Abstract
Circadian rhythm is a universal life phenomenon that plays an important role in maintaining the multiple physiological functions and regulating the adaptability to internal and external environments of flora and fauna. Circadian alignment in humans has the greatest effect on human health, and circadian misalignment is closely associated with increased risk for metabolic syndrome, cardiovascular diseases, neurological diseases, immune diseases, cancer, sleep disorders, and ophthalmic diseases. The recent description of clock proteins and related post-modification targets was involved in several diseases, and numerous lines of evidence are emerging that small molecule modulators of circadian rhythms can be used to rectify circadian disorder. Herein, we attempt to update the disclosures about the modulators targeting core clock proteins and related post-modification targets, as well as the relationship between circadian rhythm disorders and human health as well as the therapeutic role and prospect of these small molecule modulators in circadian rhythm related disease.
1. Introduction
Circadian rhythm is the result of natural selection during the long-term evolution of organisms, enabling organisms to better adapt to changes in the external environmentCitation1,Citation2. Various behaviours and physiological functions of the body show obvious circadian rhythms, such as the sleep-wake cycleCitation3,Citation4, food intake and other autonomous activitiesCitation5, as well as physiological activities including blood pressureCitation6, blood lipids, coagulation-fibrinolysis balance, heart rateCitation7,Citation8, body temperatureCitation9, locomotor activityCitation10,Citation11, hormone levelsCitation12, cell metabolismCitation13, and cell proliferationCitation14,Citation15. The generation, maintenance, and regulation of circadian rhythms depend on the synergy of the circadian clock system, circadian input system, and circadian output system at the overall level () and at the cellular level, relying on the precise regulation of the endogenous circadian clock gene network (). Any abnormalities in these intrinsic rhythms can cause disturbances in the circadian rhythm.
Figure 1. The physiological basis for the generation and maintenance of mammalian circadian rhythm. Reproduced from Chen et al.Citation19
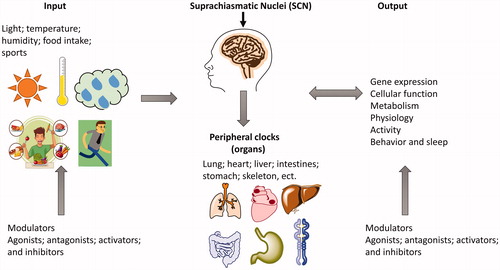
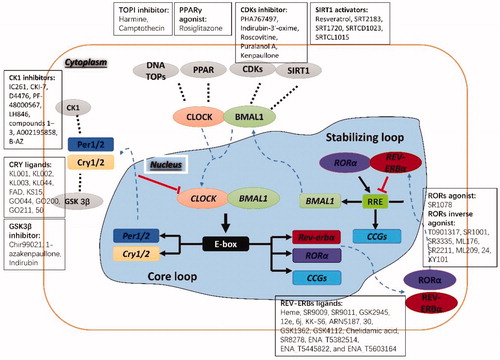
Figure 2. Molecular clock loops and their potential targets with representative small molecule modulators. CLOCK: circadian locomotor output cycles kaput; BMAL1: brain and muscle ARNT-like 1; CRY: cryptochrome; PER: period; ROR: RAR-related orphan receptor; RRE: retinoic acid receptor-related orphan receptor binding element; CCGs: clock-controlled genes; CK1: casein kinase 1; CDKs: cyclin-dependent kinases; GSK3β: glycogen synthase kinase 3β; SIRT1: silent information regulator 1; PPARγ: peroxisome proliferator-activated receptor γ; DNA TOPs: DNA topoisomerases. Reproduced from He and ChenCitation49. Copyright 2016 American Chemical Society.
The physiological basis for the generation and maintenance of circadian rhythms comprises the central and peripheral circadian clock systems, rhythm input systems, and rhythm output systems. The rhythm input system senses and transmits environmental synchronisation signals represented by light signals to the central circadian clock system. The central biological clock system acts as the circadian rhythm pacemaker through the output system to transmit the generated rhythm signals to the periphery, and cooperates with the endogenous biological clock system of the peripheral organs to maintain the physiological activity of the bodyCitation16 (). The circadian clock system is composed of the central circadian clock and the peripheral circadian clock. In mammals, the apex of this system is the suprachiasmatic nuclei (SCN) master pacemaker, which is considered the central or master clockCitation17. The SCN integrates the environmental time information (primarily light) via the retina to revamp or entrain its phase, and then mastermind other oscillators in extra-SCN brain regions and peripheral organsCitation18,Citation19. The rhythm output system is regulated by SCN, which can regulate gene expression, cellular function, metabolism, physiology, activity, behaviour, and sleep-wake cyclesCitation20. Additionally, the rhythm output systems in turn can affect the SCN master pacemakerCitation19,Citation20. For example, the arrhythmic food intake, excessive exercises, and sleep/circadian disorders affect SCN by remodel clock-controlled circuitCitation21–24.
Circadian rhythm production and maintenance are regulated by circadian clock genes. The molecular mechanism of the mammalian circadian clock is produced by a cell-autonomous feedback loopCitation25–27. The periodic oscillation of circadian rhythm depends on the precise regulation of the circadian clock gene and the clock-controlled gene regulatory network, including transcriptional-translational feedback loopsCitation28,Citation29 and the non-transcription mechanism of post-translational modificationCitation30,Citation31. As shown in , the transcriptional-translational feedback loops include a core loop and a secondary stabilisation loop.
In mammals, the transcription factors circadian locomotor output cycles kaput (CLOCK) and brain and muscle ARNT-like 1 (BMAL1) form a heterodimer, which binds to E-box enhancers to activate the target gene transcription of circadian clock gene Period (including Per1 and Per2) and Cryptochrome (including Cry1 and Cry2). When PER and CRY proteins accumulate to a certain extent, they could be further transferred from the cytoplasm to the nucleus, and the PER/CRY heterodimer as a negative regulator directly interacts with CLOCK/BMAL1 to inhibit its transcriptional activityCitation32. In the stabilisation loop, the CLOCK/BMAL1 heterodimer can also induce the expression of nuclear receptors REV-ERBα and RORα. As a negative regulator, REV-ERBα can bind to the retinoic acid receptor-related orphan receptor binding element (RRE) (sequence AGGTCA) in the BMAL1 promoter region and block the transcription of BMAL1Citation33. Conversely, RORα can be used as a positive regulator to bind to the RRE of the BMAL1 promoter region to promote the transcription of BMAL1, thereby forming an auxiliary loop for the transcription and translation oscillations of the circadian clock geneCitation34. However, beyond that, post-translational modifications (phosphorylation/dephosphorylation, acetylation/deacetylation, etc.) and degradation (ubiquitination/proteasome pathway) of various circadian proteins enable fine-tuning of the transcriptional-translational feedback loops (such as adjusting the expression phase and the period of oscillation), so it can also play an important role in the cyclical cycle of circadian rhythmsCitation30,Citation31. For example, PER and CRY proteins can be phosphorylated by casein kinase 1ε (CK1ε)/casein kinase 1ε (CK1δ), which affects the increase in the continuous length of the cycleCitation35. Silent information regulator 1 (SIRT1) regulates the expression of the clock gene BMAL1, Cry1, and Per2 by interacting with the CLOCK/BMALI complex and catalysing the deacetylation and degradation of the PER proteinCitation36.
Circadian alignment in humans has great effect on human health, and circadian misalignment has been involved in metabolic syndromeCitation37,Citation38, cardiovascular diseasesCitation39,Citation40, acute lung injury and inflammationCitation41, cancerCitation42–44, neurological diseasesCitation45,Citation46, and immune diseasesCitation47,Citation48. While accumulating evidence indicates that small molecule modulators of circadian rhythms can be used to rectify circadian disorderCitation18,Citation49–51, in this review, we pay attention to the recent progress of small molecule modulators targeting core clock proteins (such as CRYs, REV-ERBs, and RORs) and related post-modification targets (such as casein kinase 1 (CK1), cyclin-dependent kinases (CDKs), glycogen synthase kinase 3 (GSK3), cdc2-like kinase 1 (CLK1), breakpoint cluster region-Abelson tyrosine kinase (BCR-ABL), and silent information regulator 1 (SIRT1)), as well as the relationship between circadian rhythm disorders and human health and the therapeutic role and prospect of these small molecules in circadian rhythm related disease.
2. Overview of modulators targeting circadian rhythms
As mentioned above, circadian rhythms are associated with a variety of biological functions and biological dysfunctions. Efforts to develop initial modulators have focussed on the circadian clock, and modulators including endogenous and synthetic compounds have been discovered. The identified modulators can be classified into two broad categories, which are targeting core clock proteins and other or unknown targets.
