Binding affinity of 8 and 17 with PPK1
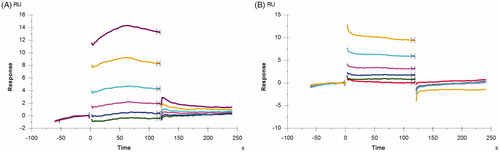
To further investigate the interactions between the compounds and PPK1, the SPR assay was used to detect the binding affinity of the compounds with PPK1. In the binding assays, PPK1 was immobilised in a sensor chip and the concentration of the compound in the running buffer used to evaluate their binding affinity to PPK1 was in the range of 6.25–200 μM. Sensorgrams of 8 and 17 obtained were shown in . The concentration-dependent binding responses were observed in both 8 and 17. The dissociation constant (KD) was obtained with the Biacore evaluation software 3.0. The KD values show that 8 and 17 have a similar binding affinity with PPK1. The KD values estimated were 85.8 and 84.6 μM, respectively.
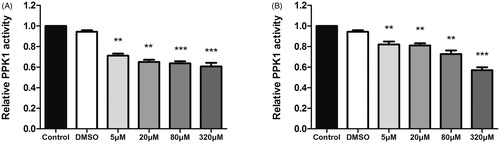
2.7. 8. And 17 inhibit the enzymatic activity of PPK1
Since PPK1 plays an important role in the synthesis of poly P and the binding affinity assay also shows that 8 and 17 may directly interact with PPK1, an enzymatic activity assay was used to determine whether the compounds can affect the enzymatic activity of PPK1. As shown in , 8 and 17 can inhibit the enzymatic activity of PPK1 in a dose-dependent manner. The results showed that compound 8 or 17 at 5 μM effectively reduced the PPK1 activity compared to the control and DMSO treated samples. These results suggest that 8 and 17 may prevent UPEC infection by interfering with the biological function of PPK1.
2.8. Molecular docking study
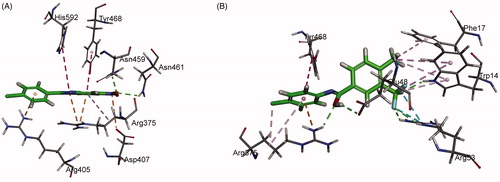
To have a better understanding of 8 and 17 interacting with PPK1 and facilitate the rational design of potential drugs based on their molecular scaffolds in the future, a molecular docking study was conducted using the Discovery Studio. As shown in , the top-scoring pose of compound 8 displays two hydrogen bonds between the nitro group of the molecule and amino acid residues Asn459/Asn461 of PPK1. An attractive charge interaction was predicted between the nitro group of 8 and Asp407 of PPK1. In addition, the benzimidazole and phenyl ring of the small molecule establishes hydrophobic interactions with the side chains of Arg375, Arg405, Tyr468, and His592. On the other hand, compound 17 establishes five conventional hydrogen bonds with Trp14, Glu48, Arg53, and Arg375 of PPK1. Arg53 was found to interact with the trifluoromethyl group of 17 via two halogen bonds. Moreover, a number of hydrophobic interactions can be found between the molecule and the side chains of Trp14, Phe17, Arg375, and Tyr468 ().
3. Conclusions
In summary, two potential PPK1-targeting small molecules were discovered through virtual screening. Their antibacterial activity and possible mode of action were thoroughly investigated. Antimicrobial tests confirmed that these compounds are able to reduce effectively the invasion ability to normal cells and resistance to oxidative stress of uropathogenic E. coli (CFT073) without any inhibitory effect on the growth of this strain. In addition, 8 and 17 can inhibit the biofilm formation of CFT073. Moreover, in a CFT073 infectious mice model, 8 and 17 were found to reduce bacterial burdens in the bladder and prevent urinary tract infection. The results of binding assay may prove that 8 and 17 directly interact with PPK1 with binding affinities (KD) of 85.8 and 84.6 μM, respectively. The enzymatic activity assays revealed that 8 and 17 effectively inhibit the biological function of PPK1. The molecular docking study predicted that several hydrophobic and hydrogen bond interactions could be established between 8 (or 17) and the amino acid residues of PPK1. Based on the results obtained, 8 and 17 may be good candidates for further development as the effective antibacterial agents for uropathogenic E. coli and urinary tract infections.
