Abstract
We report the biochemical and structural characterisation of a beta-carbonic anhydrase (β-CA) from Trichomonas vaginalis, a unicellular parasite responsible for one of the world’s leading sexually transmitted infections, trichomoniasis. CAs are ubiquitous metalloenzymes belonging to eight evolutionarily divergent groups (α, β, γ, δ, ζ, η, θ, and ι); humans express only α-CAs, whereas many clinically significant pathogens express only β- and/or γ-CAs. For this reason, the latter two groups of CAs are promising biomedical targets for novel antiinfective agents. The β-CA from T. vaginalis (TvaCA1) was recombinantly produced and biochemically characterised. The crystal structure was determined, revealing the canonical dimeric fold of β-CAs and the main features of the enzyme active site. The comparison with the active site of human CA enzymes revealed significant differences that can be exploited for the design of inhibitors selective for the protozoan enzyme with respect to the human ones.
Introduction
The widespread use of antiinfectives has had a profound impact on global health, causing the development of antiinfective-resistant pathogens. For this reason, new drugs with different mechanisms of action are necessary.
Trichomonas vaginalis is a flagellated protozoan parasite that resides in the urogenital tract of men and womenCitation1. It causes trichomoniasis, one of the most common sexually transmitted infections in the worldCitation2. Based on a World Health Organisation investigation in 2016, it can be stated that 156 million new trichomoniasis cases emerge every yearCitation3,Citation4. This incidence accounts for almost half of the total sexually transmitted infection acquisitions. In women, trichomoniasis usually affects the vagina, but it can also spread to the urethraCitation5. The infection typically causes a variety of mild to severe symptomsCitation1, with 10–50% of women showing no symptomsCitation3 and 5–15% of cases remaining undetectable upon examinationCitation6. The majority of men infected with T. vaginalis are asymptomaticCitation3. Mild or nonexistent symptoms make the detection of trichomoniasis particularly challenging, and the infection may go totally unnoticed. The increasing interest in this infection is related to data reporting the relationship between trichomoniasis and other critical pathologies. In fact, it has been shown that infected subjects could exhibit increased susceptibility to human immunodeficiency virus (HIV) acquisition and/or transmissionCitation7, while in pregnant women, trichomoniasis could induce several complications, such as preterm delivery or premature membrane ruptureCitation8. Previous studies have suggested that there is an association between T. vaginalis and the risk of cervical neoplasiaCitation9. In addition, new remarkable results have recently been reported on a protein encoded by this infective parasite that is able to drive inflammation and cell proliferation, thus activating molecular pathways that are involved in the promotion and progression of prostate cancerCitation10. As a consequence, the diagnosis and effective treatment of T. vaginalis infection have become an extremely important goal for global health in both women and men.
A single dose of metronidazole (MET) has been the main treatment against trichomoniasis for nearly five decadesCitation1. However, the treatment quite often involves multiple rounds of METCitation1, which can lead to a lack of drug compliance and increased antibiotic resistance as the organism adapts and becomes refractory towards the medication. The first reported MET-resistant T. vaginalis dates back to 1981Citation11. Since then, rare reported cases of MET-resistant strains have emergedCitation12. However, the clinical resistance of T. vaginalis will likely increase in the future, thus posing a real threat unless novel therapies are discovered.
Alternative approaches for the treatment of trichomoniasis can be developed through the identification of new molecular targets. Among these, carbonic anhydrases (CAs), ubiquitous metalloenzymes present in organisms from all kingdoms of lifeCitation13,Citation14, have recently emerged. CAs catalyse reversible CO2 hydration to bicarbonate and proton. This simple reaction plays an essential role in several physiological processes of microorganism life, such as photosynthesis, CO2 transport, pH regulation, and biosynthetic reactionsCitation13. CAs are divided into eight genetic families: α, β, γ, δ, ζ, η, θ, and ιCitation14–17, which can vary in terms of amino acid sequence, oligomeric state, kinetics, and inhibition and activation profilesCitation13. α-CAs are the only isoforms present in humans, whereas many pathogens have been discovered with only β- and/or γ-CA genes in their genome. Based on this observation, these enzymes have been introduced as potential and novel antiinfective drug targets. Indeed, effective inhibitors targeting the active site and thus hindering CA function have been discovered through the production and characterisation of pathogen-specific β- and/or γ-CAsCitation18–24.
The analysis of the T. vaginalis genome revealed the presence of two β-CA genes (TVAG_005270 and TVAG_268150), which encode two proteins, TvaCA1 and TvaCA2, respectively, that share a very high amino acid sequence identity (approximately 72%)Citation25. With the aim of identifying new targets for the development of innovative drugs against trichomoniasis, we started our studies on TvaCA1. In this paper, we report the cloning, expression, kinetic and structural characterisation of this enzyme. Our results indicate that TvaCA1 represents a novel potential target for antimicrobial therapy against trichomoniasis.