2.1. Small molecule modulators of core clock proteins
2.1.1. Modulators for CRYs
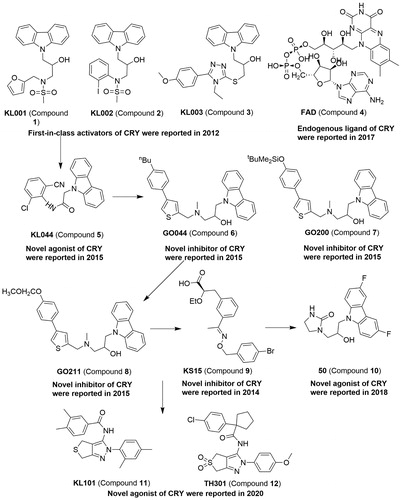
Compound 1 ( and ), the first-in-class small molecules, comprise carbazole derivative and an activator of cryptochromes (CRYs) Citation52. The carbazole derivatives, such as compound 1–3, can specifically interact with CRY1 and CRY2 and cause period lengthening and amplitude reduction in a dose-dependent manner in stable U2OS reporter cell lines harbouring Bmal1-dLuc or Per2-dLuc. Compound 1 can inhibit glucagon-induced gluconeogenesis by stabilising the CRYs. The co-crystal structure of murine CRY2 PHR core domain (1–512) with compound 1 has been reported, and shows that compound 1 can be readily located in the compound 4 (Flavin adenine dinucleotide, FAD)-binding pocket of CRY2Citation53. Compound 4 was also proven to be an endogenous ligand which can stabilise CRY proteins by competing with F-Box and leucine rich repeat protein 3 (FBXL3), thus lengthening the circadian periodCitation54. The complex of small molecule and protein is vital in understanding the binding mode and further improving the potency for acting as a modulator against protein. Therefore, the highly active compound 2-(9H-carbazol-9-yl)-N-(2-chloro-6-cyanophenyl)acetamide (compound 5) was disclosed under structure–activity relationship analysis and CRY2-compound 1 complex structureCitation55. Compound 5 can lengthen the circadian period, repress Per2 activity, and stabilise CRY better than compound 1. More interestingly, another group discovered a series of compound 1 derivatives, compounds 6–8, which can shorten the period by targeting cryptochrome in the mammalian circadian clockCitation56. Unfortunately, no physiological effects were reported by subsequent studies. The novel derivative of 2-ethoxypropanoic acid, compound 9, can inhibit the target CRY1 and CRY2Citation57. Compound 9 can enhance E-box-mediated transcription and attenuate the rhythm without affecting the period. Recently, the potent compound 1–(3-(3,6-difluoro-9H-carbazol-9-yl)-2-hydroxypropyl)imidazolidin-2-one (compound 10) significantly enhanced glucose clearance at 100 mg/kg in an oral glucose tolerance testCitation58. Furthermore, the compound N-(2–(2,4-dimethylphenyl)-2,6-dihydro-4H-thieno[3,4-c]pyrazol-3-yl)-3,4-dimethylbenzamide (compound 11) as a selective agonist for CRY1 and 1–(4-chlorophenyl)-N-(2–(4-methoxyphenyl)-5,5-dioxido-2,6-dihydro-4H-thieno[3,4-c]pyrazol-3-yl)cyclopentane-1-carboxamide (compound 12) as moderately selective agonist for CRY2 than CRY1 were reported by using human U2OS cells with a Bmal1 promoter-luciferase (Bmal1-dLuc) reporterCitation59. The X-ray crystal structures of CRY1 in complex with compound 11 and compound 12 show that these molecules were located in the FAD-binding pocket. As a useful tool for high selectivity against CRY isoform, the compound 11 and compound 12 were proved to facilitate brown adipocyte differentiation. Altogether, the modulators including the agonist or inhibitor of CRYs may be useful tools to treat circadian clock-related diseases through its action on CRY (see and ).
Table 1. Modulators targeting CRYs.
2.1.2. Modulators for REV-ERBs
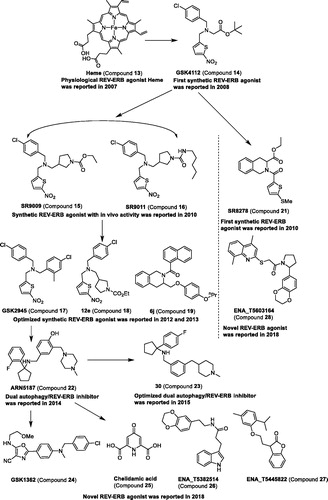
Endogenous ligands for REV-ERBs. In 2007, compound 13 was confirmed as a physiological ligand of nuclear receptors REV-ERBα (encoded by nuclear receptor subfamily 1, group D, member 1 (NR1D1)) and REV-ERBβ (Nuclear receptor subfamily 1, group D, member 2 (NR1D2)) by two research groups, Rastinejad et al. and Lazar et alCitation60,Citation61. Multiple biochemical and biophysical methods were used to demonstrate the association of compound 13 with ligand-binding domains of REV-ERB receptors, including mutation studies, transcriptional repressor function and repression of target gene transcription, ultraviolet-visible spectroscopy, mass spectrometry, isothermal titration calorimetry (ITC), and circular dichroism. Soon afterward, the crystal structure of REV-ERBβ in complex with compound 13 was also reportedCitation62,Citation63. All the results disclosed suggest that compound 13 can bind the REV-ERBs and is indeed a physiological ligand of nuclear receptors REV-ERBs. In mammalian cells, compound 13 can cause the recruitment of the co-repressor nuclear receptor corepressor-1 (NCoR) by targeting REV-ERB, giving rise to the repression of target genes including BMAL1 (also known as ARNTL)Citation60. Moreover, by targeting the REV-ERBα, compound 13 can suppress the expression of hepatic gluconeogenic gene and the output of glucoseCitation61. These findings would facilitate the development of small molecule modulators against REV-ERBs to treat diseases related to the dysfunctional disorder of metabolism and the mammalian clock.
Synthetic ligands for REV-ERBs. In 2008, the compound 1,1-dimethylethyl N-[(4-chlorophenyl)methyl]-N-[(5-nitro-2-thienyl)methyl]glycinate was reported by using REV-ERBα–NCoR fluorescence resonance energy transfer (FRET) assay, which showed an EC50 value of 250 nMCitation64. This compound was the first agonist of REV-ERBα and was competitive with compound 13. In subsequent studies, this compound was successively named SR6452 or GSK4112 (compound 14) ( and )Citation65,Citation66. Compound 14 can induce adipocyte differentiation in 3T3-L1 cells, enhance the recruitment of nuclear receptor co-repressor (NCoR) to REV-ERBα, and inhibit expression of the circadian target gene Bmal1. In addition, similar to compound 13, compound 14 also repressed the expression of gluconeogenic genes in liver cells and reduced glucose output in primary hepatocytesCitation66. These studies suggest that compound 14 may be used to treat diabetes or to modulate the circadian rhythm.
Table 2. Modulators targeting REV-ERBs.
Although compound 14 was used as a probe to investigate the pharmacological effects in in vitro, it has a poor pharmacokinetic profile with rapid clearance (Cint > 1.0 ml min−1 mg−1 protein) in rat liver microsomes and lower oral bioavailability (F ≤ 1% in mice)Citation66. Therefore, a series of analogues of compound 14 were synthesised by medicinal chemists to explore the applicable pharmacokinetics and pharmacodynamics used in in vivo studies.
The analogues of compound 14, the potent compounds 15–16, were disclosed by Burris et al.Citation67, which were the first REV-ERB agonists with in vivo activity. Compounds 15–16 can generate loss of locomotor activity during the subject dark phase and 1–3-h delay in the onset of nocturnal locomotor activity. The two compounds can alter the expression of the core clock genes, including Per2, Bmal1, Clock, Cry2, and Npas2. The ability of REV-ERB agonists in modulating the circadian behaviour of C57BL/6 mice may be used as a drug to treat sleep disorders and jet lag. Indeed, compound 15 was found to be able to induce wakefulness and reduce paradoxical sleep-rapid eye movement (REM) and slow-wave sleepCitation77–79.
As previously reported, the double-knockout REV-ERBα and REV-ERBβ mice can also markedly alter metabolic effectsCitation80. The administration of the agonist of REV-ERBα and REV-ERBβ, compound 16, gives rise to increase in energy expenditure and weight lossCitation67. In obese mice, including diet-induced obese mice and genetic model of obesity (OB/OB mice), REV-ERB agonist treatment results in a decrease in fat mass and plasma lipids. Recently, a study investigated further the metabolic profile of the nuclear receptor REV-ERB agonist. The results of the experiment show that the enzymatic isoforms mainly involved in the compound 15 phase I biotransformation pathways are cytochrome P450 3A4 (CYP3A4), cytochrome P450 3A5 (CYP3A5), cytochrome P450 2C19 (CYP2C19), and cytochrome P450 2D6 (CYP2D6)Citation81.
With the further study of REV-ERB agonist, compound 15 was associated with heart failureCitation82, cancerCitation83,Citation84, atherosclerosisCitation85, chikungunya and O’nyong’nyong virusCitation86, and autoimmune diseaseCitation87. However, Lazar et al.Citation88 discovered that compound 15 can decrease cell viability, rewire cellular metabolism, and alter gene transcription in hepatocytes and embryonic stem cells lacking both REV-ERBα and REV-ERBβ, which means that the effects of compound 15 cannot be used solely as surrogate for REV-ERB activity. Therefore, more efforts are needed to explore its mechanism of action. Highly selective compounds also need to be developed urgently.
According to published papers in the same period as compound 15, Kamenecka et al. also conducted structure-activity relationship analysis on compound 14. Compounds 18–19 show slightly better plasma and brain exposure as compound 14, but they displayed the best CNS exposure with brain penetration of 100% or 67%, respectivelyCitation69,Citation70. The analogue of compound 14, compound 17, was reported by Tomkinson et al.Citation68 Compound 17 shows > 1000-fold selectivity over liver X receptor α (LXRα) and is a potent agonist with REV-ERBα activity (EC50 = 0.05 μM), which may be the best compound with high selectivity and may serve as a pharmacological toolbox to investigate the biology of REV-ERBα. Recently, the novel small molecular compound 20 was disclosed, which can reinforce REV-ERBα activity by acting in a RORE-dependent manner, though not by the same mechanism as known REV-ERB agonists. It may also provide a new way of exploring the REV-ERB modulatorCitation71.