4. Materials and methods
4.1. Virtual screening workflow in silico
The workflow was carried out by using several modules embedded in Schrödinger Suite 2019 (www.schrodinger.com). The crystal structure of PPK1 (PDB ID: 1XDP) was prepared using the Protein Preparation Wizard module, which assigned bond orders, added hydrogen atoms, and removed water molecules. The PKU-CNCL (http://www.pkucncl.com/) and SPECS (http://www.specs.net/) chemical library were prepared previously using the LigPrep module with default settings. The library was further screened by the Glide module based on the 3 D structure of residues in the designated ATP binding pocket in PPK1.
All investigated structures in the database were docked into the defined pocket using the rigid docking model following a three-step procedure to estimate binding affinities. The first step was conducted using the standard-precision (SP) scoring function and the top 10% of the hits were retained to the extra-precision (XP) screening. The top 3% were then qualified for the final calculation of their MM-GBSA. At last, only 44 molecules were manually selected based on structural clusters to identify their biological activity.
4.2. Surface plasmon resonance (SPR) studies
SPR measurements were performed on a Biacore T200 system (GE Healthcare) using a CM7 sensor chip. The PPK1 protein was overexpressed and purified as previous reportCitation24, and immobilised on the sensor chip via using an amine coupling kit (GE Healthcare) with a response around 15000 RU. Solutions of tested compounds were prepared with a concentration of 100 μM with running buffer (10 mM PBS-P+, pH7.4, 137 mM NaCl, 2.7 mM KCl, 0.05% P20). The samples were injected at a flow rate of 100 µL/min for 60 s of the association phase, followed with 60 s of dissociation phase at 25 °C. The sensor chip was regenerated with Glycine-HCl (pH1.5) between consecutive measurements. The final graphs were obtained by subtracting blank sensorgrams from PPK1 sensorgrams. Data were analysed with Biacore evaluation software 3.0.
4.3. Bacterial growth curve
The bacterial cultures in the exponential phase of growth were inoculated in fresh LB broth to initial OD600 of 0.05 and then exposed to 100 or 200 μM of compounds and DMSO (vehicle control), respectively. The bacterial cultures were incubated at 37 °C without shaking and the OD600 was obtained at 0, 2, 4, 6, 8, 10, 12, 14, 16, 18, and 20 h. Growth curves were generated by plotting OD vs. time points in increasing time increments through graphing software (GraphPad Prism 5).
4.4. Invasion assay
To evaluate the impact of 8 and 17 on the invasion ability of CFT073 on uroepithelial cells, invasion assays were performed. △ppk1 strain was prepared as our previous studyCitation25. Liquid cultures of CFT073 and △ppk1 were grown aerobically in LB broth supplemented with different concentrations of compounds 8 and 17 (5, 20, 40, and 80 μM) and DMSO (vehicle control) at 37 °C for 18 h. 5637 cells and SV-HUC-1 cells (1 × 105 cells/well) were seeded respectively into 24-well polystyrene plates and were allowed to adhere overnight. After being washed with PBS, the cells were respectively infected with CFT073 and △ppk1 under each condition for 1.5 h. Cells were washed with PBS and then incubated in experimental medium containing gentamicin (100 μg/mL) for 1 h to kill extracellular bacteria. Finally, cells were washed with PBS and lysed with 0.5% Triton X-100. The released intracellular bacteria were enumerated by plating on LB agar plates. Each assay was conducted in triplicate and repeated at least three times. All values represent the means of triplicate determinations. Invasion% = (Bacteria after invasion/Bacteria before invasion) × 100%. Data are represented as mean ± SEM. Significant differences are marked by asterisks (*p < 0.05 vs. wild-type + 0 μM compounds group; **p < 0.01 vs. wild-type + 0 μM compounds group).
4.5. Oxidative stress resistance assay
Liquid cultures of CFT073 and △ppk1 were inoculated in fresh LB broth and were exposed to compounds (5, 20, 40, and 80 μM) and DMSO (vehicle control) with shaking. After 4 h incubation at 37 °C, the bacterial cultures were stimulated by hydrogen peroxide (50 mmol/L) for 30 min. And then, the cultures were serially diluted and plated onto LB agar plates. After incubation at 37 °C for 18 h, colonies were counted. The survival rates were calculated by dividing the number of colonies after stimulation by the number of colonies before stimulation. All these experiments were performed in triplicate at least three times. Survival rates = (Bacteria after stimulation/Bacteria before stimulation) × 100%. Data are represented as mean ± SEM. Significant differences are marked by asterisks (*p < 0.05 vs. wild-type + 0 μM compounds group; **p < 0.01 vs. wild-type + 0 μM compounds group).
