Abstract
A potential microtubule destabilising series of new indolizine derivatives was synthesised and tested for their anticancer activity against a panel of 60 human cancer cell lines. Compounds 11a, 11b, 15a, and 15j showed a broad spectrum of growth inhibitory activity against cancer cell lines representing leukaemia, melanoma and cancer of lung, colon, central nervous system, ovary, kidney, breast, and prostate. Among them, compound 11a was distinguishable by its excellent cytostatic activity, showing GI50 values in the range of 10–100 nM on 43 cell lines. The less potent compounds 15a and 15j in terms of GI50 values showed a high cytotoxic effect against tested colon cancer, CNS cancer, renal cancer and melanoma cell lines and only on few cell lines from other types of cancer. In vitro assaying revealed tubulin polymerisation inhibition by all active compounds. Molecular docking showed good complementarity of active compounds with the colchicine binding site of tubulin.
Graphical Abstract
Introduction
Recognised as key dynamic structural components in cells, microtubules play an important role in cellular shape organisation, intracellular movement, cell division, and mitosisCitation1,Citation2. Thus, they have been considered an attractive target for the development of new antiproliferative agents in the past few yearsCitation3–7.
Commonly, agents targeting tubulin are divided function of the site they interact with tubulin. The three major sites of tubulin are: the paclitaxel binding site (compounds showing microtubule stabilising effects), vinblastine binding site, and colchicine binding site (compounds showing inhibition of tubulin polymerisation)Citation1,Citation3–5,Citation8. The microtubule-targeting strategy in the drug development field is validated by the use of microtubule-targeting agents such as paclitaxel, docetaxel, vinblastine, colchicine, combretastatin A-4 (CA-4), nocodazole, and many others in cancer chemotherapyCitation1,Citation8–10. However, these compounds also present many drawbacks (high toxicity that induces many side effects, low oral bioavailability, and development of drug resistance) that limit their efficiencyCitation11–13. Therefore, there is a huge demand of novel antimitotic agents to overcome the abovementioned inconveniences.
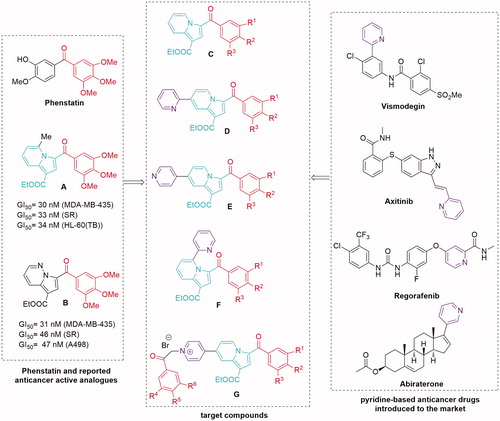
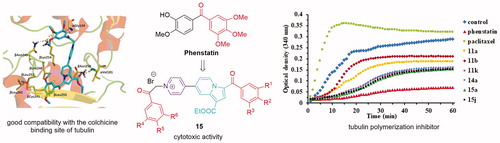
Among the large number of microtubule-targeting agents with diverse scaffolds investigated during last decadesCitation1,Citation2,Citation11, Phenstatin () stands out as one of the simplest molecules that significantly inhibit tubulin polymerisation by binding to the colchicine site of tubulinCitation14,Citation15. Phenstatin is also known for its outstanding antitumor activities on a wide variety of human cancer cellsCitation14,Citation15. Its biological properties are comparable to CA-4, currently investigated in clinical trialsCitation16, but in contrast to CA-4, Phenstatin has a greater pharmacological potential due to improved metabolic stability and requires an easier synthesis for large-scale productionCitation17. In the process of drug discovery, this kind of compounds are lead scaffolds for the development of improved bioactive analogues, and Phenstatin continues to be a source of inspiration for researchers in designing new potential anticancer drugsCitation18–25.
To increase the structural diversity, the combination of various types of bioactive moieties and rings has become a practical strategy in the field of medicinal chemistry. Thus, our study was inspired by several reported anticancer active Phenstatin analogues having the 3′-hydroxy-4′-methoxyphenyl ring replaced with an indolizine moietyCitation21 (compound A, ) or a pyrrolo[1,2-b]pyridazine groupCitation24 (compound B, ).
At the same time, the pyridine ring, present in many natural products and even genetic material, has been noted for its role in many biological processes as well as in cancer pathogenesis, which makes it a privileged scaffold in anticancer agents discoveryCitation26. Thus, there are reports of a variety of monosubstituted pyridines with cytotoxic activityCitation26–29, and there are also several pyridyl-containing drugs introduced on the market for their antitumor propertiesCitation26 (, right).
In our continuous effortsCitation22,Citation24 to discover more effective microtubule destabilising agents, we have applied a structural combination strategy to design and synthesise a new series of indolizine-based Phenstatin analogues and evaluated their anticancer activity. Thus, we considered unsubstituted indolizines (at the pyridine ring) (compounds C, ), as well as several pyridyl-substituted indolizines (compounds D, E, and F, ) in order to investigate a possible beneficial influence of this group to the binding properties of generated compounds due to the lone pair electrons in this moiety. In order to increase biological activity, we also designed an indolizine series that contains two 3,4,5-trimethoxybenzoyl groups (compounds G, ). Furthermore, we modified the 3,4,5-trimethoxyphenyl ring of Phenstatin for each series, replacing it with either a 3,5-dimethoxyphenyl, 3,4-dimethoxyphenyl, or a 4-bromophenyl ring. The structure–activity relationships (SARs), effects on tubulin assembly and theoretical binding interactions were also explored in our study.
Materials and methods
Chemistry
All commercially available reagents and solvents employed were used without further purification. Melting points were recorded on a A. Krüss Optronic Melting Point Meter KSPI and are uncorrected. Analytical thin-layer chromatography was performed with commercial silica gel plates 60 F254 (Merck, Darmstadt, Germany) and visualised with UV light (λmax=254 or 365 nm). The NMR spectra were recorded on an Avance III 500 MHz spectrometer (Bruker, Vienna, Austria) operating at 500 MHz for 1H and 125 MHz for 13C. Chemical shifts were reported in delta (δ) units, part per million (ppm), and coupling constants (J) in Hz. The following abbreviations were used to designate chemical shift multiplicities: s = singlet, d = doublet, t = triplet, q = quartet, m = multiplet, and bs = broad singlet. Infrared (IR) data were recorded as films on potassium bromide (KBr) pellets on an FT-IR (Shimadzu, Kyoto, Japan) Prestige 8400s spectrophotometer or a Jasco 660 plus FTIR spectrophotometer. Analyses indicated by the symbols of the elements or functions were within ±0.4% of the theoretical values.
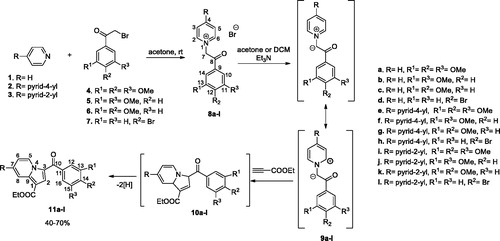
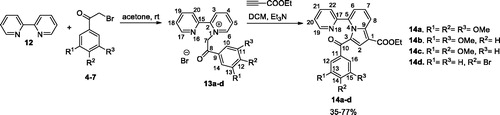
General procedure for synthesis of monoquaternary salts 8a–l and 13a–d
The corresponding heterocycle (pyridine 1, 4,4′-bipyridine 2, 2,4′-bipyridine 3, or 2,2′-bipyridine 12) (1 mmol, 1 equiv.) was dissolved in 5–7 ml acetone. Then, reactive halide (2-bromo-1-(3,4,5-trimethoxyphenyl)ethanone 4, 2-bromo-1-(3,5-dimethoxyphenyl) ethanone 5, 2-bromo-1-(3,4-dimethoxyphenyl) ethanone 6, or 2-bromo-1-(4-bromophenyl) ethanone 7) (1.1 mmol, 1.1 equiv.) was added and the resulted mixture was stirred overnight at room temperature (r.t.). The formed precipitate was filtered and washed with diethyl ether to give the desired product which was used in the next reaction without any further purification.
General procedure for preparation of compounds 11a–l and 14a–d
The cycloimmonium salt (8a–l and 13a–d) (1 mmol, 1 equiv.) and ethyl propiolate (1.1 mmol, 1.1 equiv.) were added to dichloromethane (DCM) and the obtained suspension was stirred at r.t. Then, a solution of triethylamine (TEA) (3 mmol, 3 equiv.) in DCM (3 ml) was added drop-wise over 1 h (magnetic stirring) and the resulting mixture was then stirred overnight at r.t. Methanol (10 ml) was added and the resulting solid was collected by filtration and washed with 5 ml methanol. The product was then purified by crystallisation from DCM/methanol (1/1, v/v) and/or column chromatography using DCM/methanol (99.5/0.5, v/v).
General procedure for preparation of compounds 15a–j
The monoindolizine 11 (1 mmol, 1 equiv.) and reactive halide derivative (4, 5, or 6) (2 mmol, 2 equiv.) were suspended in acetone (10 ml) and the resulted reaction mixture was refluxed under magnetical stirring overnight. The resulting precipitate was collected by filtration and then washed with acetone. Compounds 15a–i were further purified by crystallisation from chloroform/methanol (1:1, v/v).
Ethyl 3-(3,4,5-trimethoxybenzoyl)indolizine-1-carboxylate (11a). Beige solid, yield 51%, mp 197–198 °C. IR ν (cm−1): 1701, 1622, 1584, 1526, 1485, 1350, 1223, 1202, 1130, 1055. 1H NMR (500 MHz, CDCl3): δ 1.40 (t, J = 7.0 Hz, 3H, CH3), 3.92 (s, 6H, 2 × OMe), 3.95 (s, 3H, OMe), 4.38 (q, J = 7.0 Hz, 2H, CH2), 7.08 (s, 2H, H12, H16), 7.10 (t, J = 7.0 Hz, 1H, H6), 7.46 (t, J = 7.5 Hz, 1H, H7), 7.88 (s, 1H, H2), 8.40 (d, J = 8.5 Hz, 1H, H8), 9.91 (d, J = 6.5 Hz, 1H, H5). 13C NMR (125 MHz, CDCl3): δ 14.7 CH3, 56.52 × OMe, 60.3 CH2, 61.2 OMe, 106.5 C1, 107.8 C12, C16, 115.4 C6, 119.7 C8, 122.5 C3, 127.8 C7, 128.9 C2, 129.3 C5, 135.2 C11, 140.1 C9, 141.4 C14, 153.2 C13, C15, 164.2 COO, 184.8 C10. Anal. Calcd. for C21H21NO6: C, 65.79; H, 5.52; N, 3.65. Found: C, 65.74; H, 5.52; N, 3.70.