The first antagonist of REV-ERBα is compound 21 ( and ). Compound 21 is derived from compound 14 based on the tertiary amine scaffold. In HepG2 cells, compound 21 could increase the expression of either glucose 6-phosphatase (G6Pase) or phosphoenolpyruvate carboxykinase (PEPCK) mRNA expression by blocking the action of the endogenous agonistCitation72. Compound 21 also caused significant increases in the expression levels of growth/differentiation factor Growth and differentiation factor 10 (GDF10) and Growth and differentiation factor 15 (GDF15) in uterine endometrial stromal cells (UESCs). These results show that cellular oscillators may serve an important role of regulating the expression of downstream genes during the differentiation of UESCsCitation89.
Although the pharmacokinetic properties of small molecular compound 21 is poorCitation72, which has also been confirmed by our groupCitation90, compound 21 serves as a useful probe to explore the REV-ERB function by others. In vesicular stomatitis virus (VSV)-induced encephalitis model, administration of compound 21 increased C-C motif chemokine ligand 2 (CCL2) mRNA expression and decreased mice survival, which is associated with neuroprotective effects and lifetimeCitation91. The molecular connection between the circadian timing system and mood regulation was identified by Kim et al.Citation92 The circadian nuclear receptor REV-ERBα is associated with bipolar disorder, as it influences midbrain dopamine production and mood-related behaviour in mice. Treatment with compound 21 induced mania-like behaviour in association with a central hyperdopaminergic state. The evidence suggests that targeting REV-ERBα may be beneficial to the treatment of circadian rhythm-related affective disorders. Compound 21 could slow the progression of muscular dystrophy by increasing lean mass and muscle function and decreasing muscle fibrosis and muscle protein degradation in C57BL/10ScSn-Dmdmdx/J (mdx) miceCitation93. This research suggests that the antagonist compound 21 of REV-ERB may be a profound agent for the treatment of Duchenne muscular dystrophy (DMD). In conclusion, these results suggest that compound 21 is a unique chemical tool. However, it must be clearly recognised that poor pharmacokinetic properties of compound 21 also limit the further development of the compound. It is urgent to discover novel and potent compounds against REV-ERBs.
The novel dual autophagy/REV-ERB inhibitor compound 22 was revealed in 201473. Compound 22 can relieve the clock transcriptional repression mediated by REV-ERB and enhance the expression of REV-ERB target genes, Bmal1, Per1, and phosphoenolpyruvate carboxykinase (PEPCK), in BT-474 cells. It can also block autophagy by disrupting the lysosomal function and preventing autophagolysosome final maturation. Although the potency of compound 22 is under micromolar range, this compound provides an uncloaking the new measures to treat cancers. Therefore, Grimaldi et al.74 carried out structure–activity relationship (SAR) studies of compound 22 and finally obtained the potent compound 23 (1–(4-Fluorophenyl)-N-[[3-[(1-methyl-4-piperidyl)methyl]phenyl]methyl]cyclopentanamine) with 15-fold greater REV-ERBβ-inhibitory and cytotoxic activities compared to compound 22.
Recently, a novel oxazole inverse agonist of REV-ERB, compound 24, was discovered by Ray et al.Citation75 based on fluorescence resonance energy transfer (FRET) assay. Compound 24 showed a high selectivity over 20 nuclear receptors, which can reverse the degradation of REV-ERBα protein mediated by inflammatory stimuli. Subsequently, Gul et al.76 established a mammalian cell-based two-hybrid assay system and found compound 25 as a novel agonist of REV-ERB. In addition, three other compounds against REV-ERB, compounds 26–28 (), were found using this method. Compound 28 was confirmed as an antagonist, and compounds 26–27 were confirmed as agonists. Although the three compounds showed a poor selectivity against other targets, these compounds present a new kind of scaffold and can be used as a profound hit to reveal a drug-like compound.
2.1.3. Modulators for RORs
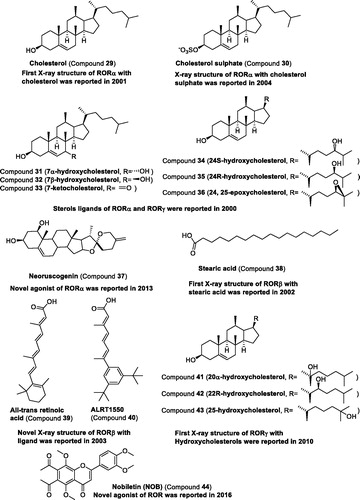
Natural ligands for RORs. In 2002, the first ligand of RORα, compound 29, was proved by X-ray structure (PDB entry 1N83). It is present in the ligand-binding pocket (LBP) and is important in designing the ligand targeting RORsCitation94. The analogue of compound 29, compound 30, can also bind to RORα as confirmed by the crystal structure (PDB entry 1S0X)Citation95. Other sterols including oxysterols as ROR inverse agonists and neoruscogenin as RORα agonist were found and reviewed in other papersCitation96,Citation97. The representative structure of sterols (compounds 31–37) is presented in to analyse the structure for researchers. In 2001, the first ligand of RORβ, compound 38, was proved by X-ray structure (PDB entry 1K4W)Citation98. This crystal structure of compound 38 and the ligand-binding domain (LBD) of the rat RORβ shed new light on the development of ligands against RORs. Subsequently, the crystal structure of the complex between compound 38 and RORβ (PDB entry 1N4H) was solved by Schüle groupCitation99. They also solved the crystal structure of the complex between synthetic analog compound 40 and RORγ (PDB entry 1NQ7). All these two-crystal structures present similar results, namely, the compound 39 and analogs were binding to the RORβ ligand-binding domain (LBD). Hydroxycholesterols (compounds 41–43) were binding to the RORγ LBD using the same method in 2010, with accession codes 3KYT (RORγ/Compound 41), 3L0J (RORγ/Compound 42), and 3L0L (RORγ/Compound 43), respectivelyCitation100. Recently, the natural compound 44 as an agonist for the ROR was reported by using ClockΔ19/+ cells with PER2::Luc reporterCitation101,Citation102. The potent natural compound and all these crystal structures of the complex between natural ligand and ROR have inspired researchers to search for potent and selective small molecule modulators targeting RORs ().
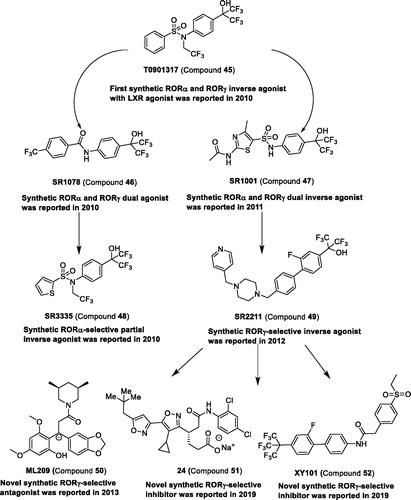
Synthetic ligands for RORs. In 2010, using cell-based GAL4-NR LBD cotransfection assay, Griffin et al.Citation103 found the first RORα/γ inverse agonist compound 45, which was also the agonist of the liver X receptor (LXR)Citation104. Compound 45 was binding to RORα/γ but not to RORβ. This compound provided the scaffold to further exploit the potent and selective ligands targeting ROR. A compound with multiple targets is not an ideal tool to disclose the function of protein. Therefore, the core scaffold of compound 45 was optimised, and a round of agonists or inverse agonists against RORα/γ, RORα, and RORγ were reported. These compounds have been reviewed elsewhereCitation96,Citation97. The representative compounds can be found in and to systematically review the research studies.
Table 3 Representative modulators targeting RORs.
The first synthetic RORα-selective partial inverse agonist compound 48 based on the core scaffold of compound 45Citation103 and compound 46Citation105 was reported in 2010Citation107. Compound 48 can inhibit the constitutive transactivation activity of RORα with an IC50 of 480 nM, but it cannot inhibit the activity of LXRα, RORβ, and RORγ. Compound 48 can suppress hepatic gluconeogenesis and improve glucose homeostasis in vivo, suggesting that compound 48 may be a potential tool to treat type 2 diabetes.
After structure–activity relationship (SAR) studies of compound 47, the potent and selective inverse agonist compound 49 targeting RORγ was obtained, which can reduce the conformational mobility of RORγ LBD. The other potent and selective agonists, inverse agonists, or inhibitors of RORγ were reviewed elsewhereCitation50,Citation112. Recently, 4-(isoxazol-3-yl) butanoic acid derivatives as high selective inhibitors of RORγ were reported. The potent compound 51 showed commendable anti-inflammatory effects in a mouse dermatitis model. A novel compound 52, 2–(4-(ethylsulfonyl)phenyl)-N-(2′-fluoro-4′-(1,1,1,3,3,3-hexafluoro-2-(trifluoromethyl)propan-2-yl)-[1,1′-biphenyl]-4-yl)acetamide, in complex with the RORγ ligand binding domain (LBD), was reportedCitation111. Compound 52 possess good metabolic stability and pharmacokinetic profile, and shows a significant tumour growth inhibition in vivo.