Materials and methods
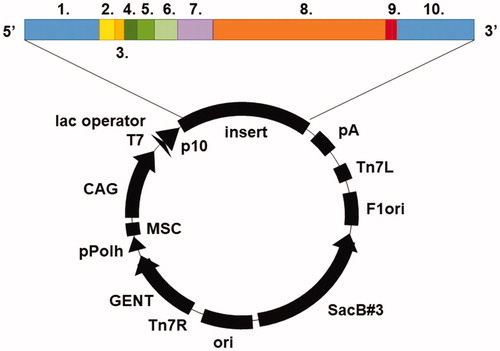
Protein expression
The TvaCA1 gene sequence was retrieved from Universal Protein Resource Database UniProt (protein entry: A2ENQ8). The destination vector was pBVboostFGCitation26, and the subcloned insert was composed of Gateway-compatible recombination sites (attL1, attL2), Shine-Dalgarno and Kozak sequences, a 6xHis-tag with surrounding spacer regions (MSTT and ATAIPTTCitation27), TvaCA1, and a thrombin cleavage site (LVPRGSCitation28) (). Gene synthesis and subcloning were performed by GeneArt (Thermo Fisher Scientific, Germany). TvaCA1 was expressed recombinantly in E. coli (OneShot® BL21 Star™ (DE3) Chemically Competent Cells, #C601003, Thermo Fisher Scientific, Finland). Transformation was performed according to the Thermo Fisher Scientific OneShot® BL21(DE3) Competent Cells manual (part no. 28–0182). Cells were cultured in Luria-Bertani (LB) medium supplemented with 10 mg/mL gentamicin (1:1000, v/v) at 37 °C until an optical density (OD595) of 0.4–0.6 was reached. Expression of the protein was induced by adding 1 M isopropyl β-D-1-thiogalactopyranoside (IPTG) 1:1000 (v/v), after which the culturing was continued overnight at 37 °C. The cells were harvested by centrifugation at 5000 × g for 15 min at 4 °C.
Protein purification
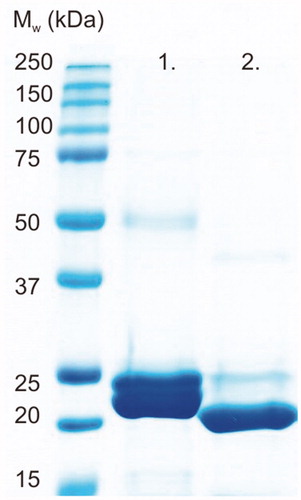
Harvested cells were mechanically disrupted in 50 mM Na2HPO4, 0.5 M NaCl and 50 mM imidazole buffer pH 8.0 (binding buffer (BB)) with an EmulsiFlex-C3 homogeniser (AVESTIN, Canada). The lysate was centrifuged at 13000 × g for 20 min at 4 °C. The supernatant was diluted with Ni2+-NTA agarose affinity chromatography resin (Macherey-Nagel GmbH Co., Germany) and BB (1:≥3 (vol/vol)). The suspension was incubated for 2 h at RT with gentle agitation, followed by overnight incubation at 4 °C without agitation. Subsequently, the resin was washed generously with BB and packed into a chromatography column with an EMD Millipore™ vacuum filtering flask (#XX1004705, Merck, Finland) and filter paper. The protein was eluted from the resin with 50 mM Na2HPO4, 0.5 M NaCl and 350 mM imidazole (pH 7.0). The 6xHis-tag was removed by thrombin (#RECOMT, Sigma-Aldrich, Finland) according to the Thrombin CleanCleave™ Kit manual (Sigma-Aldrich, Finland), and the tag was separated from the core protein by Ni2+-NTA affinity chromatography. The yield of the protein was determined by a NanoDrop One (Thermo Fisher Scientific, Finland). The quality of the purified protein was analysed by SDS-PAGE using a 12% (w/v) polyacrylamide gel and visualised with PageBlue Protein staining solution (Thermo Fisher Scientific, #24620, Finland). The obtained polypeptide bands of the SDS-PAGE gel were excised and identified using tandem mass spectrometry (Meilahti Clinical Proteomics Core Facility, University of Helsinki, Finland). The sample for crystallisation trials was further purified on a Phenomenex Biosep SEC-S2000 300 × 7.8 mm column in the following running buffer: 50 mM Tris-HCl, 150 mM NaCl, 1.0 mM DTT, pH 8.0. Pooled fractions were concentrated on a 5000 MWCO polyethersulfone membrane (Vivaspin 2, Vivascience Sartorius group, VS0211). The quality of purified protein was analysed by 15% SDS-PAGE, and the protein was detected by blue staining solution (Coomassie Brilliant Blue R-250 #1610400).