4.6. Biofilm inhibition assay
Biofilms of the bacterial cultures can form on 1 × 1 cm glass piece kept inside the 6-well polystyrene plates. Compounds (5, 20, 40, and 80 μM) and DMSO (vehicle control) were supplemented to the wells containing the glass slides and the plates were incubated for 24 h to allow for biofilms formation at 37 °C. After incubation, the biofilms that were grown on the glass pieces were washed with PBS and stained with 0.01% acridine orange (AO). And then the biofilms were analysed with a confocal laser scanning microscope (LSM 800, Carl Zeiss 200, Germany). The viable bacteria with intact cell membranes appear fluorescent green. The images were acquired from random positions on the surfaces of the samples.
4.7. Animal efficacy studies
Prior to the establishment of the model, the mice bladders were gently squeezed to empty the urine. Mice were anaesthetised and inoculated via transurethral catheterisation with 50 μL bacterial suspension (∼1 × 108 to 2 × 108 CFU in total) in PBS. And then mice were transurethrally inoculated either with 50 mg/kg compounds or DMSO (control). Healthy control mice (blank) were injected with PBS and DMSO without bacteria into the bladder. Mice were killed 48 h posttreatment, and bladders were excised. The bladders were cut into two sections along the long axis. One was homogenised using a rotor-stator homogeniser, and bladder tissue homogenates were serially diluted and plated onto LB agar plates for quantification. The other was embedded in paraffin, cut into 4 μm thickness sections, and stained with Hematoxylin-Eosin (HE). A light microscope was used to observe the morphological changes and inflammatory cells infiltration of bladders.
4.8. Binding affinity study
The sensor chips and samples in this study were prepared as Section 4.2. Different concentrations (0, 6.25, 12.5, 25, 50, 100, and 200 μm) of 8 or 17 in running buffer were injected at a flow rate of 100 µL/min for 120 s of association phase, followed with 120 s of dissociation phase at 25 °C. The final graphs were obtained by subtracting blank sensorgrams from PPK1 sensorgrams. Data were analysed with Biacore evaluation software 3.0 in a steady state affinity model.
4.9. Enzymatic activity assay of PPK1
Five micrograms PPK1 proteins were incubated with 1 mM ATP in 500 μL of reaction buffer containing 50 mM Hepes-KOH (pH 7.2), 40 mM ammonium sulphate, 4 mM MgCl2, 2 mM creatine phosphate and 20 μg/mL creatine kinase. After incubation for 45 min at 37 °C, the reaction was stopped by adding 40 mM EDTA. The poly P was detected using DAPI. One unit of enzyme is defined as the amount incorporating 1 μmol of phosphate into poly P per min. PPK1 activity was calculated as follows:
V is the total volume of the reaction, and T is the reaction time.
4.10. Molecular docking
Molecular docking of 8 and 17 with PPK1 protein was carried out using the CDocker protocol of Discovery Studio (BIOVIA, San Diego, CA). Ligands were drawn and then optimised for docking through the small molecules’ tools of DS. The 2.5 Å crystal structure of the E. coli PPK1 (PDB code 1XDP)Citation8 was used to predict the potential binding pose of 8 and 17. The ATP binding area was defined as the binding pocket of these two compounds.
4.11. Statistical analysis
Data were expressed as mean values ± standard error of mean (SEM). All statistical tests were carried out through SPSS 16.0. Data were evaluated by analysis of variance (One-way ANOVA) and Mann–Whitney U test. Graphs were generated using GraphPad Prism 5. p-Value <0.05 was considered statistically significant (*), while p-Value <0.01 were considered as extremely significant (**).
4.12. Preparation and characterisation of 8 and 17
Please refer to the supplemental information.
Supplemental Material
Download PDF (844.2 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Gill CM, Hughes MA, LaPlante KL. A review of nonantibiotic agents to prevent urinary tract infections in older women. J Am Med Dir Assoc 2020;21:46–54.
- Malik RD, Wu YR, Christie AL, et al. Impact of allergy and resistance on antibiotic selection for recurrent urinary tract infections in older women. Urology 2018;113:26–33.