Ethyl 3-(3,5-dimethoxybenzoyl)indolizine-1-carboxylate (11b). Beige solid, yield 50%, mp 178–180 °C. IR ν (cm−1): 1699, 1628, 1593, 1528, 1458, 1356, 1200, 1155, 1045. 1H NMR (500 MHz, CDCl3): δ 1.40 (t, J = 7.0 Hz, 3H, CH3), 3.86 (s, 6H, 2 × OMe), 4.38 (q, J = 7.0 Hz, 2H, CH2), 6.66 (s, 1H, H14), 6.93 (s, 2H, H12, H16), 7.09 (t, J = 7.0 Hz, 1H, H6), 7.46 (t, J = 7.5 Hz, 1H, H7), 7.87 (s, 1H, H2), 8.40 (d, J = 8.0 Hz, 1H, H8), 9.95 (d, J = 7.0 Hz, 1H, H5). 13C NMR (125 MHz, CDCl3): δ 14.7 CH3, 55.82 × OMe, 60.3 CH2, 103.8 C14, 106.5 C1, 107.0 C12, C16, 115.5 C6, 119.7 C8, 122.5 C3, 127.9 C7, 129.2 C5, 129.4 C2, 140.2 C9, 142.0 C11, 160.8 C13, C15, 164.2 COO, 185.3 C10. Anal. Calcd. for C20H19NO5: C, 67.98; H, 5.42; N, 3.96. Found: C, 68.00; H, 5.40; N, 3.90.
Ethyl 3-(3,4-dimethoxybenzoyl)indolizine-1-carboxylate (11c). Beige solid, yield 56%, mp 196–198 °C. IR ν (cm−1): 1703, 1610, 1514, 1483, 1261, 1211, 1138, 1047; 1H NMR (500 MHz, CDCl3): δ 1.40 (t, J = 7.0 Hz, 3H, CH3), 3.96 (s, 3H, OMe), 3.98 (s, 3H, OMe), 4.38 (q, J = 7.0 Hz, 2H, CH2), 6.96 (d, J = 8.5 Hz, 1H, H15), 7.07 (t, J = 7.0 Hz, 1H, H6), 7.41–7.45 (overlapped signals, 2H, H7, H12), 7.48 (d, J = 8.0 Hz, 1H, H16), 7.86 (s, 1H, H2), 8.39 (d, J = 8.5 Hz, 1H, H8), 9.88 (d, J = 7.0 Hz, 1H, H5). 13C NMR (125 MHz, CDCl3): δ 14.7 CH3, 56.2 OMe, 56.3 OMe, 60.2 CH2, 106.2 C1, 110.3 C15, 111.8 C12, 115.2 C6, 119.7 C8, 122.5 C3, 123.6 C16, 127.5 C7, 128.5 C2, 129.2 C5, 132.6 C11, 139.9 C9, 149.2 C13, 153.4 C14, 164.3 COO, 184.6 C10. Anal. Calcd. for C20H19NO5: C, 67.98; H, 5.42; N, 3.96. Found: C, 67.64; H, 5.40; N, 3.93.
Ethyl 3-(4-bromobenzoyl)indolizine-1-carboxylate (11d)Citation30. Beige solid, yield 50%, mp 130–132 °C. IR ν (cm−1): 3086, 2978, 1697, 1612, 1522, 1479, 1339, 1219, 1140, 1043. 1H NMR (500 MHz, CDCl3): δ 1.41 (t, J = 7.2 Hz, 3H, CH3), 4.38 (q, J = 7.2 Hz, 2H, CH2), 7.10 (dt, J = 6.8; 1.2 Hz, 1H, H6), 7.47 (m, 1H, H7), 7.65 (d, J = 8.4 Hz, 2H, H12, H16), 7.70 (d, J = 8.4 Hz, 2H, H13, H15), 7.75 (s, 1H, H2), 8.40 (d, J = 8.8 Hz, 1H, H8), 9.93 (d, J = 6.8 Hz, 1H, H5). 13C NMR (125 MHz, CDCl3): δ 14.5 CH3, 60.2 CH2, 106.5 C1, 115.4 C6, 119.5 C8, 122.1 C3, 126.2 C14, 127.9 C7, 128.8 C2, 129.1 C5, 130.5 C13, C15, 131.6 C12, C16, 140.0 C9, 138.6 C11, 163.9 COO, 184.1 C10. Anal. Calcd. for C18H14BrNO3: C, 58.08; H, 3.79; N, 3.76. Found: C, 58.04; H, 3,80; N, 3.80.
Ethyl 7-(pyridin-4-yl)-3-(3,4,5-trimethoxybenzoyl)indolizine-1-carboxylate (11e). Beige solid, yield 70%, mp 194–196 °C. IR ν (cm−1): 1701, 1622, 1206, 1580, 1528, 1471, 1352, 1206, 1132, 1051. 1H NMR (500 MHz, CDCl3): δ 1.42 (as, 3H, CH3), 3.93 (s, 6H, 2 x OMe), 3.96 (s, 3H, OMe), 4.41 (bs, 2H, CH2), 7.10 (s, 2H, H12, H16), 7.37 (as, 1H, H6), 7.69 (as, 2H, 2 × Hpy), 7.91 (s, 1H, H2), 8.75 (overlapped signals, 3H, 2 × Hpy, H8), 9.96 (as, 1H, H5). 13C NMR (125 MHz, CDCl3): δ 14.7 CH3, 56.52 × OMe, 60.5 CH2, 61.2 OMe, 107.6 C1, 106.9 C12, C16, 113.7 C6, 117.3 C8, 121.62 × CHpy, 122.8 C3, 129.0 C2, 129.6 C5, 134.9 C14, 137.0 C7, 139.9 C9, 141.6 C11, 145.3 Cpy, 150.82 × CHpy, 153.2 C13, C15, 164.1 COO, 184.9 C10. Anal. Calcd. for C26H24N2O6: C, 67.82; H, 5.25; N, 6.08. Found: C, 67.84; H, 5.20; N, 6.09.
Ethyl 3-(3,5-dimethoxybenzoyl)-7-(pyridin-4-yl)indolizine-1-carboxylate (11f). Beige solid, yield 55%, mp 208–210 °C. IR ν (cm−1): 1717, 1684, 1595, 1514, 1468, 1362, 1246, 1207, 1159, 1045. 1H NMR (500 MHz, CDCl3): δ 1.42 (t, J = 7.0 Hz, 3H, CH3), 3.86 (s, 6H, 2 × OMe), 4.40 (q, J = 7.0 Hz, 2H, CH2), 6.68 (s, 1H, H14), 6.95 (s, 2H, H12, H16), 7.36 (ad, J = 7.0 Hz, 1H, H6), 7.67 (as, 2H, 2 × Hpy), 7.90 (s, 1H, H2), 8.74 (overlapped signals, 3H, 2 × Hpy, H8), 9.99 (ad, J = 7.0 Hz, 1H, H5). 13C NMR (125 MHz, CDCl3): δ 14.7 CH3, 55.8 2 × OMe, 60.4 CH2, 104.0 C14, 107.1 C12, C16, 107.6 C1, 113.8 C6, 117.3 C8, 121.4 2 × CHpy, 122.8 C3, 129.4 C2, 129.7 C5, 137.0 C7, 139.9 C9, 141.6 C11, 145.3 Cpy, 150.82 × CHpy, 160.8 C13, C15, 164.0 COO, 184.4 C10. Anal. Calcd. for C25H22N2O5: C, 69.76; H, 5.15; N, 6.51. Found: C, 69.74; H, 5.10; N, 6.50.
Ethyl 3-(3,4-dimethoxybenzoyl)-7-(pyridin-4-yl)indolizine-1-carboxylate (11g). Beige solid, yield 56%, mp 192–195 °C. IR ν (cm−1): 3079, 2979, 1691, 1596, 1514, 1347, 1266, 1219, 1141, 1020. 1H NMR (500 MHz, CDCl3): δ 1.42 (t, J = 7.0 Hz, 3H, CH3), 3.97 (s, 3H, OMe), 3.99 (s, 3H, OMe), 4.41 (q, J = 7.0 Hz, 2H, CH2), 6.98 (d, J = 8.5 Hz, 1H, H15), 7.35 (d, J = 7.0 Hz, 2H, H6), 7.45 (s, 1H, H12), 7.51 (d, J = 7.0 Hz, 1H, H16), 7.66 (d, J = 5.0 Hz, 2H, 2 × Hpy), 7.89 (s, 1H, H2), 8.75 (overlapped signals, 3H, 2 × Hpy, H8), 9.93 (d, J = 7.0 Hz, 1H, H5). 13C NMR (125 MHz, CDCl3): δ 14.7 CH3, 56.2 OMe, 56.3 OMe, 60.4 CH2, 107.3 C1, 110.3 C15, 111.7 C12, 113.5 C6, 117.3 C8, 121.4 2 × CHpy, 123.0 C3, 123.7 C16, 128.7 C5, 129.5 C2, 132.2 C11, 136.6 C7, 139.6 C9, 145.5 Cpy, 149.3 C13, 150.8 2 × CHpy, 152.6 C14, 164.2 COO, 184.7 C10; Anal. Calcd. for C25H22N2O5: C, 69.76; H, 5.15; N, 6.51. Found: C, 69.72; H, 5.11; N, 6.55.
Ethyl 3-(4-bromobenzoyl)-7-(pyridin-4-yl)indolizine-1-carboxylate (11h). Yield 65%. All spectral data are in agreement with the literatureCitation31.