Light scattering
Light scattering methods used to determine the Mw of TvaCA1 included SLS and DLS combined with SEC. All measurements were performed after His-tag removal. The instrumentation, which simultaneously measured both LS data, consisted of a Malvern Zetasizer (microV) (Malvern Instruments Ltd., Worcestershire, UK) and a liquid chromatography instrument (CBM-20A, Shimadzu Corporation, Kyoto, Japan) equipped with an autosampler (SIL-20A) and UV–VIS (SPD-20A) and fluorescence detectors (RF-20Axs). UV absorption intensity at 280 nm was used for the determination of the protein concentration. Acquired data were processed with Lab Solution Version 5.51 (Shimadzu Corporation) and OmniSec 4.7 (Malvern Instruments Ltd., Worcestershire, UK) software. Two samples of TvaCA1 (total of 0.4 mg, in PBS) were injected into a Superdex 200 5/150 column (GE Healthcare, Uppsala, Sweden) equilibrated with 50 mM NaH2PO4 and 500 mM NaCl (pH 8) buffer. Measurements were performed within a thermostable chamber at 20 °C, with a flow rate of 0.1 ml/min. The molecular weight of TvaCA1 was determined in two independent ways: first, based on elution time by using a standard curve calculated according to the elution profiles of standard proteins (SEC analysis: alcohol dehydrogenase 150 kDa, b-amylase 200 kDa, bovine serum albumin 66 kDa and CA 29 kDa (Sigma-Aldrich, Inc., St. Louis, MO, USA)), and second, by calibrating the light-scattering detector based on the monomeric peak of BSA and using the light-scattering intensity (SLS) to determine the protein size. The protein concentration was determined with A280.
Kinetics
An Applied Photophysics stopped-flow instrument was used for assaying CA-catalyzed CO2 hydration activity. Phenol red (at a concentration of 0.2 mM) was used as a pH indicator, working at the absorbance maximum of 557 nm, with 20 mM Hepes (pH 7.5) as buffer and 20 mM Na2SO4 (for maintaining constant ionic strength), following the initial rates of the CA-catalyzed CO2 hydration reaction for a period of 10 − 100 s. The CO2 concentrations ranged from 1.7 to 17 mM for the determination of the kinetic parameters and AAZ inhibition constant. Six traces of the initial 5 − 10% of the reaction were used to determine the initial velocity. The uncatalyzed rates were determined in the same manner and subtracted from the total observed rates. A stock solution of the inhibitor (0.1 mM) was prepared in distilled − deionized water, and dilutions up to 0.01 nM were prepared thereafter with distilled − deionized water. Inhibitor (I) and enzyme (E) solutions were preincubated together for 15 min at room temperature prior to the assay to allow formation of the E − I complex. The inhibition constant was obtained by nonlinear least squares methods using PRISM 3 and represents the means from at least three different determinations.
Crystallographic studies
TvaCA1 crystals were obtained at 20 °C by the hanging drop vapour diffusion method. The search for initial crystallisation conditions was performed using Crystal Screen, Crystal Screen 2 and Index from Hampton ResearchCitation29. The wells contained 500 μL of precipitant solution, and the drops were prepared by mixing 1 μL of enzyme solution (11 mg/ml) in 50 mM Tris-HCl, pH 8.0, with 1 μL of the reservoir solution. Good conditions for crystallisation were achieved using a precipitant buffer consisting of 30% (w/v) PEG 4000, 0.2 M sodium acetate, 0.1 M Tris-HCl, pH 8.5. Crystals appeared in the drops within 48 h and grew in approximately one week to maximum dimensions of 0.2 × 0.2 × 0.15 mm3. Complete X-ray diffraction data were collected at 100 K with a copper rotating anode generator developed by Rigaku and a Rigaku Saturn CCD detector. Prior to cryogenic freezing, crystals were transferred to the precipitant solution with the addition of 15% (v/v) glycerol. Diffraction data were processed and scaled using the programme HKL2000 (HKL Research)Citation30. Crystals belonged to the space group P212121 with unit cell dimensions of a = 47.3 Å, b = 77.3 Å and c = 90.7 Å. The Matthews coefficient (VM = 2.08 Å3/Da) indicated that the asymmetric unit contained two molecules, with a solvent content of 41%. Data collection statistics are reported in .
Table 1. Data collection and refinement statistics
The structure of TvaCA1 was solved by the molecular replacement technique using the programme AMoReCitation31 and the crystallographic structure of the β-CA from the archaeon M. thermoautotrophicum (PDB code 1G5C) as a model templateCitation32. Refinement of the structure was initially performed with the CNS programmeCitation33,Citation34, and model building was performed using OCitation35. However, since the electronic density maps were poorly defined both in the enzyme N-terminal region and in the loop encompassing the residues 95–104 of chain B, Auto-Rickshaw was used for rounds of automated model buildingCitation36,Citation37. This approach allowed the complete reconstruction of the model for both monomers, reducing the Rwork and Rfree values to 0.265 and 0.329, respectively. Many cycles of manual rebuilding and positional and temperature factor refinement were then performed using the REFMAC 5.8 programmeCitation38 in CCP4iCitation39. All refinement cycles were performed with the application of noncrystallographic symmetry restraints. The final model contained 64 solvent molecules and presented crystallographic Rwork and Rfree values (in the 41.9–2.48 Å resolution range) of 0.198 and 0.258, respectively. The refinement statistics are summarised in . Coordinates and structure factors have been deposited in the Protein Data Bank (accession code 6Y04).