- Mody L, Juthani-Mehta M. Urinary tract infections in older women: a clinical review. JAMA 2014;311:844–54.
- Kalas V, Hibbing ME, Maddirala AR, et al. Structure-based discovery of glycomimetic FmlH ligands as inhibitors of bacterial adhesion during urinary tract infection. Proc Natl Acad Sci USA 2018;115:E2819–28.
- Ronald AR, Nicolle LE, Stamm E, et al. Urinary tract infection in adults: research priorities and strategies. Int J Antimicrob Agents 2001;17:343–8.
- Kallen AJ, Welch HG, Sirovich BE. Current antibiotic therapy for isolated urinary tract infections in women. Arch Intern Med 2006;166:635–9.
- McGann P, Snesrud E, Maybank R, et al. Escherichia coli harboring mcr-1 and blaCTX-M on a novel IncF plasmid: first report of mcr-1 in the United States. Antimicrob Agents Chemother 2016;60:4420–1.
- Willyard C. The drug-resistant bacteria that pose the greatest health threats. Nature 2017;543:15.
- Theuretzbacher U, Piddock LJV. Non-traditional antibacterial therapeutic options and challenges. Cell Host Microbe 2019;26:61–72.
- Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol 2015;13:269–84.
- Marston HD, Dixon DM, Knisely JM, et al. Antimicrobial resistance. JAMA 2016;316:1193–204.
- Seufferheld MJ, Alvarez HM, Farias ME. Role of polyphosphates in microbial adaptation to extreme environments. Appl Environ Microbiol 2008;74:5867–74.
- Zhu Y, Huang W, Lee SS, Xu W. Crystal structure of a polyphosphate kinase and its implications for polyphosphate synthesis. EMBO Rep 2005;6:681–7.
- Tiwari P, Gosain TP, Singh M, et al. Inorganic polyphosphate accumulation suppresses the dormancy response and virulence in Mycobacterium tuberculosis. J Biol Chem 2019;294:10819–32.
- Peng L, Luo WY, Zhao T, et al. Polyphosphate kinase 1 is required for the pathogenesis process of meningitic Escherichia coli K1 (RS218). Future Microbiol 2012;7:411–23.
- Akiyama M, Crooke E, Kornberg A. The polyphosphate kinase gene of Escherichia coli. Isolation and sequence of the ppk gene and membrane location of the protein. J Biol Chem 1992;267:22556–61.
- Haakenson CL, Crooke E. Role of polyphosphates in enhancing survival after DNA damage in Escherichia coli. Faseb J 2007;21:A626.
- Albi T, Serrano A. Inorganic polyphosphate in the microbial world. Emerging roles for a multifaceted biopolymer. World J Microbiol Biotechnol 2016;32:27.
- Bashatwah RM, Khanfar MA, Bardaweel SK. Discovery of potent polyphosphate kinase 1 (PPK1) inhibitors using structure-based exploration of PPK1Pharmacophoric space coupled with docking analyses. J Mol Recognit 2018;31:e2726.
- Saha SB, Verma V. in silico analysis of Escherichia coli polyphosphate kinase (PPK) as a novel antimicrobial drug target and its high throughput virtual screening against PubChem library. Bioinformation 2013;9:518–23.
- Campos F, Alvarez JA, Ortiz-Severin J, et al. Fluorescence enzymatic assay for bacterial polyphosphate kinase 1 (PPK1) as a platform for screening antivirulence molecules. Infect Drug Resist 2019;12:2237–42.
- Varas M, Valdivieso C, Mauriaca C, et al. Multi-level evaluation of Escherichia coli polyphosphate related mutants using global transcriptomic, proteomic and phenomic analyses. Biochim Biophys Acta Gen Subj 2017;1861:871–83.
- Rashid MH, Rumbaugh K, Passador L, et al. Polyphosphate kinase is essential for biofilm development, quorum sensing, and virulence of Pseudomonas aeruginosa. Proc Natl Acad Sci USA 2000;97:9636–41.
- Zhu Y, Lee SS, Xu W. Crystallization and characterization of polyphosphate kinase from Escherichia coli. Biochem Biophys Res Commun 2003;305:997–1001.
- Luo S, Peng L, Pan JY, Wu XM. Construction of uropathogenic Escherichia coli strain with ppk1 gene deletion and study on its biological properties. Chin J Microbiol Immunol 2013;33:531–6.