Ethyl 7-(pyridin-2-yl)-3-(3,4,5-trimethoxybenzoyl)indolizine-1-carboxylate (11i). Yellow solid, yield 41%, mp 202–203 °C. IR ν (cm−1): 2994, 2926, 1700, 1620, 1580, 1480, 1352, 1204, 1136, 780. 1H NMR (500 MHz, CDCl3): δ 1.43 (t, J = 7.0 Hz, 3H, CH3), 3.94 (s, 6H, 2 × OMe), 3.97 (s, 3H, OMe), 4.42 (q, J = 7.0 Hz, 2H, CH2), 7.11 (s, 2H, H12, H16), 7.35 (dd, J = 7.5; 5.0 Hz, 1H, Hpy), 7.85 (dt, J = 8.0; 2.0 Hz, 1H, Hpy), 7.90 (s, 1H, H2), 7.92 (dd, J = 7.5; 2.0 Hz, 1H, H6), 7.97 (d, J = 8.0 Hz, 1H, Hpy), 8.77 (d, J = 4.5 Hz, 1H, Hpy), 9.01 (bs, 1H, H8), 9.95 (d, J = 7.0 Hz, 1H, H5). 13C NMR (125 MHz, CDCl3): δ 14.7 CH3, 55.62 × OMe, 60.5 CH2, 61.2 OMe, 107.0 C12, C16, 108.0 C1, 113.9 C6, 118.1 C8, 122.5 CHpy, 123.2 C3, 124.3 CHpy, 128.9 C2, 129.6 C5, 134.9 C11, 138.9 C7, 139.5 C9, 140.2 CHpy, 141.7 C14, 147.9 CHpy, 153.2 Cpy, 153.3 C13, C15, 164.1 COO, 184.9 C10. Anal. Calcd. for C26H24N2O6: C, 67.82; H, 5.25; N, 6.08. Found: C, 67.85; H, 5.17; N, 6.13.
Ethyl 3-(3,5-dimethoxybenzoyl)-7-(pyridin-2-yl)indolizine-1-carboxylate (11j). Yellow solid, yield 40%, mp 165–166 °C. IR ν (cm−1): 2934, 1696, 1587, 1448, 1352, 1209, 1152, 1044, 774. 1H NMR (500 MHz, CDCl3): δ 1.43 (t, 3H, J = 7.0 Hz, CH3), 3.87 (s, 6H, 2 × OMe), 4.41 (q, 2H, J = 7.0 Hz, CH2), 6.68 (t, J = 2.5 Hz, 1H, H14), 6.96 (d, J = 2.5 Hz, 2H, H12, H16), 7.34 (dd, J = 7.0; 5.0 Hz, 1H, Hpy), 7.84 (dt, J = 7.5; 2.0 Hz, 1H, Hpy), 7.89 (s, 1H, H2), 7.92 (dd, J = 7.5; 2.0 Hz, 1H, H6), 7.97 (d, J = 8.0 Hz, 1H, Hpy), 8.77 (d, J = 4.5 Hz, 1H, Hpy), 9.00 (d, J = 1.0 Hz, 1H, H8), 9.99 (d, J = 7.5 Hz, 1H, H5). 13C NMR (125 MHz, CDCl3): δ 14.7 CH3, 55.82 × OMe, 60.4 CH2, 103.9 C14, 107.0 C12, C16, 107.5 C1, 114.0 C6, 116.8 C8, 121.0 CHpy, 122.8 C3, 123.6 CHpy, 129.4 C2, 129.5 C5, 137.3 CHpy, 138.6 C7, 140.3 C9, 141.9 C11, 150.1 CHpy, 154.4 Cpy, 160.8 C13, C15, 164.2 COO, 185.6 C10. Anal. Calcd. for C25H22N2O5: C, 69.76; H, 5.15; N, 6.51. Found: C, 69.79; H, 5.10; N, 6.53.
Ethyl 3-(3,4-dimethoxybenzoyl)-7-(pyridin-2-yl)indolizine-1-carboxylate (11k). Yellow solid, yield 44%, mp 199–200 °C. IR ν (cm−1): 2974, 2931, 1699, 1608, 1517, 1472, 1426, 1347, 1267, 1199, 773. 1H NMR (500 MHz, CDCl3): δ 1.44 (t, J = 7.0 Hz, 3H, CH3), 3.98 (s, 3H, OMe), 4.00 (s, 3H, OMe), 4.42 (q, J = 7.0 Hz, 2H, CH2), 6.99 (d, J = 8.5 Hz, 1H, H15), 7.46 (bs, 1H, H12), 7.33 (dd, J = 7.0; 5.0 Hz, 1H, Hpy), 7.85 (t, J = 7.5 Hz, 1H, Hpy), 7.88 (s, 1H, H2), 7.90 (dd, J = 7.5; 1.5 Hz, 1H, H6), 7.97 (d, J = 8.0 Hz, 1H, Hpy), 8.77 (d, J = 4.5 Hz,1H, Hpy), 7.52 (dd, J = 8.0; 1.5 Hz, 1H, H16), 9.00 (bs, 1H, H8), 9.93 (d, J = 7.5 Hz, 1H, H5). 13C NMR (125 MHz, CDCl3): δ 14.7 CH3, 56.2 OMe, 56.3 OMe, 60.3 CH2, 107.2 C1, 110.3 C15, 111.8 C12, 113.8 C6, 116.8 C8, 121.0 CHpy, 123.0 C3, 123.5 CHpy, 123.7 C16, 128.8 C2, 129.2 C5, 132.5 C11, 137.2 CHpy, 138.2 C7, 140.0 C9, 149.2 C13, 150.1 CHpy, 152.5 C14, 154.5 Cpy, 164.3 COO, 184.6 C10. Anal. Calcd. for C25H22N2O5: C, 69.76; H, 5.15; N, 6.51. Found: C, 69.78; H, 5.09; N, 6.53.
Ethyl 3-(4-bromobenzoyl)-7-(pyridin-2-yl)indolizine-1-carboxylate (11l). Yellow solid, yield 40%, mp 197–198 °C. IR ν (cm−1): 2928, 1694, 1620, 1526, 1479, 1342, 1200, 1079. 1H NMR (500 MHz, CDCl3): δ 1.43 (t, J = 7.0 Hz, 3H, CH3), 4.41 (q, J = 7.0 Hz, 2H, CH2), 7.34 (dd, J = 7.5; 4.5 Hz, 1H, Hpy), 7.68 (d, J = 8.0 Hz, 2H, H12, H16), 7.73 (d, J = 8.0 Hz, 2H, H13, H15), 7.80 (s, 1H, H2), 7.84 (dt, J = 7.5; 2.0 Hz, 1H, Hpy), 7.94 (dd, J = 7.5; 2.0 Hz, 1H, H6), 7.97 (d, J = 8.0 Hz, 1H, Hpy), 8.77 (d, J = 4.5 Hz, 1H, Hpy), 9.01 (d, J = 1.0 Hz, 1H, H8), 9.98 (d, J = 7.5 Hz, 1H, H5). 13C NMR (125 MHz, CDCl3): δ 14.7 CH3, 60.6 CH2, 108.5 C1, 114.0 C6, 118.3 C8, 122.7 CHpy, 123.0 C3, 124.4 CHpy, 126.8 C14, 129.1 C2, 129.7 C5, 130.7 C13, C15, 132.0 C12, C16, 138.5 C11, C7, 139.5 C9, 140.2 CHpy, 147.6 CHpy, 153.0 Cpy, 164.0 COO, 184.5 C10. Anal. Calcd. for C23H17BrN2O3: C, 61.48; H, 3.81; N, 6.23. Found: C, 61.49; H, 3.78; N, 6.26.
Ethyl 5-(pyridin-2-yl)-3-(3,4,5-trimethoxybenzoyl)indolizine-1-carboxylate (14a). Cream solid, yield 35%, mp 161–163 °C. IR ν (cm−1): 2984, 1686, 1639, 1585, 1520, 1419, 1350, 1234, 1130, 1059, 752. 1H NMR (500 MHz, CDCl3): δ 1.40 (t, J = 7.0 Hz, 3H, CH3), 3.87 (s, 6H, 2 × OMe), 3.95 (s, 3H, OMe), 4.38 (q, J = 7.0 Hz, 2H, CH2), 7.08 (d, J = 6.0 Hz, 1H, H6), 7.13 (s, 2H, H12, H16), 7.20 (dd, J = 6.5; 4.5 Hz, 1H, H20), 7.46 (dd, J = 9.0; 7.0 Hz, 1H, H7), 7.64 (s, 1H, H2), 7.72 (d, J = 8.0 Hz, 1H, H22), 7.91 (t, J = 7.5 Hz, 1H, H21), 8.27 (d, J = 4.0 Hz, 1H, H19), 8.47 (d, J = 8.5 Hz, 1H, H8). 13C NMR (125 MHz, CDCl3): δ 14.7 CH3, 56.5 2 x OMe, 60.2 CH2, 61.1 OMe, 105.9 C1, 107.1 C12, C16, 118.1 C6, 120.2 C8, 122.2 C22, 124.0 C20, 126.5 C7, C3, 127.3 C2, 133.3 C11, 138.6 C5, 138.7 C21, 141.4 C9, 141.8 C14, 148.5 C19, 153.0 C13, C15, 155.2 C17, 164.4 COO, 183.4 C10. Anal. Calcd. for C26H24N2O6: C, 67.82; H, 5.25; N, 6.08. Found: C, 67.81; H, 5.23; N, 6.11.
Ethyl 3-(3,5-dimethoxybenzoyl)-5-(pyridin-2-yl)indolizine-1-carboxylate (14b). Yellow solid, yield 77%, mp 149–150 °C. IR ν (cm−1): 2974, 1715, 1630, 1595, 1421, 1188, 1155, 754. 1H NMR (500 MHz, CDCl3): δ 1.39 (t, J = 7.0 Hz, 3H, CH3), 3.81 (s, 6H, 2 × OMe), 4.37 (q, J = 7.0 Hz, 2H, CH2), 6.66 (bs, 1H, H14), 7.00 (d, J = 2.0 Hz, 2H, H12, H16), 7.09 (d, J = 7.0 Hz, 1H, H6), 7.18 (dd, J = 6.5; 5.5 Hz, 1H, H20), 7.47 (dd, J = 8.5; 7.5 Hz, 1H, H7), 7.65 (s, 1H, H2), 7.71 (d, J = 8.0 Hz, 1H, H22), 7.88 (t, J = 7.5 Hz, 1H, H21), 8.30 (d, J = 4.5 Hz, 1H, H19), 8.47 (d, J = 8.5 Hz, 1H, H8). 13C NMR (125 MHz, CDCl3): 14.7 CH3, 55.82 × OMe, 60.2 CH2, 105.1 C14, 105.9 C1, 107.3 C12, C16, 118.2 C6, 120.2 C8, 122.0 C22, 123.9 C20, 126.5 C3, 126.7 C7, 127.8 C2, 138.5 C21, 138.7 C5, 140.2 C11, 141.6 C9, 148.5 C19, 155.2 C17, 160.7 C13, C15, 164.4 COO, 183.6 C10. Anal. Calcd. for C25H22N2O5: C, 69.76; H, 5.15; N, 6.51. Found: C, 69.78; H, 5.13; N, 6.54.