Results
Protein production and purification
Recombinant TvaCA1 containing a His-tag and a thrombin cleavage site () was expressed in Escherichia coli and purified by affinity chromatography, with a yield of approximately 15 mg of purified protein/L of culture. Cleavage of the tag was carried out by thrombin treatment, followed by Protino® nickel-nitrilotriacetic acid (Ni2+-NTA) purification, and monitored by sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) ().
Light scattering
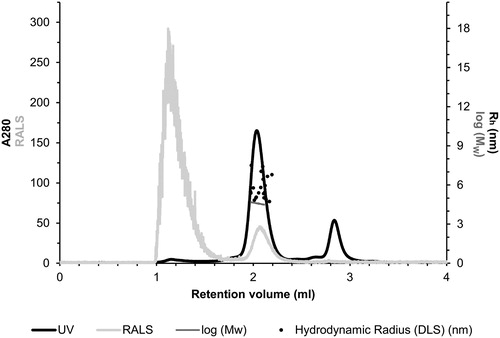
The quaternary structure of the purified TvaCA1 was investigated by size exclusion chromatography combined with static light scattering/dynamic light scattering (SEC-SLS/DLS). Based on UV absorption at 280 nm (, black curve), the main peak was eluted at 2.04 ml. First, the Mw of the eluted TvaCA1 was determined using the measured light scattering intensity, and the concentration was determined using UV absorption, resulting in an estimated Mw of 39.7 ± 0.4 kDa (, horizontal dark grey line across the main peak). Second, the Mw was calculated based on elution time by using a Mw standard curve and the elution profile of standard proteins, resulting in an estimation of 39.2 ± 0.7 kDa. Both calculations indicated that in our experimental conditions, the native protein is dimeric.
Kinetics
The kinetic parameters of TvaCA1 are presented in and compared with those of human CA isoforms I (hCA I) and II (hCA II).
Table 2. Kinetic data of TvaCA1. For comparison, kinetic parameters of hCA I, hCA II, and other representative β-CA enzymes are shown.
Structural characterization
The structure of TvaCA1 was investigated by X-ray crystallography. Before crystallisation experiments, an additional purification step was performed. In particular, TvaCA1 was purified by SEC to gain a purity level above 98%. Pooled samples were concentrated to 11 mg/mL and used for crystallisation trials. Crystals were obtained with the hanging-drop vapour diffusion method, using PEG 4000 as the precipitant. The crystals belonged to the space group P212121, with two molecules per asymmetric unit, and diffracted to a 2.48 Å resolution. The structure was solved by molecular replacement using the β-CA from Methanobacterium thermoautotrophicum (MtCab; PDB code 1G5C; 36.3% sequence identity) as the initial modelCitation32 and refined with the CNS 1.3Citation33,Citation34 and REFMAC 5.8 programmesCitation38 to Rwork and Rfree values of 19.8% and 25.7%, respectively. The refined structure presented a good geometry, with r.m.s.d. from ideal bond lengths and angles of 0.004 Å and 1.0°, respectively. The refinement statistics are summarised in .
TvaCA1 shows the typical α/β-fold observed for other β-CAsCitation32,Citation43–54, consisting of a central mixed five-stranded β-sheet surrounded by several α-helices (). In agreement with light-scattering experiment results, the two molecules in the asymmetric unit form a tightly associated dimer characterised by a buried surface area of approximately 4366 Å2 () and many hydrogen bonds and van der Waals interactions at the interface between subunits.
Figure 4. (A) Ribbon representation of the TvaCA1 monomer. (B) Enlarged view of the active site, showing Zn2+ coordination. (C) σA-weighted |2Fo-Fc| electron density map (contoured at 1.0 σ) relative to zinc ion coordination site.
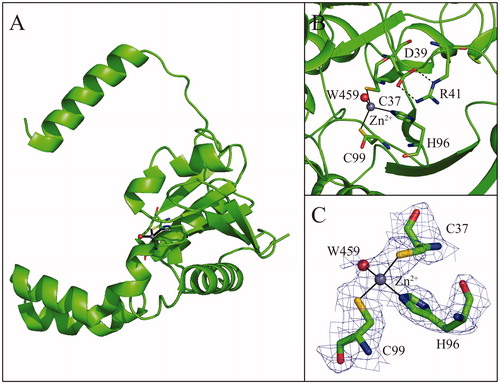
Figure 5. Dimeric structure of TvaCA1, with one monomer coloured in magenta and the other in green. The catalytic zinc ions are depicted as yellow spheres.