2.2. Small molecule modulators with other or unknown targets
Compounds targeting other proteins including kinase, epigenetic proteins, and others can also alter circadian characteristics. All these compounds are summarised as follows.
2.2.1. Modulators for kinases
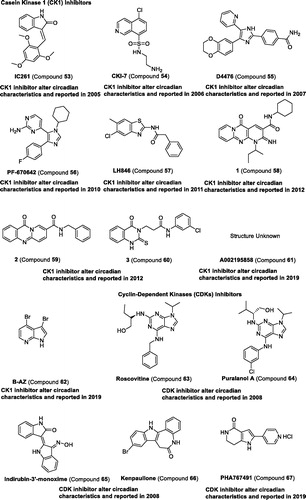
Casein kinase 1 (CK1). The casein kinase family comprises seven distinct genes encoding CK1 isoforms (α, α2, γ1, γ2, γ3, δ, and ε) in mammalsCitation113. CK1δ and CK1ε have been discovered to regulate the circadian clock, and their substrates are proved to be PER1, PER2, BMAL1, and CRYsCitation114. CK1ε-selective inhibitor compound 53 can increase in period length, leading to about 1.2-h in synchronised Rat-1 (mPer1::luc) cellsCitation115. Afterward, compounds 54–60 ( and ) were also proven to lengthen the period in cultured cells and were reviewed in other papers Citation49,Citation116. Recently, compound 61 was identified as a regulator to increase period length in mammalian cells and larval zebrafish assayCitation117. Compound 62 lengthens the period through CK1 inhibitionCitation118. All these studies reveal that the role of CK1 is important in the regulation of circadian rhythmCitation119.
Figure 7. Development and structure of synthetic modulators targeting kinases.
Table 4 Representative modulators targeting kinases.
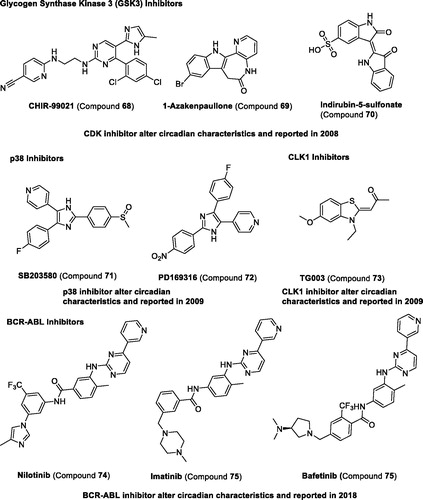
Cyclin-dependent kinases (CDKs). The cyclin-dependent kinase family comprises 11 distinct genes encoding CDK isoforms (1–11)113. CDK5 has been reported to directly phosphorylate CLOCKCitation126, inhibitor compound 63 targets CDK1, CDK2, and CDK5, and compound 64 targets CDK2, CDK4, and CDK5 can lengthen the circadian periodCitation123. However, the multi-target inhibitors, compounds 65–66 targeting both CDK and GSK3, were proved to shorten the circadian period. Recently, compound 67, an inhibitor of CDK7 and CDK9, has been reported to increase period length in mammalian cellsCitation124.
Other kinases. As other studies in the literature have reported, glycogen synthase kinase 3β (GSK3β) can also regulate the circadian clock, which can phosphorylate CLOCK, PER, REV-ERB, and CRY proteinsCitation114. The selective GSK3β inhibitors, compounds 68–70, have been reported to shorten the circadian periodCitation123. Other kinase inhibitors including compounds 71–72 targeting p38 and compound 73 targeting CLK1 have been reported to increase period lengthCitation125. Recently, compounds 74–76 selective BCR-ABL tyrosine kinase inhibitor were found to shorten the circadian periodCitation51 ( and ).
2.2.2. Modulators for epigenetic proteins and others
Silent information regulator 1 (SIRT1) has been found to contribute to circadian control, which regulates circadian clock gene expression through PER2 deacetylationCitation36,Citation127. SIRT1 activator compound 77 is involved in physiological rhythms and clock gene expressionCitation128. The potent SIRT1 activators, compounds 78–81, also show that they can reduce circadian expression, lengthen period, and reduce amplitudeCitation128. Recently, SIRT6 was also found to regulate circadian rhythms via Per2Citation129. However, the small molecules of SIRT6 have not been tested by researchers.
In a recent study, peroxisome proliferator-activated receptor γ (PPARγ) was involved in regulating the expression of Bmal1 and REV-ERBα, and its agonist compound 82 can induce expression of Bmal1131. Compounds 83–84, DNA topoisomerase (TOP) inhibitors, were also found to enhance the circadian expression and lengthen the circadian periodCitation132. Recently, the androgen antagonist and oestrogen activator compound 85 was found to shorten the circadian periodCitation51 ( and ).
Figure 8. Development and structure of synthetic modulators targeting epigenetic proteins and others.
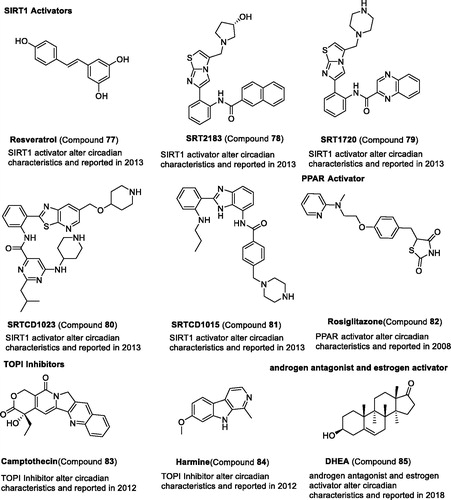
Table 5. Representative modulators targeting epigenetic proteins.
3. Implications in circadian rhythm-related diseases
Circadian rhythm plays a very important role in the normal maintenance of organisms, but physical and psychological influences including jet lag, shift work, and diseases can cause a misalignment of the intrinsic oscillators. Jet lag occurs in individuals travelling across multiple time zones, who may suffer from some symptoms including disruption of sleep, gastrointestinal disturbances, decreased vigilance and attention span, a general feeling of malaise, and an increased risk of cancer and heart diseaseCitation133,Citation134. Shift work is apparent among people employed in factories or social event firms and work from 7 pm to 9 amCitation135. Shift work has become a common phenomenon in society, and was found to be involved in cancer, cardiovascular disease, depression, and infertility. Jet lag and shift work induce rhythm disorder, which can cause a mass of psychological, nervous system, mental health, and physical health problemsCitation135. Beyond all that, diseases are closely related to circadian rhythms. Diseases can cause disturbances in circadian rhythms, and disorder in circadian rhythms, in turn, further aggravate the severity of the diseaseCitation136,Citation137. This section will focus on the relationship between disease and circadian rhythm disorders ().
3.1. Metabolic diseases
Circadian rhythm has been associated with homeostasis and physiology, which is closely related to physical healthCitation138. Numerous lines of evidence are emerging that circadian dysfunctions are closely associated with increased risk for metabolic disease such as obesity and diabetesCitation136,Citation139. Evidence that the circadian rhythm is associated with energy homeostasis, glucose homeostasis, and lipid homeostasis has been found. Homozygous Clock mutant mice can lead to type 2 diabetes mellitus, with metabolic syndromes of hyperleptinemia, hyperlipidaemia, hepatic steatosis, and hyperglycaemia, with insufficient compensatory insulin production. Clock mutant animals can induce obesity, hyperphagia, reduced energy expenditure, adiposity, as well as dysregulation of glucose and lipid metabolismCitation140. The core clock genes Clockmut or Bmal1−/− depress and abolish gluconeogenesisCitation141. Clockmut also induced hypertriglyceridaemia in animal modelsCitation140. REV-ERBα knockout mice also displayed altered lipid and bile metabolismCitation142. Subsequent studies have shown that double knockout mice (REV-ERBα and REV-ERBβ) have disorganised lipid homeostatic gene networksCitation80. The other core circadian rhythm gene ROR also turns out to be related to the regulation of energy homeostasis and several lipid and glucose metabolic genesCitation143. Mutant RORα mice (also known as staggerer mice) display hypo-α-lipoproteinemiaCitation144. Recent studies have shown that RORα accommodates peripheral glucose tolerance, torpor, and hepatic lipid metabolism by regulating the expression of fibroblast growth factor 21 (FGF21)Citation145,Citation146. All of these pieces of evidence suggest that the circadian rhythm is associated with metabolism and that clock proteins can be as drug targets to treat metabolic diseases.
Many small molecule modulators of circadian proteins have been found to be useful in metabolic diseases. The CRYs activator compound 1 has been shown to inhibit glucagon-induced gluconeogenesis, which may provide a foundation for the treatment of diabetesCitation53,Citation147. Aside from the ligands of CRYs, the ligands of circadian nuclear receptors REV-ERB and RORs also demonstrated that they can be conducive to regulate metabolism in vivo. Compounds 15–16 as agonists of REV-ERBα and REV-ERBβ proved highly effective in the improvement of the metabolic profile in obese miceCitation67. Recently, Chen et al.Citation101 identified that compound 44 as an agonist for ROR can potently protect against metabolic syndrome and remodel the circadian and metabolic gene expression in diet-induced obese mice. Subsequently, they demonstrate that compound 44 can serve as a potential drug to treat the metabolic disorders and age-related decline by regulating cholesterol and bile acid metabolismCitation148 and overcome the metabolic challenge by enhancing mitochondrial respiration in skeletal muscleCitation149. Therefore, with an in-depth study of the mechanism for clock proteins and the discovery of selective and potent small molecule modulators, it is believed that in the near future, the ligands of CRYs, REV-ERBs, or RORs will provide first-class treatment for metabolic diseases such as obesity and diabetes.