Ethyl 3-(3,4-dimethoxybenzoyl)-5-(pyridin-2-yl)indolizine-1-carboxylate (14c). Yellow solid, yield 35%. IR ν (cm−1): 2933, 1719, 1676, 1624, 1593, 1514, 1417, 1269, 1224, 1024, 766. 1H NMR (500 MHz, CDCl3): δ 1.39 (t, J = 7.0 Hz, 3H, CH3), 3.98 (s, 3H, OMe), 4.00 (s, 3H, OMe), 4.38 (q, J = 7.0 Hz, 2H, CH2), 6.95 (d, J = 8.5 Hz, 1H, H15), 7.08 (d, J = 6.0 Hz, 1H, H6), 7.19 (dd, J = 6.5; 4.5 Hz, 1H, H20), 7.42 (bs, 1H, H12), 7.47 (dd, J = 8.5; 7.0 Hz, 1H, H7), 7.48 (d, J = 8.0 Hz, 1H, H16), 7.64 (s, 1H, H2), 7.71 (d, J = 8.0 Hz, 1H, H22), 7.90 (t, J = 7.5 Hz, 1H, H21), 8.28 (d, J = 4.0 Hz, 1H, H19), 8.47 (d, J = 8.5 Hz, 1H, H8). 13C NMR (125 MHz, CDCl3): δ 14.6 CH3, 56.2 OMe, 56.3 OMe, 60.2 CH2, 106.0 C1, 110.3 C15, 111.9 C12, 118.2 C6, 120.2 C8, 122.1 C22, 123.5 C16, 124.0 C20, 126.5 C3, 126.6 C7, 127.6 C2, 132.5 C11, 138.7 C5, 138.6 C21, 141.5 C9, 148.5 C19, 149.1 C13, 153.3 C14, 155.2 C17, 164.3 COO, 184.5 C10. Anal. Calcd. for C25H22N2O5: C, 69.76; H, 5.15; N, 6.51. Found: C, 69.79; H, 5.12; N, 6.55.
Ethyl 3-(4-bromobenzoyl)-5-(pyridin-2-yl)indolizine-1-carboxylate (14d)Citation30. Yellow crystals, yield 40%, mp 229–231 °C. IR ν (cm−1): 2925, 1725, 1694, 1650, 1586, 1419, 1342, 1070, 750. 1H NMR (500 MHz, DMSO-d6): δ 1.31 (t, J = 7.0 Hz, 3H, CH3), 4.29 (q, J = 7.0 Hz, 2H, CH2), 7.24 (dd, J = 7.0; 4.5 Hz, 1H, H20), 7.35 (dd, J = 7.5; 1.0 Hz, 1H, H6), 7.42 (s, 1H, H2), 7.67–7.70 (overlapped signals, 3H, H12, H16, H7), 7.74 (d, J = 8.5 Hz, 2H, H13, H15), 7.81 (d, J = 8.0 Hz, 1H, H22), 7.93 (dt, J = 7.5; 1.5 Hz, 1H, H21), 8.13 (d, J = 4.0 Hz, 1H, H19), 8.38 (dd, J = 9.0; 1.0 Hz, 1H, H8). 13C NMR (125 MHz, DMSO-d6): δ 14.5 CH3, 59.8 CH2, 104.4 C1, 118.3 C6, 119.0 C8, 122.2 C22, 124.3 C20, 125.1 C2, 126.2 C3, 126.8 C14, 127.7 C7, 131.2 C12, C16, 131.8 C13, C15, 136.2 C11, 138.4 C21, 138.8 C5, 140.4 C9, 148.3 C19, 154.2 C17, 163.3 COO, 182.2 C10. Anal. Calcd. for C23H17BrN2O3: C, 61.48; H, 3.81; N, 6.23. Found: C, 61.51; H, 3.77; N, 6.25.
4-(1-(Ethoxycarbonyl)-3-(3,4,5-trimethoxybenzoyl)indolizin-7-yl)-1-(2-oxo-2-(3,4,5-trimethoxy phenylethyl)pyridin-1-ium bromide (15a). Orange solid, yield 70%, mp 188 °C. IR ν (cm−1): 2986, 2941, 1713, 1640, 1583, 1530, 1508, 1465, 1412, 1346, 1323, 1207, 1130. 1H NMR (500 MHz, CDCl3): δ 1.42 (t, J = 7.0 Hz, 3H, CH3), 3.92 (s, 6H, 2 × OMe), 3.93 (s, 3H, OMe), 3.96 (s, 6H, 2 × OMe), 3.97 (s, 3H, OMe), 4.42 (q, J = 7.0 Hz, 2H, CH2), 7.09 (s, 2H, H12, H16), 7.26 (s, 2H, H23), 7.47–7.49 (3H, overlapped signals, H6, H26, H30), 7.89 (s, 1H, H2), 8.35 (bs, 2H, H18, H22), 8.89 (bs, 1H, H8), 9.35 (bs, 2H, H19, H21,), 9.92 (d, J = 7.0 Hz, 1H, H5). 13C NMR (125 MHz, CDCl3): δ 14.7 CH3, 56.62 × OMe, 57.22 × OMe, 60.9 CH2, 61.1 OMe, 61.2 OMe, 66.4 C23, 106.9 C26, C30, 107.0 C12, C16, 109.8 C1, 112.4 C6, 119.9 C8, 123.9 C3, 124.1 C18, C22, 128.4 C2, 128.6 C25, 130.0 C5, 130.9 C7, 134.2 C11, 138.2 C9, 142.1 C14, 144.4 C28, 147.0 C19, C21, 153.3 C13, C15, 153.6 C27, C29, 153.9 C17, 163.5 COO, 185.0 C10, 189.3 C24. Anal. Calcd. for C37H37BrN2O10: C, 59.28; H, 4.98; N, 3.74. Found: C, 59.32; H, 4.87; N, 3.79.
1-(2-(3,5-Dimethoxyphenyl)-2-oxoethyl)-4-(1-(ethoxycarbonyl)-3-(3,4,5-trimethoxybenzoyl) indolizin-7-yl)pyridin-1-ium bromide (15b). Orange solid, yield 65%, mp 175 °C. IR ν (cm−1): 2928, 1700, 1638, 1529, 1457, 1351, 1207, 1016. 1H NMR (400 MHz, DMSO-d6): δ 1.37 (t, J = 6.8 Hz, 3H, CH3), 3.80 (s, 3H, OMe), 3.86 (s, 12H, 4 × OMe), 4.37 (q, J = 6.8 Hz, 2H, CH2), 6.50 (s, 2H, H23), 6.95 (s, 1H, H28), 7.15 (s, 2H, H12, H16), 7.21 (s, 2H, H26, H30), 7.84 (s, 1H, H2), 7.96 (bs, H6), 8.80 (bs, 2H, H18, H22), 8.95 (bs, 1H, H8), 9.11 (bs, 2H, H19, H21), 9.87 (bs, 1H, H5). 13C NMR (100 MHz, DMSO-d6): δ 14.3 CH3, 55.82 × OMe, 56.23 × OMe, 60.3 CH2, 65.9 C23, 106.1 C26, C28, C30, 106.7 C12, C16, 108.2 C1, 113.7 C6, 119.0 C8, 123.1 C3, 124.8 C18, C22, 127.9 C2, 129.2 C5, 132.0 C7, 133.9 C11, 135.4 C25, 137.8 C9, 141.0 C14, 146.6 C19, C21, 152.5 C17, 152.7 C13, C15, 160.9 C27, C29, 162.8 COO, 184.1 C10, 190.6 C24. Anal. Calcd. for C36H35BrN2O9: C, 60.09; H, 4.90; N, 3.89. Found: C, 60.12; H, 4.87; N, 3.94.
1-(2-(3,4-Dimethoxyphenyl)-2-oxoethyl)-4-(1-(ethoxycarbonyl)-3-(3,4,5-trimethoxybenzoyl) indolizin-7-yl)pyridin-1-ium bromide (15c). Orange solid, yield 50%, mp 236–239 °C. IR ν (cm−1): 2925, 1699, 1638, 1582, 1523, 1460, 1345, 1272, 1205. 1H NMR (400 MHz, DMSO-d6): δ 1.37 (t, J = 6.8 Hz, 3H, CH3), 3.80 (s, 3H, OMe), 3.86 (s, 9H, 3 × OMe), 3.92 (s, 3H, OMe), 4.37 (q, J = 6.8 Hz, 2H, CH2), 6.47 (s, 2H, H23), 7.16 (s, 2H, H12, H16), 7.24 (bs, 1H, H29), 7.55 (bs, 1H, H26), 7.81 (bs, 1H, H30), 7.85 (s, 1H, H2), 7.96 (bs, 1H, H6), 8.80 (bs, 2H, H18, H22), 8.97 (bs, 1H, H8), 9.11 (bs, 2H, H19, H21), 9.89 (bs, 1H, H5). 13C NMR (100 MHz, DMSO-d6): δ 14.3 CH3, 55.8 OMe, 56.0 2 × OMe, 56.22 × OMe, 60.2 CH2, 65.4 C23, 110.3 C26, 106.7 C12, C16, 108.2 C1, 111.3 C29, 113.7 C6, 118.9 C8, 123.2 C3, 123.4 C30, 124.7 C18, C22, 126.2 C25, 127.9 C2, 129.3 C5, 132.0 C7, 133.9 C11, 137.8 C9, 141.0 C14, 146.6 C19, C21, 148.9 C27, 152.4 C28, 152.7 C13, C15, C17, 162.8 COO, 184.1 C10, 189.0 C24. Anal. Calcd. for C36H35BrN2O9: C, 60.09; H, 4.90; N, 3.89. Found: C, 60.10; H, 4.85; N, 3.93.