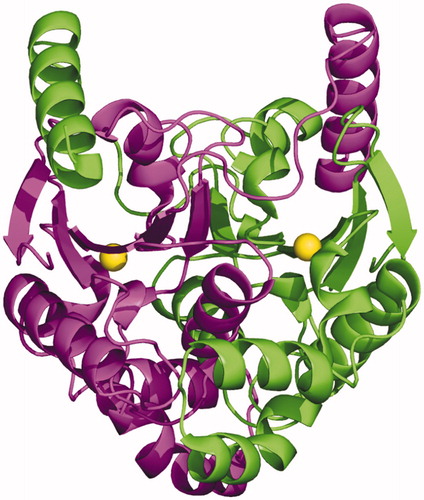
The dimer formation produces an extended β-sheet core consisting of ten β–strands, where the N-terminal helix of each monomer extends around the other monomer (). There are two active sites per dimer, which are located in clefts at the dimeric interface. Each active site contains a zinc ion on the bottom, which is coordinated by three protein residues, Cys37, His96 and Cys99. In one of the two active sites, a water molecule is clearly visible in the fourth coordination position (), whereas in the other active site, this water molecule is not visible, probably due to the lower quality of the electron density maps in this region.
Since the first crystallographic structure of a β-CA from the red alga Porphyridium purpureum in 2000Citation48, 18 other distinct β-CA structures have been reported, belonging to different kingdoms, i.e. plants, fungi, archaeon bacteria and eubacteria ()Citation32,Citation43–55. Although the sequence alignment of TvaCA1 with all these proteins does not show a very high sequence identity, substantial conservation of the three-dimensional structure is observed, with the highest similarity detected with MtCabCitation32, as determined by the DALI serverCitation58. The main structural differences can be observed in the loops connecting the central β-strands and in the N- and C-terminal regions.
Table 3. β-CAs whose crystal structure has been determined
Discussion
Compelling data in the literature indicate that interference with CA activity in various protozoan parasites causes impairment of parasite growth and virulence, which in turn leads to a significant antiinfective effectCitation59–61. These data, together with the observation that β-CAs are not present in humans, indicate the latter enzymes as excellent targets for the development of new antiparasitic drugs. However, despite their growing importance, only a few papers on the kinetics and inhibition profiles of β-CAs have been publishedCitation62–64, and no crystal structures of a protozoan CA have so far been reported. Here, we illustrate a full biochemical characterisation of TvaCA1, together with its crystallographic structure, thus providing the first detailed characterisation of a protozoan β–CA. In detail, TvaCA1 was expressed in E. coli, purified with high yield and kinetically characterised, showing a significant catalytic efficiency comparable to that of known prokaryotic β-CAs, such as Salmonella entericaCitation41 and Legionella pneumophilaCitation42 (). Inhibition experiments showed that similarly to CAs from other sources, TvaCA1 is inhibited from the well-known CA inhibitor acetazolamide. Light scattering analysis indicated a dimeric quaternary structure. This finding is in agreement with previous reports on β-CAs, which always show a dimeric structure that in some cases can arrange in higher oligomers, such as tetramers, hexamers or octamers (see ). Accordingly, the crystallographic structure of the enzyme shows the typical dimeric arrangement of β-CAs, characterised by a central β-sheet consisting of 8–10 strands surrounded by several helices.
As observed for other β-CAs so far structurally characterised (), the TvaCA1 active site is located in a narrow cavity spanning from the protein surface to the catalytic zinc ion. To date, two different subclasses of β-CA enzymes have been identified, which differ in the structural organisation of the active site. Type I β-CAs show a catalytic zinc ion coordination sphere consisting of three protein residues (two Cys and one His) and a water molecule. This kind of coordination was termed “open”, indicating the possibility of performing the catalytic reaction. In type II β-CAs, instead of the water molecule, the metal ion is coordinated to a fourth protein ligand, an Asp residue, resulting in a “closed” metal coordination sphere ()Citation65,Citation66. Our crystallographic analysis clearly indicates that TvaCA1 belongs to the type I subclass. Indeed, even if the water molecule in the fourth coordination position is clearly visible only in one active site of the dimer (see ), the aspartic acid residue, which is coordinated to the zinc ion in Type II β-CAs (Asp39 in the TvaCA1 sequence), is in both TvaCA1 active sites, very well defined in the electron density maps and far from the catalytic metal, leaving the active site in the open conformation.
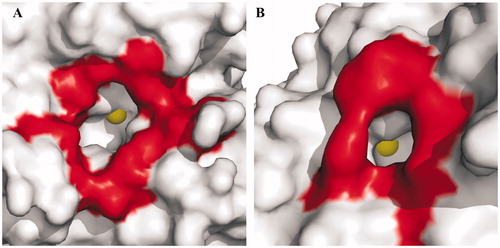
The comparison between the TvaCA1 catalytic cavity and that of human CAs showed significant differences in dimensions (), being the latter much larger and more accessible. This finding is particularly important for the development of drugs against trichomoniasis, since these differences can be exploited for the design of inhibitors selective for the protozoan enzyme with respect to the human CAs, which represent an off target. Further studies are currently underway to test this hypothesis.