3.2. Sleep disorders
Sleep plays a very important role in the biological process of all creatures; it is regulated by circadian rhythm and homeostatic mechanismsCitation150. Normal circadian rhythms play an irreplaceable role in sleep. Circadian misalignments such as jet lag, shift work, and sleep deprivation have resulted in sleep disordersCitation134,Citation135. Kiessling’s group and Yamaguchi’s group identified that the different organs of mice showed heterogeneity entrainment kinetics in an experimental paradigm for jet lagCitation151,Citation152. The rhythm gene has been linked to sleep disorders. Mutations in both PER2 (PER2 S662G) and CSNK1D (CK1δ T44A) have been involved in familial advanced sleep phase syndrome (FASPS)Citation153. Recent studies indicate that the core clock gene expression has a close association with sleep apnoea (SA). Canales et al.Citation154 identified that the Per3 expression of SA was lower than that in the normal group. Pharmacological treatment targeting the mammalian clock has been shown to have beneficial effects on sleep architectureCitation78. Compound 16 as an agonist of REV-ERBα and REV-ERBβ displays increase in wakefulness and reduction of paradoxical sleep-rapid eye movement (REM) sleep and slow-wave sleep in vivoCitation67,Citation78. Therefore, the REV-ERB ligands may be beneficial in treating sleep disorders.
3.3. Ophthalmic diseases
As widely appreciated, light has profoundly influenced the mammalian circadian rhythm. Light is mainly received by intrinsically photosensitive retinal ganglion cells (ipRGCs)Citation155. A large number of studies show that the knockout of the rhythm gene affects retinal processing of light informationCitation156–160. The circadian rhythm is involved in ophthalmic diseases including glaucoma, macular degeneration, cataract, retinitis pigmentosa, diabetic retinopathy, and optic nerve atrophy. Evidence is accumulating that glaucoma directly damages the light input into the circadian system and causes optic nerve dysfunctionCitation161,Citation162. Recently, a mass of transcripts of nocturnal rodents and diurnal primates with daily and circadian oscillations were presented by RNA Sequencing (RNA-Seq) technologyCitation163–165. Panda et al.Citation163 identified that around 4–12% of the transcripts are rhythmic in the cornea, optic nerve head, retina, and retinal pigment epithelium for young male baboons (Papio anubis). Recently, we also disclosed that 3% and 24% of the transcripts are rhythmic in the murine extraorbital lacrimal glands and murine corneaCitation164,Citation165. In addition, FitzGerald et al. identified that structural modification of the cornea and the lens was observed in Bmal1 knockout miceCitation157,Citation158. Moreover, rhythm disorders can further aggravate diabetic retinopathy in per2 knockout miceCitation160. In previous studies, we found that compound 21 as an antagonist of REV-ERBα can enhance corneal wound healingCitation166. Therefore, the small molecule modulators of circadian proteins provide a potential solution for the treatment of ophthalmic diseases.
3.4. Other diseases
The impact of the circadian system on immune diseasesCitation166,Citation167, mood disordersCitation168,Citation169, neuropsychiatric diseasesCitation18,Citation170, agingCitation171,Citation172, renal diseases (such as hypertension, chronic kidney disease, renal fibrosis, and kidney stones)Citation173,Citation174, and cancerCitation175,Citation176 has been reviewed by others. As described in section 2, small molecule modulators of circadian proteins supply pharmacological tools to treat these diseases. For example, the REV-ERB ligand compound 14 can regulate innate immune responses by repressing interleukin 6 (il6)Citation92. Interestingly, Kim et al. identified that the pharmacological inhibition of REV-ERBα activity produces mania-like behaviour. The mice showed more hyperactive behaviour after the administration of REV-ERBα antagonist compound 21Citation177. The REV-ERBα agonist may be useful for mood regulation.
4. Perspectives and concluding remarks
In this review, we detailed all aspects of the physiological basis, molecular clock loops, biological function, potential targets, and small molecule modulators of circadian rhythm. The generation, maintenance, and regulation of circadian rhythms depend on the synergy of the circadian clock system, circadian input system, and circadian output system at the overall level. In particular, the circadian clock system is composed of the central circadian clock and the peripheral circadian clock. The apex of this system is the SCN master pacemaker in mammals. The periodic oscillation of circadian rhythm depends on the precise regulation of the circadian clock gene and the clock-controlled gene regulatory network, including transcriptional-translational feedback loops and the non-transcription mechanism of post-translational modification.
Extensive research has been performed on the relationship between circadian clock disorder and disease. Circadian clock genes knockout has confirmed that circadian misalignment is involved in metabolic syndrome, cardiovascular diseases, acute lung injury and inflammation, neurological diseases, immune diseases, cancer, mood disorders, sleep disorders, and ophthalmic diseases. As summarised in this article, circadian rhythms are important for human health, which suggests that the development of small molecules is imminent and could be used to treat circadian rhythm related diseases.
More importantly, a large number of small molecule modulators of circadian rhythm have been discovered, and most modulators have potential therapeutic effects on disease. In order to identify hits of the circadian clock, hundreds of thousands of compounds have been filtered by cell-based high-throughput circadian assays. The effectiveness of chemical biology approaches contributed to the discovery of the small molecule modulators of circadian rhythmCitation178. In recent years, with the emergence and popularisation of some new technologies, biophysical methods (such as differential scanning fluorimetry, differential scanning calorimetry, isothermal titration calorimetry, and surface plasmon resonance) and computer-aided drug design will help in the discovery of more modulators targeting clock proteins. It is believed that in the near future, small molecule modulators will be a useful tool in the treatment of circadian rhythm related diseases.
Author contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Takahashi JS. Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet 2017;18:164–79.
- Bell-Pedersen D, Cassone VM, Earnest DJ, et al. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Genet 2005;6:544–56.
- Lopez R, Barateau L, Dauvilliers Y. [Normal organization of sleep and its changes during life]. Rev Prat 2019;69:537–45.
- Abbott SM, Reid KJ, Zee PC. Circadian Rhythm Sleep-Wake Disorders. Psychiatr Clin North Am 2015;38:805–23.
- Challet E. The circadian regulation of food intake. Nat Rev Endocrinol 2019;15:393–405.
- Smolensky MH, Hermida RC, Portaluppi F. Circadian mechanisms of 24-hour blood pressure regulation and patterning. Sleep Med Rev 2017;33:4–16.
- Massin MM, Maeyns K, Withofs N, et al. Circadian rhythm of heart rate and heart rate variability. Arch Dis Child 2000;83:179–82.
- Sim SY, Joo KM, Kim HB, et al. Estimation of Circadian Body Temperature Rhythm Based on Heart Rate in Healthy, Ambulatory Subjects. IEEE J Biomed Health Inform 2017;21:407–15.
- Touitou Y, Mauvieux B, Reinberg A, et al. Disruption of the circadian period of body temperature by the anesthetic propofol. Chronobiol Int 2016;33:1247–54.
- Refinetti R, Kenagy GJ. Circadian rhythms of body temperature and locomotor activity in the antelope ground squirrel, Ammospermophilus leucurus. J Therm Biol 2018;72:67–72.
- Yamaguchi Y, Suzuki T, Mizoro Y, et al. Mice genetically deficient in vasopressin V1a and V1b receptors are resistant to jet lag. Science 2013;342:85–90.
- Huang Y, Xu C, He M, et al. Saliva cortisol, melatonin levels and circadian rhythm alterations in Chinese primary school children with dyslexia. Medicine (Baltimore) 2020;99:e19098.
- Reinke H, Asher G. Crosstalk between metabolism and circadian clocks. Nat Rev Mol Cell Biol 2019;20:227–41.
- Shostak A. Circadian Clock, Cell Division, and Cancer: From Molecules to Organism. Int J Mol Sci 2017;18:873.
- Gaucher J, Montellier E, Sassone-Corsi P. Molecular Cogs: Interplay between Circadian Clock and Cell Cycle. Trends Cell Biol 2018;28:368–79.
- Hirota T, Fukada Y. Resetting Mechanism of Central and Peripheral Circadian Clocks in Mammals. Zool. Sci 2004;21:359–68.
- Ralph MR, Foster RG, Davis FC, et al. Transplanted suprachiasmatic nucleus determines circadian period. Science 1990;247:975–8.
- Cha HK, Chung S, Lim HY, et al. Small Molecule Modulators of the Circadian Molecular Clock With Implications for Neuropsychiatric Diseases. Front Mol Neurosci 2018;11:496.
- Chen Z, Yoo SH, Takahashi JS. Development and therapeutic potential of small-molecule modulators of circadian systems. Annu Rev Pharmacol Toxicol 2018;58:231–52.
- Froy O, Miskin R. The interrelations among feeding, circadian rhythms and ageing. Prog Neurobiol 2007;82:142–50.
- Saito M, Murakami E, Suda M. Circadian rhythms in disaccharidases of rat small intestine and its relation to food intake. Biochim Biophys Acta 1976;421:177–79.