4-(3-(3,5-Dimethoxybenzoyl)-1-(ethoxycarbonyl)indolizin-7-yl)-1-(2-oxo-2-(3,4,5-trimethoxyphenyl)ethyl)pyridin-1-ium bromide (15d). Orange solid, yield 84%, mp 176–178 °C. IR ν (cm−1): 2913, 1696, 1683, 1639, 1590, 1528, 1454, 1402, 1352, 1207, 1159, 1130. 1H NMR (500 MHz, DMSO-d6): δ 1.36 (t, J = 7.0 Hz, 3H, CH3), 3.81 (s, 3H, OMe), 3.84 (s, 6H, 2 × OMe), 3.92 (s, 6H, 2 × OMe), 4.37 (q, J = 7.0 Hz, 2H, CH2), 6.52 (s, 2H, H23), 6.83 (t, J = 2.0 Hz, 1H, H14), 6.93 (d, J = 2.0 Hz, 2H, H12, H16), 7.39 (s, 2H, H26, H30), 7.76 (s, 1H, H2), 7.97 (dd, J = 7.5; 2.0 Hz, H6), 8.81 (d, J = 7.0 Hz, 2H, H18, H22), 8.97 (bs, 1H, H8), 9.10 (d, J = 6.5 Hz, 2H, H19, H21), 9.92 (d, J = 7.5 Hz, 1H, H5). 13C NMR (125 MHz, DMSO-d6): δ 14.4 CH3, 55.6 2 × OMe, 56.4 2 × OMe, 60.3 CH2, 60.4 OMe, 65.7 C23, 103.5 C14, 106.0 C26, C30, 106.8 C12, C16, 108.2 C1, 113.9 C6, 119.0 C8, 123.1 C3, 124.8 C18, C22, 128.1 C2, 128.8 C25, 129.3 C5, 132.2 C7, 138.0 C9, 140.8 C11, 143.2 C28, 146.6 C19, C21, 152.4 C17, 153.1 C27, C29, 160.4 C13, C15, 162.8 COO, 184.6 C10, 189.7 C24. Anal. Calcd. for C36H35BrN2O9: C, 60.09; H, 4.90; N, 3.88. Found: C, 60.03; H, 4.81; N, 4.84.
4-(3-(3,5-Dimethoxybenzoyl)-1-(ethoxycarbonyl)indolizin-7-yl)-1-(2-(3,5-dimethoxyphenyl)-2-oxoethyl)pyridin-1-ium bromide (15e). Orange solid, yield 75%, mp 149–150 °C. IR ν (cm−1): 2940, 1709, 1640, 1603, 1465, 1425, 1348, 1207, 1159, 1062, 758. 1H NMR (500 MHz, DMSO-d6): δ 1.36 (t, J = 7.0 Hz, 3H, CH3), 3.84 (s, 6H, 2 × OMe), 3.87 (s, 6H, 2 × OMe), 4.37 (q, J = 7.0 Hz, 2H, CH2), 6.48 (s, 2H, H23), 6.83 (t, J = 2.0 Hz, 1H, H14), 6.93 (d, J = 2.0 Hz, 2H, H12, H16), 6.95 (t, J = 2.0 Hz, 1H, H28), 7.21 (d, J = 2.0 Hz, 2H, H26, H30), 7.75 (s, 1H, H2), 7.97 (dd, J = 7.5; 1.5 Hz, H6), 8.80 (d, J = 6.5 Hz, 2H, H18, H22), 8.96 (bs, 1H, H8), 9.10 (d, J = 6.5 Hz, 2H, H19, H21), 9.91 (d, J = 7.5 Hz, 1H, H5). 13C NMR (DMSO-d6, 125 MHz): δ 14.4 CH3, 55.62 × OMe, 55.82 × OMe, 60.2 CH2, 65.9 C23, 103.5 C14, 106.1 C26, C30, 106.2 C28, 106.8 C12, C16, 108.2 C1, 113.8 C6, 119.0 C8, 123.1 C3, 124.8 C18, C22, 128.1 C2, 129.3 C5, 132.2 C7, 135.4 C25, 138.0 C9, 140.8 C11, 146.6 C19, C21, 152.5 C17, 160.4 C13, C15, 160.9 C27, C29, 162.8 COO, 184.6 C10, 190.5 C24. Anal. Calcd. for C35H33BrN2O8: C, 60.96; H, 4.82; N, 4.06. Found: C, 60.93; H, 4.77; N, 4.13.
4-(3-(3,5-Dimethoxybenzoyl)-1-(ethoxycarbonyl)indolizin-7-yl)-1-(2-(3,4-dimethoxyphenyl)-2-oxoethyl)pyridin-1-ium bromide (15f). Orange solid, yield 60%, mp 220–223 °C. IR ν (cm−1): 2924, 2851, 1696, 1640, 1596, 1524, 1463, 1401, 1349, 1200, 1157. 1H NMR (500 MHz, DMSO-d6): δ 1.36 (t, J = 7.0 Hz, 3H, CH3), 3.84 (s, 6H, 2 × OMe), 3.86 (s, 3H, OMe), 3.92 (s, 3H, OMe), 4.37 (q, J = 7.0 Hz, 2H, CH2), 6.45 (s, 2H, H23), 6.83 (bs, 1H, H14), 6.93 (d, J = 2.0 Hz, 2H, H12, H16), 7.24 (d, J = 8.5 Hz, 1H, H29), 7.54 (bs, 1H, H26), 7.76 (s, 1H, H2), 7.79 (dd, J = 8.0; 1.0 Hz, 1H, H30), 7.97 (dd, J = 7.5; 1.5 Hz, H6), 8.79 (d, J = 6.5 Hz, 2H, H18, H22), 8.97 (bs, 1H, H8), 9.09 (d, J = 6.5 Hz, 2H, H19, H21), 9.92 (d, J = 7.5 Hz, 1H, H5). 13C NMR (125 MHz DMSO-d6): δ 14.4 CH3, 55.72 × OMe, 55.8 OMe, 56.1 OMe, 60.3 CH2, 65.6 C23, 103.6 C14, 106.9 C12, C16, 108.0 C1, 110.1 C26, 111.6 C29, 113.7 C6, 119.0 C8, 123.2 C3, 123.5 C30, 124.8 C18, C22, 126.3 C25, 128.2 C2, 129.3 C5, 132.3 C7, 138.0 C9, 140.8 C11, 146.7 C19, C21, 149.0 C27, 152.4 C28, 154.3 C17, 160.5 C13, C15, 162.9 COO, 184.9 C10, 189.1 C24. Anal. Calcd. for C35H33BrN2O8: C, 60.96; H, 4.82; N, 4.06; Found: C, 60.93; H, 4.80; N, 4.13.
4-(3-(3,4-Dimethoxybenzoyl)-1-(ethoxycarbonyl)indolizin-7-yl)-1-(2-oxo-2-(3,4,5-trimethoxyphenyl)ethyl)pyridin-1-ium bromide (15g). Orange solid, yield 70%, mp 132–137 °C. IR ν (cm−1): 2924, 1698, 1640, 1619, 1456, 1410, 1344, 1265, 1206, 1124. 1H NMR (500 MHz, CDCl3): δ 1.44 (t, J = 7.0 Hz, 3H, CH3), 3.93 (s, 3H, OMe), 3.97 (s, 6H, 2 × OMe), 4.00 (s, 6H, 2 × OMe), 4.42 (q, J = 7.0 Hz, 2H, CH2), 7.23 (s, 2H, H23), 6.95 (d, J = 8.5 Hz, 1H, H15), 7.42–7.47 (3H, overlapped signals, H12, H16, H6), 7.49 (bs, 2H, H26, H30), 7.84 (s, 1H, H2), 8.32 (d, J = 5.5 Hz, 2H, H18, H22), 8.84 (bs, 1H, H8), 9.32 (d, J = 5.0 Hz, 2H, H19, H21), 9.87 (d, J = 7.5 Hz, 1H, H5). 13C NMR (125 MHz, CDCl3): δ 14.7 CH3, 56.1 OMe, 56.22 × OMe, 56.92 × OMe, 60.8 CH2, 66.4 C23, 106.7 C26, C30, 109.3 C1, 110.1 C15, 111.7 C12, C6, 119.4 C8, 123.7 C16, C3, 123.8 C18, C22, 128.1 C25, 128.4 C2, 129.7 C5, 131.5 C11, 130.4 C7, 137.8 C9, 144.1 C28, 146.9 C19, C21, 149.3 C13, 152.9 C1, 153.3 C17, 153.4 C27, C29, 163.5 COO, 184.3 C10, 189.4 C24. Anal. Calcd. for C36H35BrN2O9: C, 60.09; H, 4.90; N, 3.89. Found: C, 60.13; H, 4.89; N, 3.93.
4-(3-(3,4-Dimethoxybenzoyl)-1-(ethoxycarbonyl)indolizin-7-yl)-1-(2-(3,5-dimethoxyphenyl)-2-oxoethyl)pyridin-1-ium bromide (15h). Orange solid, yield 62%, mp 138–139 °C. IR ν (cm−1): 2925, 1698, 1641, 1596, 1519, 1458, 1342, 1264, 1204, 1017. 1H NMR (500 MHz, CDCl3): δ 1.45 (t, J = 7.0 Hz, 3H, CH3), 3.86 (s, 6H, 2 × OMe), 3.98 (s, 3H, OMe), 4.00 (s, 3H, OMe), 4.42 (q, J = 7.0 Hz, 2H, CH2), 6.71 (s, 2H, H23), 6.63 (bs, 1H, H28), 6.94 (d, J = 8.5 Hz, 1H, H15), 7.28 (bs, 2H, H26, H30), 7.43–7.45 (3H, overlapped signals, H12, H16, H6), 7.81 (s, 1H, H2), 8.32 (bs, 2H, H18, H22), 8.82 (bs, 1H, H8), 9.35 (bs, 2H, H19, H21), 9.91 (d, J = 7.5 Hz, 1H, H5). 13C NMR (125 MHz, CDCl3): δ 14.7 CH3, 56.23 OMe, 56.24 2 x OMe, 56.3 OMe, 60.9 CH2, 66.8 C23, 106.6 C26, C30, 107.9 C28, 109.5 C1, 110.2 C15, 111.8 C12, 112.3 C6, 119.7 C8, 123.9 C16, C3, 124.0 C18, C22, 128.3 C2, 129.8 C5, 130.6 C7, 131.6 C11, 135.3 C25, 137.9 C9, 147.1 C19, C21, 149.4 C13, 153.0 C14, 153.6 C17, 161.3 C27, C29, 163.5 COO, 184.5 C10, 190.4 C24. Anal. Calcd. for C35H33BrN2O8: C, 60.96; H, 4.82; N, 4.06. Found: C, 60.95; H, 4.79; N, 4.09.