Figure 6. Surface representation of (A) hCA II, chosen as a representative hCA isoform, and (B) TvaCA1. Residues delimiting the rim of the active site cavity are coloured in red. The metal ions are shown as yellow spheres. It is evident that in hCA II, the active site rim is larger (approximately 15 Å × 14 Å) and more accessible than that in TvaCA1 (8 Å × 6.5 Å).
Taken together, data here reported demonstrate that TvaCA1 is a druggable target and that its selective inhibition is feasible, with the aim of obtaining new antitrichomoniasis drugs.
Conclusion
In this study, TvaCA1 was successfully expressed in E. coli, purified by means of affinity chromatography, tested for kinetic and inhibitory properties, and characterised by X-ray diffraction studies, thus providing the first structural characterisation of a protozoan β-CA. The enzyme was demonstrated to be a noncovalently linked dimer with a narrow cavity leading to the active site. TvaCA1 possessed significant catalytic activity for the CO2 hydration reaction and is inhibited by the CA inhibitor acetazolamide. Significant differences between the active site of TvaCA1 and that of human CAs were observed that could be exploited for the design of selective inhibitors for the protozoan enzyme.
Acknowledgements
We thank Dr. Juha Määttä for valuable advice. We also thank Ms. Aulikki Lehmus, Mr. Luca De Luca and Mr. Maurizio Amendola for their technical assistance
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Lin WC, Chang WT, Chang TY, Shin JW. The pathogenesis of human cervical epithelium cells induced by interacting with trichomonas vaginalis. PLoS One 2015;10:e0124087
- Schwebke JR, Burgess D. Trichomoniasis. Clin Microbiol Rev 2004;17:794–803.
- Rowley J, Vander Hoorn S, Korenromp E, et al. Chlamydia, gonorrhoea, trichomoniasis and syphilis: global prevalence and incidence estimates, 2016. Bull World Health Organ 2019;97:548–62. P.
- Available from: http://www.who.int/mediacentre/factsheets/fs110/en/.
- Lewis D. Trichomoniasis. Medicine 2014;42:369–71.
- Sherrard J, Ison C, Moody J, et al. United Kingdom National Guideline on the Management of Trichomonas vaginalis 2014. Int J STD AIDS 2014;25:541–9.
- McClelland RS, Sangare L, Hassan WM, et al. Infection with Trichomonas vaginalis increases the risk of HIV-1 acquisition. J Infect Dis 2007;195:698–702.
- Cotch MF, Pastorek JG, 2nd, Nugent RP, et al. Trichomonas vaginalis associated with low birth weight and preterm delivery. The Vaginal Infections and Prematurity Study Group. Sex Transm Dis 1997;24:353–60.
- Zhang ZF, Begg CB. Is Trichomonas vaginalis a cause of cervical neoplasia? Results from a combined analysis of 24 studies. Int J Epidemiol 1994;23:682–90.
- Twu O, Dessi D, Vu A, et al. Trichomonas vaginalis homolog of macrophage migration inhibitory factor induces prostate cell growth, invasiveness, and inflammatory responses. Proc Natl Acad Sci USA 2014;111:8179–84.
- Waitkins SA, Thomas DJ. Isolation of Trichomonas vaginalis resistant to metronidazole. Lancet 1981;2:590.
- Lewis DA, Habgood L, White R, et al. Managing vaginal trichomoniasis resistant to high-dose metronidazole therapy. Int J STD AIDS 1997;8:780–4.
- Supuran C.T., De Simone, G. eds. Carbonic anhydrases as biocatalysts. In: From theory to medical and industrial applications. 1st ed. The netherlands: Elsevier; 2015: 398.
- Alterio V, Di Fiore A, D’Ambrosio K, et al. Multiple binding modes of inhibitors to carbonic anhydrases: how to design specific drugs targeting 15 different isoforms? Chem Rev 2012;112:4421–68.
- Kikutani S, Nakajima K, Nagasato C, et al. Thylakoid luminal θ-carbonic anhydrase critical for growth and photosynthesis in the marine diatom Phaeodactylum tricornutum. Proc Natl Acad Sci USA 2016;113:9828–33.
- Jensen EL, Clement R, Kosta A, et al. A new widespread subclass of carbonic anhydrase in marine phytoplankton. Isme J 2019;13:2094–106.
- De Simone G, Di Fiore A, Capasso C, Supuran CT. The zinc coordination pattern in the η-carbonic anhydrase from Plasmodium falciparum is different from all other carbonic anhydrase genetic families. Bioorg Med Chem Lett 2015;25:1385–9.
- Syrjanen L, Kuuslahti M, Tolvanen M, et al. The β-carbonic anhydrase from the malaria mosquito Anopheles gambiae is highly inhibited by sulfonamides. Bioorg Med Chem 2015;23:2303–9.