- Fernández MP, Berni J, Ceriani MF. Circadian remodeling of neuronal circuits involved in rhythmic behavior. PLoS Biol 2008;6:e69–e69.
- Tomioka K, Matsumoto A. A comparative view of insect circadian clock systems. Cell Mol Life Sci 2010;67:1397–406.
- Gangwisch JE. Epidemiological evidence for the links between sleep, circadian rhythms and metabolism. Obe Rev 2009;10:37–45.
- Oishi K, Miyazaki K, Kadota K, et al. Genome-wide expression analysis of mouse liver reveals CLOCK-regulated circadian output genes. J Biol Chem 2003;278:41519–27.
- Yamazaki S, Numano R, Abe M, et al. Resetting central and peripheral circadian oscillators in transgenic rats. Science 2000;288:682–5.
- Yoo SH, Yamazaki S, Lowrey PL, et al. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc. Natl. Acad. Sci. U.S.A 2004;101:5339–46.
- Takahashi JS. Molecular components of the circadian clock in mammals. Diabetes Obes Metab 2015;17:6–11.
- Liu AC, Lewis WG, Kay SA. Mammalian circadian signaling networks and therapeutic targets. Nat Chem Biol 2007;3:630–9.
- Kojima S, Shingle DL, Green CB. Post-transcriptional control of circadian rhythms. J Cell Sci 2011;124:311–20.
- Gallego M, Virshup DM. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol 2007;8:139–48.
- Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature 2002;418:935–41.
- Preitner N, Damiola F, Zakany J, et al. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 2002;110:251–60.
- Cermakian N, Sassone-Corsi P. Multilevel regulation of the circadian clock. Nat Rev Mol Cell Biol 2000;1:59–67.
- Meng Q-J, Logunova L, Maywood ES, et al. Setting clock speed in mammals: the CK1 epsilon tau mutation in mice accelerates circadian pacemakers by selectively destabilizing PERIOD proteins. Neuron 2008;58:78–88.
- Asher G, Gatfield D, Stratmann M, et al. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 2008;134:317–28.
- Maury E, Hong H, Bass JJD. Circadian disruption in the pathogenesis of metabolic syndrome. Diabetes Metab 2014;40:338–46.
- Gale JE, Cox HI, Qian J, et al. Disruption of circadian rhythms accelerates development of diabetes through pancreatic beta-cell loss and dysfunction. J Biol Rhythms 2011;26:423–33.
- Shanmugam V, Wafi A, Al-Taweel N, et al. Disruption of circadian rhythm increases the risk of cancer, metabolic syndrome and cardiovascular disease. Global Health Sci 2013;2013:1–42.
- Jiang T, Ji S, Yang G, et al. [Research advances in relationship between biological clock and cardiovascular diseases]. Sheng Li Xue Bao 2019;71:783–91.
- Yu D, Fang X, Xu Y, et al. Rev-erbα can regulate the NF-κB/NALP3 pathway to modulate lipopolysaccharide-induced acute lung injury and inflammation. Int Immunopharmacol 2019;73:312–20.
- Fu L, Lee CC. The circadian clock: pacemaker and tumour suppressor. Nat Rev Cancer 2003;3:350–61.
- Sulli G, Lam M, Panda S. Interplay between circadian clock and cancer: new frontiers for cancer treatment. Trends Cancer 2019;5:475–94.
- Shafi AA, Knudsen KE. Cancer and the Circadian Clock. Cancer Res 2019;79:3806–14.
- Haspel J. Mind your bedtime: The circadian clock and mTOR in an orphan brain disease. Sci Transl Med 2017;9:aao2261.
- Leng Y, Musiek ES, Hu K, et al. Association between circadian rhythms and neurodegenerative diseases. Lancet Neurol 2019;18:307–18.
- Weintraub Y, Cohen S, Chapnik N, et al. Does the circadian clock have a role in the pathogenesis of inflammatory bowel disease (IBD)?. J Crohns Colitis 2018; 12:S270–S71.
- Scheiermann C, Gibbs J, Ince L, et al. Clocking in to immunity. Nat Rev Immunol 2018;18:423–37.
- He B, Chen Z. Molecular targets for small-molecule modulators of circadian clocks. Curr Drug Metab 2016;17:503–12.
- Cyr P, Bronner SM, Crawford JJ. Recent progress on nuclear receptor RORγ modulators. Bioorg Med Chem Lett 2016;26:4387–93.
- Tamai TK, Nakane Y, Ota W, et al. Identification of circadian clock modulators from existing drugs. EMBO Mol Med 2018;10:e8724.
- Hirota T, Lee JW, John PCS, et al. Identification of small molecule activators of cryptochrome. Science 2012;337:1094–97.
- Nangle S, Xing W, Zheng N. Crystal structure of mammalian cryptochrome in complex with a small molecule competitor of its ubiquitin ligase. Cell Res 2013;23:1417–19.
- Hirano A, Braas D, Fu Y-H, et al. FAD regulates CRYPTOCHROME protein stability and circadian clock in mice. Cell Rep 2017;19:255–66.
- Lee JW, Hirota T, Kumar A, et al. Development of Small-Molecule Cryptochrome Stabilizer Derivatives as Modulators of the Circadian Clock . ChemMedChem 2015;10:1489–97.
- Oshima T, Yamanaka I, Kumar A, et al. C-H Activation Generates Period-Shortening Molecules That Target Cryptochrome in the Mammalian Circadian Clock. Angew Chem Int Ed Engl 2015;54:7193–97.
- Chun SK, Jang J, Chung S, et al. Identification and validation of cryptochrome inhibitors that modulate the molecular circadian clock. ACS Chem. Biol 2014;9:703–10.
- Humphries PS, Bersot R, Kincaid J, et al. Carbazole-containing amides and ureas: Discovery of cryptochrome modulators as antihyperglycemic agents. Bioorg Med Chem Lett 2018;28:293–97.
- Miller S, Son YL, Aikawa Y, et al. Isoform-selective regulation of mammalian cryptochromes. Nat Chem Bio 2020;16:676–85.
- Raghuram S, Stayrook KR, Huang P, et al. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBalpha and REV-ERBbeta. Nat Struct Mol Biol 2007;14:1207–13.
- Yin L, Wu N, Curtin JC, et al. Rev-erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science 2007;318:1786–89.
- Pardee KI, Xu X, Reinking J, et al. The structural basis of gas-responsive transcription by the human nuclear hormone receptor REV-ERBbeta. PLoS Biol 2009;7:e43.
- Matta-Camacho E, Banerjee S, Hughes TS, et al. Structure of REV-ERBβ ligand-binding domain bound to a porphyrin antagonist. J Biol Chem 2014;289:20054–66.
- Meng QJ, McMaster A, Beesley S, et al. Ligand modulation of REV-ERBalpha function resets the peripheral circadian clock in a phasic manner. J Cell Sci 2008;121:3629–35.
- Kumar N, Solt LA, Wang Y, et al. Regulation of adipogenesis by natural and synthetic REV-ERB ligands. Endocrinology 2010;151:3015–25.
- Grant D, Yin L, Collins JL, et al. GSK4112, a small molecule chemical probe for the cell biology of the nuclear heme receptor Rev-erbα. ACS Chem Biol 2010;5:925–32.
- Solt LA, Wang Y, Banerjee S, et al. Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists. Nature 2012;485:62–8.
- Trump RP, Bresciani S, Cooper AW, et al. Optimized chemical probes for REV-ERBα. J. Med. Chem 2013;56:4729–37.
- Shin Y, Noel R, Banerjee S, et al. Small molecule tertiary amines as agonists of the nuclear hormone receptor Rev-erbα. Bioorg Med Chem Lett 2012;22:4413–17.
- Noel R, Song X, Shin Y, et al. Synthesis and SAR of tetrahydroisoquinolines as Rev-erbα agonists. Bioorg Med Chem Lett 2012;22:3739–42.
- Lee J, Lee S, Chung S, et al. Identification of a novel circadian clock modulator controlling BMAL1 expression through a ROR/REV-ERB-response element-dependent mechanism. Biochem. Biophys. Res. Commun 2016;469:580–86.
- Kojetin D, Wang Y, Kamenecka TM, et al. Identification of SR8278, a synthetic antagonist of the nuclear heme receptor REV-ERB. ACS Chem Biol 2011;6:131–34.
- De Mei C, Ercolani L, Parodi C, et al. Dual inhibition of REV-ERBβ and autophagy as a novel pharmacological approach to induce cytotoxicity in cancer cells. Oncogene 2015;34:2597–2608.
- Torrente E, Parodi C, Ercolani L, et al. Synthesis and in vitro anticancer activity of the first class of dual inhibitors of REV-ERBβ and autophagy. J. Med. Chem 2015;58:5900–15.
- Pariollaud M, Gibbs J, Hopwood T, et al. Circadian clock component REV-ERBα controls homeostatic regulation of pulmonary inflammation. J Clin Invest 2018;128:2281–96.
- Hering Y, Berthier A, Duez H, et al. Development and implementation of a cell-based assay to discover agonists of the nuclear receptor REV-ERBα. J Biol Methods 2018;5:e94
- Amador A, Huitron-Resendiz S, Roberts AJ, et al. Pharmacological targeting the REV-ERBs in sleep/wake regulation. PloS One 2016;11:e0162452.