4-(3-(3,4-Dimethoxybenzoyl)-1-(ethoxycarbonyl)indolizin-7-yl)-1-(2-(3,4-dimethoxyphenyl)-2-oxoethyl)pyridin-1-ium bromide (15i). Orange solid, yield 80%, mp 247–250 °C. IR ν (cm−1): 2972, 1702, 1684, 1595, 1518, 1413, 1337, 1268, 1213, 1019. 1H NMR (500 MHz, DMSO-d6): δ 1.37 (t, J = 7.0 Hz, 3H, CH3), 3.85 (s, 3H, OMe), 3.87 (s, 3H, OMe), 3.90 (s, 3H, OMe), 3.92 (s, 3H, OMe), 4.38 (q, J = 7.0 Hz, 2H, CH2), 6.47 (s, 2H, H23), 7.18 (d, J = 8.5 Hz, 1H, H15), 7.24 (d, J = 8.5 Hz, 1H, H29), 7.43 (bs, 1H, H12), 7.52 (d, J = 8.5 Hz, 1H, H16), 7.54 (bs, 1H, H26), 7.79 (s, 1H, H2), 7.80 (d, J = 8.0 Hz, 1H, H30), 7.93 (J = 7.0 Hz, d, H6), 8.79 (d, J = 6.5 Hz, 2H, H18, H22), 8.96 (bs, 1H, H8), 9.10 (d, J = 6.5 Hz, 2H, H19, H21), 9.84 (d, J = 7.5 Hz, 1H, H5). 13C NMR (125 MHz, DMSO-d6): δ 14.4 CH3, 55.6 OMe, 55.7 OMe, 55.8 OMe, 56.0 OMe, 60.2 CH2, 65.4 C23, 107.9 C1, 110.3 C26, 111.0 C15, 111.3 C29, 111.7 C12, 113.4 C6, 119.0 C8, 123.4 C3, C30, 123.6 C16, 124.6 C18, C22, 126.3 C25, 127.3 C2, 129.2 C5, 131.0 C11, 131.8 C7, 137.7 C9, 146.6 C19, C21, 148.7 C13, 148.9 C27, 152.4 C28, 152.6 C14, 154.4 C17, 162.9 COO, 183.8 C10, 189.1 C24. Anal. Calcd. for C35H33BrN2O8: C, 60.96; H, 4.82; N, 4.06. Found: C, 60.90; H, 4.78; N, 4.11.
4-(3-(4-Chlorobenzoyl)-1-(ethoxycarbonyl)indolizin-7-yl)-1-(2-(4-methoxy phenyl)-2-oxoethyl) pyridin-1-ium bromide (15j). Yield 93%. All spectral data are in agreement with the literatureCitation32.
Anticancer activity
The compounds were tested against a panel of 60 human cancer cell lines at the National Cancer Institute (NCI) (Rockville, MD). The cytotoxicity experiments were performed using a 48 h exposure protocol which consisted of a sulphorhodamine B assayCitation33–35.
Tubulin polymerisation assay
Microtubule assembly was studied using the tubulin polymerisation assay kit (Cytoskeleton Inc., Denver, CO, Cat. # BK006P), according to the manufacturer's instructionsCitation36,Citation37.
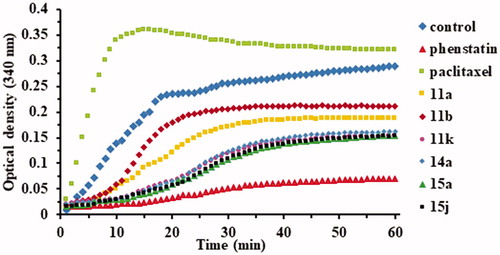
The polymerisation was monitored using FLUOstar Omega multi-mode microplate reader (BMG LABTECH, Ortenberg, Germany). The first step before the analysis is the pre-warming of the plate to 37 °C for 30 min. Plate temperature is essential for high polymerisation activity and reproducible results. The tubulin polymerisation buffer will be composed of general tubulin buffer, tubulin glycerol buffer, and GTP stock, at 4 °C. Another volume of 500 µl of general tubulin buffer is necessary for dilutions, at r.t. Ten microlitres of GTB will be pipetted into each well. Two of the wells will remain with GTB only, as controls. Into the rest, 10 µl of compounds of ×10 strength or Phenstatin, also of ×10 strength will be added. The final concentration of the compounds and Phenstatin will be 10 µM. The plate will be incubated at 37 °C for 2 min. Meanwhile, proceed to the dilution of 10 µl of the Paclitaxel Stock solution with 190 µl of r.t. general tubulin buffer, to be used in quantities of 10 µl of this per well. The tubulin will be defrosted until r.t., placed on ice and diluted with 420 TP cold buffer, reaching a final concentration of 3 mg/ml in 80 mM PIPES (piperazine-N,N′-bis(2-ethanesulfonic acid) sesquisodium salt), pH = 6.9, 2 mM MgCl2, 0.5 mM EGTA (ethylene glycol-bis (β-amino-ethyl ether) N,N,N′,N′-tetra-acetic acid, 1 mM GTP and 10.2% glycerol. The diluted tubulin will be used immediately by adding 120 µl into each of the wells with a multichannel pipette. The absorbance was measured at 340 nm for 1 h at 1 min intervals using a plate reader at 37 °C. Representative experiment (n = 3) is shown in .
Molecular modelling
Flexible-ligand docking experiments were performed as previously reportedCitation24, with slight modifications. Briefly, the 3D structures of the ligands were constructed in Avogadro v1.2.0Citation38 and were energetically optimised in the MMFF94 force field until a local energy minimum was achieved. Autodock VinaCitation39 was used for all docking experiments, using a 22 × 22 × 22 Å3 gridbox centred on the colchicine binding site of the α,β-tubulin heterodimer (PDB: 4O2B)Citation40. The co-crystallised colchicine ligand and water molecules were removed during protein preparation for docking, and the target protein was kept rigid. Twenty poses were generated for each ligand, which were then ranked based on theoretical binding energy. The best ranked models were visually inspected in order to assess the consistency of the generated docking solutions relative to the docking poses of the known inhibitors colchicine and Phenstatin. In order to evaluate the quality of the docking protocol, colchicine was extracted from the crystal structure and re-docked into the binding site. RMSD between re-docked ligand and co-crystallised conformation was computed in PyMOL. Visual inspection, molecular graphics and analyses were made in the PyMOL Molecular Graphics System, Version 1.8.2 (Schrödinger, LLC, New York, NY) and Discovery Studio Visualiser Version 20.1.0.19295 (Dassault Systemes, BIOVIA Corp., San Diego, CA).
Results and discussion
Chemistry
The pyridinium salts 8a–l and 13a–d were prepared through the direct reaction of pyridine 1, 4,4′-bipyridine 2, 2,4′-bipyridine 3, or 2,2′-bipyridine 12, respectively, with 2-bromo-acetophenones 4–7 in acetone, at r.t. (Schemes 1 and 2) (for spectral data of pyridinium salts 8a–l and 13a–d see Supplementary data). In the next step, for the synthesis of the indolizine ring, we used the 1,3-dipolar cycloaddition of the pyridinium ylides generated in situ in basic medium from the salts 8a–l and 13a–d, to ethyl propiolate (Schemes 1 and 2)Citation21,Citation30,Citation31,Citation41.
Indolizines 15a–i were obtained in good yields using the substitution of halides 4–6 generated by the indolizines 11e–g (Scheme 3)Citation32,Citation42. We also synthesised the previously reported compound 15jCitation32 using a similar procedure.
The structures of all new target compounds were fully confirmed by 1H and 13C NMR, IR and elemental analyses.
Biological activity
Anticancer activity
All synthesised compounds were submitted to the NCI, and 13 compounds (11a, b, d, e, f, i, j, k, l, 14a, 14d, 15a, and 15j) were selected for single dose (10−5 M) screening against a panel of 60 human tumour cell lines, representing leukaemia, melanoma and cancer of lung, colon, central nervous system, ovary, kidney, prostate, and breastCitation33. Representative results for 11 of the compounds are summarised in .
Table 1. Results of the in vitro growth inhibition (GI %) of tested compounds against human cancer cell lines in the single-dose assayTable Footnotea.
Indolizines 11a, b showed a very good inhibition effect on almost all 60 lines, the best results being registered on leukaemia HL-60 (TB) cells, colon cancer COLO 205 cells, SNC cancer SF-539 cells, melanoma M14 and MDA-MB-435 cells, ovarian cancer cell OVCAR-3, renal cancer A498 and RXF393 and breast cancer MDA-MB-468 cells. Compound 11a also exhibited a cytotoxic effect on all these lines, the best one being registered on melanoma MDA-MB-435 cells.
Interestingly, the substitution of indolizine heterocycle at position 7 with a pyrid-4-yl or pyrid-2-yl ring resulted in the loss of the activity, compounds 11e–f and 11i, 11j, and 11l (data not shown for compound 11l) presenting almost no inhibition effect on the tested cell lines. As an exception, compound 11k selectively inhibited the growth of NCI-H522 and NCI-H460 non-small cell lung cancer, SK-OV-3 ovarian cancer cells and T-47D breast cancer cells.
Substitution of the indolizine heterocycle at position 5 with a pyrid-2-yl ring also led to a loss of growth inhibition effect. Thus, compounds 14a () and 14d showed no inhibition on tested cancer cells (data not shown for compound 14d).