- Vullo D, Del Prete S, Di Fonzo P, et al. Comparison of the sulfonamide inhibition profiles of the beta- and gamma-carbonic anhydrases from the pathogenic Bacterium Burkholderia pseudomallei. Molecules 2017;22:421.
- Supuran CT. Bortezomib inhibits bacterial and fungal β-carbonic anhydrases. Bioorg Med Chem 2016;24:4406–9.
- Innocenti A, Hall RA, Schlicker C, et al. Carbonic anhydrase inhibitors. Inhibition of the beta-class enzymes from the fungal pathogens Candida albicans and Cryptococcus neoformans with aliphatic and aromatic carboxylates. Bioorg Med Chem 2009;17:2654–7.
- Nishimori I, Minakuchi T, Kohsaki T, et al. Carbonic anhydrase inhibitors: the beta-carbonic anhydrase from Helicobacter pylori is a new target for sulfonamide and sulfamate inhibitors. Bioorg Med Chem Lett 2007;17:3585–94.
- Carta F, Maresca A, Covarrubias AS, et al. Carbonic anhydrase inhibitors. Characterization and inhibition studies of the most active beta-carbonic anhydrase from Mycobacterium tuberculosis, Rv3588c. Bioorg Med Chem Lett 2009;19:6649–54.
- Del Prete S, Vullo D, De Luca V, et al. Sulfonamide inhibition studies of the β-carbonic anhydrase from the pathogenic Bacterium Vibrio cholerae. Bioorg Med Chem 2016;24:1115–20.
- Zolfaghari Emameh R, Barker HR, Tolvanen ME, et al. Horizontal transfer of β-carbonic anhydrase genes from prokaryotes to protozoans, insects, and nematodes. Parasit Vectors 2016;9:152
- Laitinen OH, Airenne KJ, Hytonen VP, et al. A multipurpose vector system for the screening of libraries in bacteria, insect and mammalian cells and expression in vivo. Nucleic Acids Res 2005;33:e42
- Piao S, Xu Y, Ha NC. Crystallization and preliminary X-ray crystallographic analysis of MacA from Actinobacillus actinomycetemcomitans. Acta Crystallogr Sect F Struct Biol Cryst Commun 2008;64:391–3.
- Hilvo M, Baranauskiene L, Salzano AM, et al. Biochemical characterization of CA IX, one of the most active carbonic anhydrase isozymes. J Biol Chem 2008;283:27799–809.
- Jancarik J, Scott WG, Milligan DL, et al. Crystallization and preliminary X-ray diffraction study of the ligand-binding domain of the bacterial chemotaxis-mediating aspartate receptor of Salmonella typhimurium. J Mol Biol 1991;221:31–4.
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Meth Enzymol 1997;276:307–26.
- Navaza J. AMoRe: an automated package for molecular replacement. Acta Crystallographica Section A 1994;50:157–63.
- Strop P, Smith KS, Iverson TM, et al. Crystal structure of the “cab”-type beta class carbonic anhydrase from the archaeon Methanobacterium thermoautotrophicum. J Biol Chem 2001;276:10299–305.
- Brunger AT. Version 1.2 of the Crystallography and NMR system. Nature Protocols 2007;2:2728–33.
- Brunger AT, Adams PD, Clore GM, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 1998;54:905–21.
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr Sec A, Foundat Crystallogra 1991;47:110–9.
- Panjikar S, Parthasarathy V, Lamzin VS, et al. Auto-rickshaw: an automated crystal structure determination platform as an efficient tool for the validation of an X-ray diffraction experiment. Acta Crystallogr D Biol Crystallogr 2005;61:449–57.
- Panjikar S, Parthasarathy V, Lamzin VS, et al. On the combination of molecular replacement and single-wavelength anomalous diffraction phasing for automated structure determination. Acta Crystallogr D Biol Crystallogr 2009;65:1089–97.
- Murshudov GN, Skubak P, Lebedev AA, Pannu NS, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr 2011;67:355–67.
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 2011;67:235–42.
- Supuran CT. Carbonic anhydrases: novel therapeutic applications for inhibitors and activators. Nat Rev Drug Discov 2008;7:168–81.
- Nishimori I, Minakuchi T, Vullo D, et al. Inhibition studies of the β-carbonic anhydrases from the bacterial pathogen Salmonella enterica serovar Typhimurium with sulfonamides and sulfamates. Bioorg Med Chem 2011;19:5023–30.
- Supuran CT. Legionella pneumophila carbonic anhydrases: underexplored antibacterial drug targets. Pathogens 2016;5:44.
- Kimber MS, Pai EF. The active site architecture of Pisum sativum beta-carbonic anhydrase is a mirror image of that of alpha-carbonic anhydrases. Embo J 2000;19:1407–18.
- Covarrubias AS, Bergfors T, Jones TA, Högbom M. Structural mechanics of the pH-dependent activity of beta-carbonic anhydrase from Mycobacterium tuberculosis. J Biol Chem 2006;281:4993–9.