- Banerjee S, Wang Y, Solt LA, et al. Pharmacological targeting of the mammalian clock regulates sleep architecture and emotional behaviour. Nat Commun 2014;5:5759
- Amador A, Kamenecka TM, Solt LA, et al. REV-ERBβ is required to maintain normal wakefulness and the wake-inducing effect of dual REV-ERB agonist SR9009. Biochem. Pharmacol 2018;150:1–8.
- Cho H, Zhao X, Hatori M, et al. Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature 2012;485:123–27.
- Mazzarino M, Rizzato N, Stacchini C, et al. A further insight into the metabolic profile of the nuclear receptor Rev-erb agonist, SR9009. Drug Test Anal 2018;10:1670–81.
- Zhang L, Zhang R, Tien C-L, et al. REV-ERBα ameliorates heart failure through transcription repression. JCI Insight 2017;2:e95177.
- Ercolani L, Ferrari A, De Mei C, et al. Circadian clock: time for novel anticancer strategies?. Pharmacol Res 2015;100:288–95.
- Sulli G, Rommel A, Wang X, et al. Pharmacological activation of REV-ERBs is lethal in cancer and oncogene-induced senescence. Nature 2018;553:351–355.
- Sitaula S, Billon C, Kamenecka TM, et al. Suppression of atherosclerosis by synthetic REV-ERB agonist. Biochem Bioph Res Co 2015;460:566–71.
- Hwang J, Jiang A, Fikrig E. Rev-erb agonist inhibits chikungunya and O’nyong’nyong virus replication. Open Forum Infect Dis 2018; 5:ofy315.
- Chang C, Loo C-S, Zhao X, et al. The nuclear receptor REV-ERBα modulates Th17 cell-mediated autoimmune disease. Proc Natl Acad Sci USA 2019;116:18528–36.
- Dierickx P, Emmett MJ, Jiang C, et al. SR9009 has REV-ERB-independent effects on cell proliferation and metabolism. Proc Natl Acad Sci USA 2019;116:12147–52.
- Zhao L, Isayama K, Chen H, et al. The nuclear receptor REV-ERBα represses the transcription of growth/differentiation factor 10 and 15 genes in rat endometrium stromal cells. Physiol Rep 2016;4:e12663.
- Dong D, Sun H, Wu Z, et al. A validated ultra-performance liquid chromatography-tandem mass spectrometry method to identify the pharmacokinetics of SR8278 in normal and streptozotocin-induced diabetic rats. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci 2016;1020:142–47.
- Gagnidze K, Hajdarovic KH, Moskalenko M, et al. Nuclear receptor REV-ERBα mediates circadian sensitivity to mortality in murine vesicular stomatitis virus-induced encephalitis. Proc Natl Acad Sci USA 2016;113:5730–35.
- Chung S, Lee EJ, Yun S, et al. Impact of circadian nuclear receptor REV-ERBα on midbrain dopamine production and mood regulation. Cell 2014;157:858–68.
- Welch RD, Billon C, Valfort A-C, et al. Pharmacological inhibition of REV-ERB stimulates differentiation, inhibits turnover and reduces fibrosis in dystrophic muscle. Sci Rep 2017;7:17142.
- Kallen JA, Schlaeppi J-M, Bitsch F, et al. X-ray structure of the hRORalpha LBD at 1.63 A: structural and functional data that cholesterol or a cholesterol derivative is the natural ligand of RORalpha. Structure 2002;10:1697–707.
- Kallen J, Schlaeppi J-M, Bitsch F, et al. Crystal structure of the human RORalpha Ligand binding domain in complex with cholesterol sulfate at 2.2 A. J. Biol. Chem 2004;279:14033–38.
- Kojetin DJ, Burris TP. REV-ERB and ROR nuclear receptors as drug targets. Nat Rev Drug Discov 2014;13:197–216.
- Marciano DP, Chang MR, Corzo CA, et al. The therapeutic potential of nuclear receptor modulators for treatment of metabolic disorders: PPARγ, RORs, and Rev-erbs. Cell Metab 2014;19:193–208.
- Stehlin C, Wurtz JM, Steinmetz A, et al. X-ray structure of the orphan nuclear receptor RORbeta ligand-binding domain in the active conformation. Embo J 2001;20:5822–31.
- Stehlin-Gaon C, Willmann D, Zeyer D, et al. All-trans retinoic acid is a ligand for the orphan nuclear receptor ROR beta. Nat. Struct. Biol 2003;10:820–25.
- Jin L, Martynowski D, Zheng S, et al. Structural basis for hydroxycholesterols as natural ligands of orphan nuclear receptor RORgamma. Mol Endocrinol 2010;24:923–29.
- He B, Nohara K, Park N, et al. The Small Molecule Nobiletin Targets the Molecular Oscillator to Enhance Circadian Rhythms and Protect against Metabolic Syndrome. Cell Metab 2016;23:610–21.
- Chen Z, Yoo S-H, Park Y-S, et al. Identification of diverse modulators of central and peripheral circadian clocks by high-throughput chemical screening. Proc Natl Acad Sci USA 2012;109:101–06.
- Kumar N, Solt LA, Conkright JJ, et al. The benzenesulfoamide T0901317 [N-(2,2,2-trifluoroethyl)-N-[4-[2,2,2-trifluoro-1-hydroxy-1-(trifluoromethyl)ethyl]phenyl]-benzenesulfonamide] is a novel retinoic acid receptor-related orphan receptor-alpha/gamma inverse agonist. Mol Pharmacol 2010;77:228–36.
- Schultz JR, Tu H, Luk A, et al. Role of LXRs in control of lipogenesis. Genes Dev 2000;14:2831–38.
- Wang Y, Kumar N, Nuhant P, et al. Identification of SR1078, a synthetic agonist for the orphan nuclear receptors RORα and RORγ. ACS Chem Biol 2010;5:1029–34.
- Solt LA, Kumar N, Nuhant P, et al. Suppression of TH17 differentiation and autoimmunity by a synthetic ROR ligand. Nature 2011;472:491–4.
- Kumar N, Kojetin DJ, Solt LA, et al. Identification of SR3335 (ML-176): a synthetic RORα selective inverse agonist. ACS Chem. Biol 2011;6:218–22.
- Kumar N, Lyda B, Chang MR, et al. Identification of SR2211: a potent synthetic RORγ-selective modulator. ACS Chem. Biol 2012;7:672–77.
- Huh JR, Englund EE, Wang H, et al. Identification of Potent and Selective Diphenylpropanamide RORγ Inhibitors. Acs Med Chem Lett 2013;4:79–84.
- Kotoku M, Maeba T, Fujioka S, et al. Discovery of Second Generation RORγ Inhibitors Composed of an Azole Scaffold. J Med Chem 2019;62:2837–42.
- Zhang Y, Wu X, Xue X, et al. Discovery and characterization of XY101, a potent, selective, and orally bioavailable RORγ inverse agonist for treatment of castration-resistant prostate cancer. J Med Chem 2019;62:4716–30.
- Bronner SM, Zbieg JR, Crawford JJ. RORγ antagonists and inverse agonists: a patent review. Expert Opin Ther Pat 2017;27:101–12.
- Manning G, Whyte DB, Martinez R, et al. The protein kinase complement of the human genome. Science 2002;298:1912–34.
- Reischl S, Kramer AJ. Kinases and phosphatases in the mammalian circadian clock. FEBS Lett 2011;585:1393–99.
- Eide EJ, Woolf MF, Kang H, et al. Control of mammalian circadian rhythm by CKIepsilon-regulated proteasome-mediated PER2 degradation. Mol Cell Biol 2005;25:2795–807.
- Vanselow K, Vanselow JT, Westermark PO, et al. Differential effects of PER2 phosphorylation: molecular basis for the human familial advanced sleep phase syndrome (FASPS). Genes Dev 2006;20:2660–72.
- Mosser EA, Chiu CN, Tamai TK, et al. Identification of pathways that regulate circadian rhythms using a larval zebrafish small molecule screen. Sci Rep 2019;9:12405
- Ono A, Sato A, Fujimoto KJ, et al. 3,4-Dibromo-7-Azaindole Modulates Arabidopsis Circadian Clock by Inhibiting Casein Kinase 1 Activity. Plant Cell Physiol 2019;60:2360–68.
- Lee H, Lee JW. The Roles of CKI in Circadian Rhythm. Future Med Chem 2019;11:2621–24.
- Reischl S, Vanselow K, Westermark PO, et al. Beta-TrCP1-mediated degradation of PERIOD2 is essential for circadian dynamics. J Biol Rhythms 2007;22:375–86.
- Meng Q-J, Maywood ES, Bechtold DA, et al. Entrainment of disrupted circadian behavior through inhibition of casein kinase 1 (CK1) enzymes. Proc Natl Acad Sci USA 2010;107:15240–45.
- Lee JW, Hirota T, Peters EC, et al. A small molecule modulates circadian rhythms through phosphorylation of the period protein. Angew Chem Int Ed Engl 2011;50:10608–11.
- Hirota T, Lewis WG, Liu AC, et al. A chemical biology approach reveals period shortening of the mammalian circadian clock by specific inhibition of GSK-3beta. Proc Natl Acad Sci USA 2008;105:20746–51.
- Uehara TN, Mizutani Y, Kuwata K, et al. Casein kinase 1 family regulates PRR5 and TOC1 in the Arabidopsis circadian clock. Proc Natl Acad Sci USA 2019;116:11528–36.