The presence of two 3,4,5-trimethoxybenzoyl groups in compound 15a led to the best growth inhibition effect on the tested cancer cells. Compound 15a is also distinguished by the high cytotoxic activity displayed on 18 cell lines, including cell lines from each panel except prostate cancer. Of the same serious, compound 15j showed similar behaviour to 15a on most line cells, but with much lower GI % values on NCI-H522 lung cancer cells, SF-268 CNS cancer cells, MDA-MB-231/ATCC and BT-549 breast cancer cells.
As shown in , the substitution of the 3,4,5-trimethoxyphenyl ring produced different effects in series of compounds C and D (). Thus, in series C (11a–d), replacing the 3,4,5-trimethoxyphenyl ring with 3,5-dimethoxyphenyl does not alter the potency substantially, while substitution with 4-bromophenyl ring causes a dramatical loss of inhibitory properties. In series D (11i–l), the 3,4-dimethoxyphenyl ring appears to be the only one to confer selective inhibitory properties against the above mentioned cancer cell lines.
Showing the most significant growth inhibition, compounds 11a, 15a, and 15j were selected for evaluation against 60 cell lines at five concentrationsCitation33–35. Results from the NCI-60 5-dose screen are shown in .
Table 2. Results of the 5-dose in vitro human cancer cell growth inhibitionTable Footnotea for compounds 11a, 15a, and 15j and positive control Phenstatin.
All three compounds displayed good antiproliferative properties. The best candidate in terms of growth inhibition properties was compound 11a, with GI50 values < 100 nM against 47 cell lines, most notably on melanoma MDA-MB-435 (GI50<10 nM) and UACC-62 (GI50=31 nM) cells, leukaemia SR cells (GI50=23 nM), and renal cancer A498 cells (GI50=27 nM). Compound 11a displayed selective cytotoxic activity on the melanoma MDA-MB-435 cell line (LC50=20.4 µM). Even if the overall antiproliferative activity of compound 11a is lower in comparison with control Phenstatin, there are 11 cancer cell lines against which compound 11a showed better GI50 values and several other lines against which compound 11a had comparable GI50 values.
Although displaying excellent growth inhibitory effects at the 10−5 M single dose evaluation (), compounds 15a and 15j did not exhibit submicromolar GI50 values, except compound 15j with GI50=0.33 µM against leukaemia SR cell line. Notably, both compounds showed considerable cytotoxic activity against all colon cancer, CNS cancer, renal cancer, and melanoma cell lines, but also on some cell lines from non-small cell lung cancer, ovarian cancer, breast cancer, and prostate cancer, respectively. This different behaviour of compounds 15a and 15j in comparison with compounds 11a and Phenstatin, suggests different mechanisms of action.
In vitro tubulin polymerisation inhibition activity
In order to confirm if the observed anticancer activity of the above mentioned compounds is conferred by a microtubule-targeting mechanism, we evaluated the effect of the active compounds 11a, 11b, 11k, 15a, and 15j, but also of the inactive compound 14a (for comparison) on the assembly of tubulin. To confirm their influence on microtubule dynamics, two positive controls were used: paclitaxel (a tubulin stabiliser) and Phenstatin (a tubulin polymerisation inhibitor). As presented in , Paclitaxel was found to stimulate tubulin polymerisation, while Phenstatin and all six tested compounds appeared to inhibit tubulin polymerisation.
Four compounds, namely 11k, 14a, 15a, and 15j showed a similar strong inhibitory behaviour on tubulin polymerisation, superior to the one obtained for compounds 11a and 11b. The obtained data clearly indicated that all tested target compounds effectively inhibit tubulin polymerisation in vitro.
Molecular modelling
Molecular docking experiments revealed similar docking conformations of the tested compounds to previously reported anticancer pyrrolo[1,2-b]pyridazines (compounds type B, ), which are also thought to achieve anticancer activity by binding to the colchicine binding site of tubulinCitation24. The used docking protocol was validated by computing the RMSD between re-docked colchicine and its co-crystallised conformation, which was 0.16 Å in our case. Generally, an RMSD value below 2 Å (the average resolution of a crystal structure) is considered acceptableCitation43.
Compound 11a is stabilised in the colchicine binding site of tubulin through H-bonding between its carbonyl moiety and βAsn258 and through extensive hydrophobic contacts with the accommodating pocket formed by βCys241, βLeu248, βLeu255, βAla250, βAla316, βIle318, βAla354, and βIle378, similar to Phenstatin. Moreover, the indolizine heterocycle extends towards the α subunit with the help of two polar interaction partners: βLys254 and αAsn101, and is further stabilised by the nonpolar chain of βLys352, which is engaged in an H-bond with αThr179 through its ε-NH3 + group. The 4-methoxy substituent does not engage in an H-bond with βCys241, as seen in the case of Phenstatin, but the binding orientation of this compound suggests that this bond could form if the binding site would be optimised. Further molecular dynamics simulations could be performed in order to investigate the formation and stability of this H-bond. Removal of the 4-methoxy group leads to a slight shift of the methoxy-substituted ring compared to 11a, possibly to optimise hydrophobic contacts, as reflected by the similar theoretical binding energy (), but the formed H-bonds with βAsn258, βLys254, and αAsn101 are enough to maintain a conformation roughly overlapping with 11a, which could account for the observed biological activity exhibited by this compound. The bromo-substituted compound 11d was accommodated more deeply in the colchicine binding pocket, exclusively through hydrophobic interactions, having one of the lowest theoretical binding energies of all docked compounds. The absence of polar contacts upon accommodation at the tubulin binding site could explain the low biological properties exhibited by this compound.
Table 3. Binding orientation, energy, and amino acid contacts for tested compounds, as predicted by molecular docking experiments.
Docking experiments for compounds 11e, 11f, 11i, 11j, and 11k did not reveal any conformations in which the methoxy-substituted cycle would overlap with the one in the colchicine binding site as to permit a polar interaction with βCys241. Instead, this aromatic moiety was oriented towards α subunit of the binding pocket, being stabilised by H-bonds with βLys254 (11e, 11i) or αAsn101 (11j), as well as weak hydrogen bonding interactions with αSer178, βGln247 and the nonexchangeable GTP molecule. The indolizine moiety is stabilised by extensive hydrophobic contacts and, in the case of 11f, 11k, by additional amide stacking with the backbone of βLys254. The pyridyl ring is positioned deep in the colchicine binding site, away from the α/β interface, and is stabilised through hydrophobic interactions with residues in the β subunit. Since all these compounds have good binding energies, yet lack activity, it could be postulated that a polar interaction with βCys241, or at least the positioning of possible polar interaction partners in the proximity of this residue is crucial for the observed anticancer activity, as has been seen for other colchicine binding site inhibitorsCitation44–46. However, an exception can be seen at compound 11k, which showed selective activity against nine cancer cell lines (GI > 70%), and also inhibited tubulin polymerisation in vitro, but did not form a favourable contact with this residue in our docking experiments. Further mutagenesis experiments could be performed in order to describe the impact of this residue on the binding properties of the tested compounds. Despite its potent anticancer activity, the low theoretical binding score of 11a compared to 11e–k suggests that its cytotoxicity may involve other cellular targets or pathways other than the αβ-tubulin heterodimer. At the same time, since cancer cells preferentially express different β-tubulin isoforms, it would be possible that this compound binds with greater affinity to other isoforms. This aspect could be further studied in silico, as has been done for DAMA-colchicineCitation47 and other colchicine binding site microtubule depolymerising agentsCitation48.
Compound 14a was accommodated in a similar fashion to compounds 11a,b,d, preserving the interactions of the methoxy-substituted moiety with the hydrophobic pocket formed by βCys241, βLeu248, βLeu255, βAla250, βAla316, βIle318, βAla354, and βIle378, but its indolizine ring rotated as to permit the interaction between the pyrid-2-yl ring and βMet259 through pi-sulphur stacking. This rotation also led to the formation of an H-bond between the carboxylate moiety of this compound and βGln247. While the in vitro tubulin polymerisation assay results are in agreement with the docking observations, the lack of anticancer activity in the case of compound 14a remains to be elucidated.
Compound 15a occupied the colchicine binding site similar to compounds 11a,b,d and Phenstatin, engaging in H-bonds with βCys241 and βAsn258, and forming hydrophobic contacts with βLeu248, βLeu255, βAla250, βAla316, βIle318, and βAla354. The additional 3,4,5-trimethoxybenzoyl group reached towards the H10 helix of the β-tubulin subunit to form H-bonds with βThr353 and βGln336, as well as pi-anion stacking with the sidechain of βAsp329. The indolizine moiety was stabilised by amide-pi stacking with the backbone of αThr179. This compound had the one of the lowest binding energies of all tested molecules (–10.2 kcal/mol), being surpassed only by compound 15j (–10.7 kcal/mol). Interestingly, compound 15j forms a hydrophobic interaction with βCys241 and maintains many of the polar and hydrophobic contacts observed at 15a, being accommodated in the same extended binding site-spanning conformation. Additional molecular dynamics experiments should be performed in order to confirm the stability of the observed interactions, especially with βCys241.
Conclusions
Twenty-six new substituted Phenstatin analogues with an indolizine core were synthesised and submitted to NCI for anticancer activity evaluation. Thirteen compounds were selected and tested against a panel of 60 human cancer cells. Tubulin polymerisation assays and docking studies were also performed for the active compounds. Compounds 11a, 11b, 15a, and 15j showed excellent inhibitory properties on a broad range of cancer cell lines, and tubulin polymerisation assays revealed significant inhibitory effects on tubulin assembly for these compounds. This mechanism of action is further supported by docking experiments, which showed that all four compounds fit well to the colchicine binding site of tubulin. Interestingly, substitution of the indolizine heterocycle at position 7 with a pyrid-4-yl or pyrid-2-yl, or at position 5 with a pyrid-2-yl ring resulted in the loss of anticancer activity. As an exception, compound 11k showed a good inhibitory profile on tubulin polymerisation, but only selectively inhibited the growth of NCI-H522 and NCI-H460 non-small cell lung cancer, SK-OV-3 ovarian cancer cells, and T-47D breast cancer cell lines. Interestingly, inhibitory tubulin polymerisation properties, as well as a good compatibility for the colchicine binding pocket of tubulin are shown by 14a, but this compound is basically inactive against the tested cancer cells. Taken together, these results offer new SAR insights into this class of compounds and prove that using a strategy of structural combination can generate new colchicine site tubulin polymerisation inhibitors, as well as highly cytotoxic molecules against various cancer cells, which could aid the general research community in their ongoing anticancer efforts.