- Sawaya MR, Cannon GC, Heinhorst S, et al. The structure of beta-carbonic anhydrase from the carboxysomal shell reveals a distinct subclass with one active site for the price of two. J Biol Chem 2006;281:7546–55.
- Teng YB, Jiang YL, He YX, et al. Structural insights into the substrate tunnel of Saccharomyces cerevisiae carbonic anhydrase Nce103. BMC Struct Biol 2009;9:67
- Huang S, Hainzl T, Grundstrom C, et al. Structural studies of β-carbonic anhydrase from the green alga Coccomyxa: inhibitor complexes with anions and acetazolamide. PLoS One 2011;6:e28458
- Mitsuhashi S, Mizushima T, Yamashita E, et al. X-ray structure of beta-carbonic anhydrase from the red alga, Porphyridium purpureum, reveals a novel catalytic site for CO(2) hydration. J Biol Chem 2000;275:5521–6.
- Cronk JD, Endrizzi JA, Cronk MR, et al. Crystal structure of E. coli beta-carbonic anhydrase, an enzyme with an unusual pH-dependent activity. Protein Sci 2001;10:911–22.
- Cronk JD, Rowlett RS, Zhang KY, et al. Identification of a novel noncatalytic bicarbonate binding site in eubacterial beta-carbonic anhydrase. Biochemistry 2006;45:4351–61.
- Schlicker C, Hall RA, Vullo D, et al. Structure and inhibition of the CO2-sensing carbonic anhydrase Can2 from the pathogenic fungus Cryptococcus neoformans. J Mol Biol 2009;385:1207–20.
- Ferraroni M, Del Prete S, Vullo D, et al. Crystal structure and kinetic studies of a tetrameric type II β-carbonic anhydrase from the pathogenic Bacterium Vibrio cholerae. Acta Crystallogr D Biol Crystallogr 2015;71:2449–56.
- Pinard MA, Lotlikar SR, Boone CD, et al. Structure and inhibition studies of a type II beta-carbonic anhydrase psCA3 from Pseudomonas aeruginosa. Bioorg Med Chem 2015;23:4831–8.
- McGurn LD, Moazami-Goudarzi M, White SA, et al. The structure, kinetics and interactions of the β-carboxysomal β-carbonic anhydrase, CcaA. Biochem J 2016;473:4559–72.
- Lehneck R, Neumann P, Vullo D, et al. Crystal structures of two tetrameric β-carbonic anhydrases from the filamentous ascomycete Sordaria macrospora. Febs J 2014;281:1759–72.
- Kim S, Kim NJ, Hong S, et al. The structural basis of the low catalytic activities of the two minor β-carbonic anhydrases of the filamentous fungus Aspergillus fumigatus. J Struct Biol 2019;208:61–8.
- Dostal J, Brynda J, Blaha J, et al. Crystal structure of carbonic anhydrase CaNce103p from the pathogenic yeast Candida albicans. BMC Struct Biol 2018;18:14.
- Holm L. Benchmarking fold detection by DaliLite v.5. Bioinformatics 2019;35:5326–7.
- Pal DS, Mondal DK, Datta R. Identification of metal dithiocarbamates as a novel class of antileishmanial agents. Antimicrob Agents Chemother 2015;59:2144–52.
- Pal DS, Abbasi M, Mondal DK, et al. Interplay between a cytosolic and a cell surface carbonic anhydrase in pH homeostasis and acid tolerance of Leishmania. J Cell Sci 2017;130:754–66.
- D’Ambrosio K, Supuran CT, De Simone G. Are carbonic anhydrases suitable targets to fight protozoan parasitic diseases? Curr Med Chem 2018;25:5266–78.
- Syrjanen L, Vermelho AB, Rodrigues Ide A, et al. Cloning, characterization, and inhibition studies of a β-carbonic anhydrase from Leishmania donovani chagasi, the protozoan parasite responsible for leishmaniasis. J Med Chem 2013;56:7372–81.
- Zolfaghari Emameh R, Kuuslahti M, Vullo D, et al. Ascaris lumbricoides β carbonic anhydrase: a potential target enzyme for treatment of ascariasis. Parasit Vectors 2015;8:479.
- Haapanen S, Bua S, Kuuslahti M, et al. Cloning, Characterization and Anion Inhibition Studies of a beta-Carbonic Anhydrase from the Pathogenic Protozoan Entamoeba histolytica. Molecules 2018;23:3112.
- Rowlett RS. Structure and catalytic mechanism of the beta-carbonic anhydrases. Biochim Biophys Acta 2010;1804:362–73.
- Suhanovsky MM, Sheppard K, Rowlett RS, β-Carbonic anhydrases: general features and medical implications. In: Supran CT, De Simone G, editors. Carbonic anhydrases as biocatalysts. USA: Elsevier B. V.; 2015. p. 247–74.