- Isojima Y, Nakajima M, Ukai H, et al. CKIepsilon/delta-dependent phosphorylation is a temperature-insensitive, period-determining process in the mammalian circadian clock. Proc Natl Acad Sci USA 2009;106:15744–9.
- Kwak Y, Jeong J, Lee S, et al. Cyclin-dependent kinase 5 (Cdk5) regulates the function of CLOCK protein by direct phosphorylation. J Biol Chem 2013;288:36878–89.
- Nakahata Y, Kaluzova M, Grimaldi B, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 2008;134:329–40.
- Chang HC, Guarente L. SIRT1 mediates central circadian control in the SCN by a mechanism that decays with aging. Cell 2013;153:1448–60.
- Sun S, Liu Z, Feng Y, et al. Sirt6 deacetylase activity regulates circadian rhythms via Per2. Biochem Biophys Res Commun 2019;511:234–38.
- Bellet MM, Nakahata Y, Boudjelal M, et al. Pharmacological modulation of circadian rhythms by synthetic activators of the deacetylase SIRT1. Proc Natl Acad Sci USA 2013;110:3333–8.
- Wang N, Yang G, Jia Z, et al. Vascular PPARgamma controls circadian variation in blood pressure and heart rate through Bmal1. Cell Metab 2008;8:482–91.
- Onishi Y, Kawano Y. Rhythmic binding of Topoisomerase I impacts on the transcription of Bmal1 and circadian period. Nucleic Acids Res 2012;40:9482–92.
- Moore-Ede MC. Jet lag, shift work, and maladaption. Physiology 1986;1:156–60.
- Choy M, Salbu RL. Jet lag: current and potential therapies. P T 2011;36:221–31.
- Mahoney MM. Shift work, jet lag, and female reproduction. Int J Endocrinol 2010;2010:813764–64.
- Kelly RM, Healy U, Sreenan S, et al. Clocks in the clinic: circadian rhythms in health and disease. Postgrad Med J 2018;94:653–58.
- Bollinger T, Schibler U. Circadian rhythms - from genes to physiology and disease. Swiss Med Wkly 2014;144:w13984w84.
- Karatsoreos IN. Effects of circadian disruption on mental and physical health. Curr Neurol Neurosci Rep 2012;12:218–25.
- Arble DM, Ramsey KM, Bass J, et al. Circadian disruption and metabolic disease: findings from animal models. Best Pract Res Clin Endocrinol Metab 2010;24:785–800.
- Turek FW, Joshu C, Kohsaka A, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science (New York, N.Y.) 2005;308:1043–45.
- Rudic RD, McNamara P, Curtis A-M, et al. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol 2004;2:e377–e77.
- Le Martelot G, Claudel T, Gatfield D, et al. REV-ERBalpha participates in circadian SREBP signaling and bile acid homeostasis . PLoS Biol 2009;7:e1000181
- Cook DN, Kang HS, Jetten AM. Retinoic Acid-Related Orphan Receptors (RORs): Regulatory Functions in Immunity, Development, Circadian Rhythm, and Metabolism. Nucl Receptor Res 2015;2:101185.
- Hamilton BA, Frankel WN, Kerrebrock AW, et al. Disruption of the nuclear hormone receptor RORalpha in staggerer mice. Nature 1996;379:736–39.
- Kharitonenkov A, Shiyanova TL, Koester A, et al. FGF-21 as a novel metabolic regulator. J. Clin. Invest 2005;115:1627–35.
- Solt LA, Kojetin DJ, Burris TP. The REV-ERBs and RORs: molecular links between circadian rhythms and lipid homeostasis. Future Med Chem 2011;3:623–38.
- Hirota T, Lee JW, St John PC, et al. Identification of small molecule activators of cryptochrome. Science (New York, N.Y.) 2012;337:1094–97.
- Nohara K, Nemkov T, D’Alessandro A, et al. Coordinate Regulation of Cholesterol and Bile Acid Metabolism by the Clock Modifier Nobiletin in Metabolically Challenged Old Mice. Int J Mol Sci 2019;20:4281.
- Nohara K, Mallampalli V, Nemkov T, et al. Nobiletin fortifies mitochondrial respiration in skeletal muscle to promote healthy aging against metabolic challenge. Nat Commun 2019;10:3923
- Sehgal A, Mignot E. Genetics of sleep and sleep disorders. Cell 2011;146:194–207.
- Kiessling S, Eichele G, Oster H. Adrenal glucocorticoids have a key role in circadian resynchronization in a mouse model of jet lag. J Clin Invest 2010;120:2600–09.
- Yamaguchi Y, Suzuki T, Mizoro Y, et al. Mice genetically deficient in vasopressin V1a and V1b receptors are resistant to jet lag. Science (New York, N.Y.) 2013;342:85–90.
- Jones CR, Huang AL, Ptáček LJ, et al. Genetic basis of human circadian rhythm disorders. Exp Neurol 2013;243:28–33.
- Canales MT, Holzworth M, Bozorgmehri S, et al. Clock gene expression is altered in veterans with sleep apnea. Physiol Genomics 2019;51:77–82.
- LeGates TA, Fernandez DC, Hattar S. Light as a central modulator of circadian rhythms, sleep and affect. Nat Rev Neurosci 2014;15:443–54.
- Felder-Schmittbuhl M-P, Buhr ED, Dkhissi-Benyahya O, et al. Ocular Clocks: Adapting Mechanisms for Eye Functions and Health. Invest Ophthalmol Vis Sci 2018;59:4856–70.
- Kondratov RV, Kondratova AA, Gorbacheva VY, et al. Early aging and age-related pathologies in mice deficient in BMAL1, the core componentof the circadian clock. Genes Dev 2006;20:1868–73.
- Yang G, Chen L, Grant GR, et al. Timing of expression of the core clock gene Bmal1 influences its effects on aging and survival. Sci Transl Med 2016;8:324ra16
- Baba K, Ribelayga CP, Michael Iuvone P, et al. The Retinal Circadian Clock and Photoreceptor Viability. Adv Exp Med Biol 2018;1074:345–50.
- Bhatwadekar AD, Yan Y, Qi X, et al. Per2 mutation recapitulates the vascular phenotype of diabetes in the retina and bone marrow. Diabetes 2013;62:273–82.
- Leske MC, Connell AM, Wu SY, et al. Incidence of open-angle glaucoma: the Barbados Eye Studies. The Barbados Eye Studies Group. Arch Ophthalmol 2001;119:89–95.
- Jean-Louis G, Zizi F, Lazzaro DR, et al. Circadian rhythm dysfunction in glaucoma: A hypothesis. J Circadian Rhythms 2008;6:1
- Mure LS, Le HD, Benegiamo G, et al. Diurnal transcriptome atlas of a primate across major neural and peripheral tissues. Science (New York, N.Y.) 2018;359:eaao0318.
- Lu D, Lin C, Jiao X, et al. Short-term High Fructose Intake Reprograms the Transcriptional Clock Rhythm of the Murine Extraorbital Lacrimal Gland. Invest Ophthalmol Vis Sci 2019;60:2038–48.
- Jiao X, Wu M, Lu D, et al. Transcriptional Profiling of Daily Patterns of mRNA Expression in the C57BL/6J Mouse Cornea. Curr Eye Res 2019;44:1054–66.
- Xue Y, Liu P, Wang H, et al. Modulation of Circadian Rhythms Affects Corneal Epithelium Renewal and Repair in Mice. Invest Ophthalmol Vis Sci 2017;58:1865–74.
- Downton P, Early JO, Gibbs JE. Circadian rhythms in adaptive immunity. Immunology 2020; Online ahead of print.
- Mendoza J, Vanotti G. Circadian neurogenetics of mood disorders. Cell Tissue Res 2019;377:81–94.
- Hühne A, Welsh DK, Landgraf D. Landgraf D Prospects for circadian treatment of mood disorders. Ann Med 2018;50:637–54.
- Marco EM, Velarde E, Llorente R, et al. Disrupted Circadian Rhythm as a Common Player in Developmental Models of Neuropsychiatric Disorders. Curr Top Behav Neurosci 2016;29:155–81.
- Manoogian ENC, Panda S. Circadian rhythms, time-restricted feeding, and healthy aging. Ageing Res Rev 2017;39:59–67.
- Duffy JF, Zitting K-M, Chinoy ED. Aging and Circadian Rhythms. Sleep Med Clin 2015;10:423–34.
- Firsov D, Bonny O. Circadian rhythms and the kidney. Nat Rev Nephrol 2018;14:626–35.
- Ohashi N, Isobe S, Ishigaki S, Yasuda H. Circadian rhythm of blood pressure and the renin-angiotensin system in the kidney. Hypertens Res 2017;40:413–22.
- Masri S, Sassone-Corsi P. The emerging link between cancer, metabolism, and circadian rhythms. Nat Med 2018;24:1795–803.
- Reszka E, Zienolddiny S. Epigenetic Basis of Circadian Rhythm Disruption in Cancer. Methods Mol Biol 2018;1856:173–201.
- Gibbs JE, Blaikley J, Beesley S, et al. The nuclear receptor REV-ERBα mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. Proc Natl Acad Sci USA 2012;109:582–87.
- Hirota T, Kay SA. Identification of small-molecule modulators of the circadian clock. Meth. Enzymol 2015;551:267–82.