Supplemental Material
Download PDF (1.2 MB)Acknowledgements
The authors acknowledge National Cancer Institute for the anticancer evaluation of the compounds on their 60-cell panel. The testing was performed by the Developmental Therapeutics Program, Division of Cancer Treatment and Diagnosis (the URL to the Program’s website: http://dtp.cancer.gov/). We thank CERNESIM Research Center from Alexandru Ioan Cuza University of Iasi for the NMR experiments.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Jordan MA, Wilson L. Microtubules as a target for anticancer drugs. Nat Rev Cancer 2004;4:253–65.
- Seligmann J, Twelves C. Tubulin: an example of targeted chemotherapy. Future Med Chem 2013;5:339–52.
- Chen H, Li Y, Sheng C, et al. Design and synthesis of cyclopropylamide analogues of combretastatin-A4 as novel microtubule-stabilizing agents. J Med Chem 2013;56:685–99.
- Huang X, Huang R, Li L, et al. Synthesis and biological evaluation of novel chalcone derivatives as a new class of microtubule destabilizing agents. Eur J Med Chem 2017;132:11–25.
- Fu DJ, Liu SM, Li FH, et al. Antiproliferative benzothiazoles incorporating a trimethoxyphenyl scaffold as novel colchicine site tubulin polymerisation inhibitors. J Enzyme Inhib Med Chem 2020;35:1050–9.
- Levrier C, Sadowski MC, Rockstroh A, et al. 6α-Acetoxyanopterine: a novel structure class of mitotic inhibitor disrupting microtubule dynamics in prostate cancer cells. Mol Cancer Ther 2017;16:3–15.
- Lee HY, Lee JF, Kumar S, et al. 3-Aroylindoles display antitumor activity in vitro and in vivo: effects of N1-substituents on biological activity. Eur J Med Chem 2017;125:1268–78.
- Gigant B, Wang C, Ravelli RB, et al. Structural basis for the regulation of tubulin by vinblastine. Nature 2005;435:519–22.
- Bhattacharyya B, Panda D, Gupta S, Banerjee M. Anti-mitotic activity of colchicine and the structural basis for its interaction with tubulin. Med Res Rev 2008;28:155–83.
- Lin CM, Ho HH, Pettit GR, Hamel E. Antimitotic natural products combretastatin A-4 and combretastatin A-2: studies on the mechanism of their inhibition of the binding of colchicine to tubulin. Biochemistry 1989;28:6984–91.
- Li W, Sun H, Xu S, et al. Tubulin inhibitors targeting the colchicine binding site: a perspective of privileged structures. Future Med Chem 2017;9:1765–94.
- Sherbet GV. Suppression of angiogenesis and tumour progression by combretastatin and derivatives. Cancer Lett 2017;403:289–95.
- Pettit GR, Toki B, Herald DL, et al. Antineoplastic agents. 379. Synthesis of Phenstatin phosphate. J Med Chem 1998;41:1688–95.
- Pettit GR, Grealish MP, Herald DL, et al. Antineoplastic agents. 443. Synthesis of the cancer cell growth inhibitor hydroxyphenstatin and its sodium diphosphate prodrug. J Med Chem 2000;43:2731–7.
- Dumontet C, Jordan MA. Microtubule-binding agents: a dynamic field of cancer therapeutics. Nat Rev Drug Discov 2010;9:790–803.
- Li M, Tian YS. Progress in synthesis and anti-tumor activities of combretastatin A4 derivatives. J Pharm Res 2016;35:283–9.
- Le Broc-Ryckewaert D, Pommery N, Pommery J, et al. In vitro metabolism of Phenstatin: potential pharmacological consequences. Drug Metab Lett 2011;5:209–15.
- Kumar GB, Nayak VL, Sayeed IB, et al. Design, synthesis of Phenstatin/isocombretastatin-oxindole conjugates as antimitotic agents. Bioorg Med Chem 2016;24:1729–40.
- Marx MA. Small-molecule, tubulin-binding compounds as vascular targeting agents. Expert Opin Ther Pat 2002;12:769–76.
- Ojha R, Sharma S, Nepali K. Anticancer agents targeting tubulin. In: Atta-Ur-Rahman K. Zaman, eds. Topics in anticancer research. Oak Park (IL): Bentham Science; 2015.
- Ghinet A, Abuhaie CM, Gautret P, et al. Studies on indolizines. Evaluation of their biological properties as microtubule-interacting agents and as melanoma targeting compounds. Eur J Med Chem 2015;89:115–27.
- Al Matarneh CM, Amarandi RM, Craciun AM, et al. Design, synthesis, molecular modelling and anticancer activities of new fused phenanthrolines. Molecules 2020;25:527.
- Hu S, Sun W, Wang Y, Yan H. Design, synthesis and anticancer activities of halogenated Phenstatin analogs as microtubule destabilizing agent. Med Chem Res 2019;28:465–72.
- Popovici L, Amarandi RM, Mangalagiu II, et al. Synthesis, molecular modelling and anticancer evaluation of new pyrrolo[1,2-b]pyridazine and pyrrolo[2,1-a]phthalazine derivatives. J Enzyme Inhib Med Chem 2019;34:230–43.
- Huang X, Huang R, Gou S, et al. Platinum(IV) complexes conjugated with Phenstatin analogue as inhibitors of microtubule polymerization and reverser of multidrug resistance. Bioorg Med Chem 2017;25:4686–700.
- Prachayasittikul S, Pingaew R, Worachartcheewan A, et al. Roles of pyridine and pyrimidine derivatives as privileged scaffolds in anticancer agents. Mini Rev Med Chem 2017;17:869–901.
- Kassab AE, Gedawy EM. Synthesis and anticancer activity of novel 2-pyridyl hexahyrocyclooctathieno[2,3-d]pyrimidine derivatives. Eur J Med Chem 2013;63:224–30.
- Al-Majid AM, Islam MS, Atef S, et al. Synthesis of pyridine-dicarboxamide-cyclohexanone derivatives: anticancer and α-glucosidase inhibitory activities and in silico study. Molecules 2019;24:1332.
- Badr MH, Rostom SAF, Radwan MF. Novel polyfunctional pyridines as anticancer and antioxidant agents. synthesis, Biological evaluation and in silico ADME-T Study. Chem Pharm Bull 2017;65:442–54.
- Druta II, Andrei MA, Aburel PS. Synthesis of 5-(2′-pyridyl)-indolizines by the reaction of 2-(2′-pyridyl)-pyridinium-ylides with activated alkynes. Tetrahedron 1998;54:2107–12.
- Rotaru AV, Danac RP, Druta ID. Synthesis of new non‐symmetrical substituted 7,7′‐bisindolizines by the direct reaction of 4,4′‐bipyridinium‐ylides with dimethyl acetylenedicarboxylate. J Heterocycl Chem 2004;41:893–7.
- Olaru AM, Vasilache V, Danac R, Mangalagiu II. Antimycobacterial activity of nitrogen heterocycles derivatives: 7-(pyridine-4-yl)-indolizine derivatives. Part VII8–12. J Enzyme Inhib Med Chem 2017;32:1291–8.
- Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer 2006;6:813–23.
- Skehan P, Storeng R, Scudiero D, et al. New colorimetric cytotoxicity assay for anticancer-drug screening. J Natl Cancer Inst 1990;82:1107–12.
- Boyd RB. The NCI in vitro anticancer drug discovery screen: concept, implementation, and operation. In: Teicher B, ed. Anticancer drug development guide: preclinical screening, clinical trials, and approval. Totowa (NJ): Humana Press Inc.; 1997.
- Schiff PB, Fant J, Horwitz SB. Promotion of microtubule assembly in vitro by taxol. Nature 1979;277:665–7.
- Schofield AV, Gamell C, Suryadinata R, et al. Tubulin polymerization promoting protein 1 (Tppp1) phosphorylation by Rho-associated coiled-coil kinase (rock) and cyclin-dependent kinase 1 (Cdk1) inhibits microtubule dynamics to increase cell proliferation. J Biol Chem 2013;288:7907–17.
- Hanwell MD, Curtis DE, Lonie DC, et al. Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J Cheminformatics 2012;4:17.
- Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J Comput Chem 2009;31:455–461.
- Prota AE, Danel F, Bachmann F, et al. The novel microtubule-destabilizing drug BAL27862 binds to the colchicine site of tubulin with distinct effects on microtubule organization. J Mol Biol 2014;426:1848–60.
- Marangoci NL, Popovici L, Ursu EL, et al. Pyridyl-indolizine derivatives as DNA binders and pH-sensitive fluorescent dyes. Tetrahedron 2016;72:8215–22.
- Pricope G, Ursu EL, Sardaru M, et al. Novel cyclodextrin-based pH-sensitive supramolecular host-guest assembly for staining acidic cellular organelles. Polym Chem 2018;9:968–75.
- Onodera K, Satou K, Hirota H. Evaluations of molecular docking programs for virtual screening. J Chem Inf Model 2007;47:1609–18.
- Lu Y, Chen J, Wang J, et al. Design, synthesis, and biological evaluation of stable colchicine binding site tubulin inhibitors as potential anticancer agents. J Med Chem 2014;57:7355–66.
- McLoughlin EC, O’Boyle NM. Colchicine-binding site inhibitors from chemistry to clinic: a review. Pharmaceuticals 2020;13:8.
- Klejborowska G, Urbaniak A, Maj E, et al. Synthesis, biological evaluation and molecular docking studies of new amides of 4-chlorothiocolchicine as anticancer agents. Bioorg Chem 2020;97:103664.
- Kumbhar BV, Borogaon A, Panda D, Kunwar A. Exploring the origin of differential binding affinities of human tubulin isotypes αβII, αβIII and αβIV for DAMA-colchicine using homology modelling, molecular docking and molecular dynamics simulations. PLoS One 2016;11:e0156048.
- Kumbhar BV, Panda D, Kunwar A. Interaction of microtubule depolymerizing agent indanocine with different human αβ tubulin isotypes. PLoS One 2018;13:e0194934.