Abstract
Isolation and identification of natural products from plants is an essential approach for discovering drug candidates. Herein we report the characterization of three sesquiterpene lactones from a new genus Shangwua, e.g. onopordopicrin (ONP), C2, and C3, and evaluation of their pharmacological functions in interfering cellular redox signaling. Compared to C2 and C3, ONP shows the most potency in killing cancer cells. Further experiments demonstrate that ONP robustly inhibits thioredoxin reductase (TrxR), which leads to perturbation of cellular redox homeostasis with the favor of oxidative stress. Knockdown of the TrxR sensitizes cells to the ONP treatment while overexpression of the enzyme reduces the potency of ONP, underpinning the correlation of TrxR inhibition to the cytotoxicity of ONP. The discovery of ONP expands the library of the natural TrxR inhibitors, and the disclosure of the action mechanism of ONP provides a foundation for the further development of ONP as an anticancer agent.
Graphical Abstract
1. Introduction
Abundant scaffold diversity of natural products is almost beyond human imagination. Biological co-evolution endows natural products with perfect biocompatibility, resulting in natural products that can nearly match the three-dimensional configuration requirements of various protein targets in organismsCitation1. Sesquiterpenoid derived from the medicinal plants Asteraceae has attracted wide attention from researchers due to targeting functional proteins to produce antitumor, antiviral, antibacterial, and antiinflammatory pharmacological activitiesCitation2. Shangwua, a new genus of the Asteraceae family discovered in recent years, has been identified with three species, e.g. Shangwua denticulata (DC.) Raab-Straube & Yu J. Wang, Shangwua jacea (Klotzsch) Yu J. Wang & Raab-Straube, and Shangwua masarica (Lipsky) Yu J. Wang & Raab-StraubeCitation3. However, the medicinal value and secondary metabolite components for Shangwua have not been reported yet.
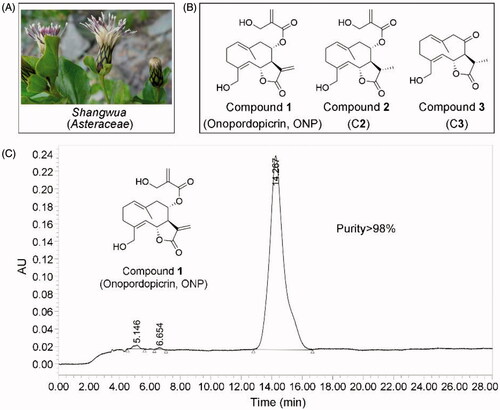
We thus isolated for the first time three sesquiterpene lactones from the whole plant of Shangwua denticulate () collected from Ji-Long County, Tibet. Compounds 1-3 () bearing the same lactone ring are onopordopicrin (Compound 1, ONP), 11β,13-dihydro-19-desoxycnicin (Compound 2, C2), and 8-oxo-15-hydroxygermacra-1(10)E, 4Z-dien-11βH-12, 6α-olide (Compound 3, C3), respectively. ONP was previously reported to harbor antibacterial and antifungalCitation4,Citation5, antiplasmodialCitation6, phytotoxicCitation7, anti-ulcerogenicCitation8, and anti-inflammatory activitiesCitation9, while the biological activities of C2 and C3 are rarely reported. However, despite ONP exhibits cytotoxicity, whether ONP is effective for inhibiting tumor growth and acting mechanism remains unclear.
Figure 1. Compounds 1-3 isolated for the first time from Shangwua, a new genus of Asteraceae. (A) Tibetan plateau and Himalayas plant Shangwua. (B) Structure of compounds 1–3, namely ONP, C2, and C3. (C) Purity analysis of ONP was analyzed by HPLC.
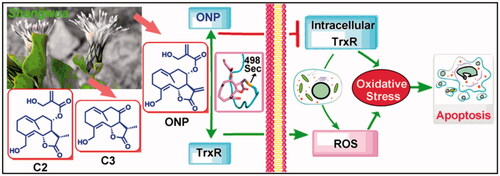
Alteration of redox homeostasis is a hallmark of cancer cellsCitation10. Thioredoxin reductase (TrxR) enzymes as a critical player in regulating cellular redox signaling are increasingly recognized as attractive targets for anticancer drugsCitation11–16. Structurally, TrxR is a selenium-dependent enzyme with unique but indispensable selenocysteine (Sec) residue at the penultimate C-terminusCitation17,Citation18, thereby providing a preferential and selective target for covalent natural moleculesCitation14,Citation19. Functionally, in addition to the key role of TrxR in redox regulationCitation20,Citation21, an ever-widening range of evidence has supported that TrxR highly enriched in many tumor types is essentially implicated with multiple steps of tumorigenesis and developmentCitation20–23, and is a potential and promising target for the current anticancer drugs discoveryCitation24–27. As our continuing efforts in discovering therapeutic molecules derived from natural products that effectively interfere with cellular redox signalingCitation28–39, we assessed herein the antitumor activity of ONP, C2 and C3, and systematically reported a new mechanism by which ONP targets intracellular TrxR inhibition to cause oxidative stress-mediated tumor cell apoptosis. Mechanistically, the β-carbocation of the lactone ring in ONP covalently bound to the redox-active Sec residue at the C-terminus of TrxR, thereby leading to a decrease in TrxR activity. Importantly, carcinoma cells bearing high levels of oxidative stress and overexpressing TrxR under pathological conditions were selectively killed by ONP, showing a perfect antitumor effect. Moreover, the silence of TrxR expression by RNA interference enhanced the observed cytotoxicity of ONP while overexpression of the enzyme reduced the potency of ONP, thereby supporting that TrxR is essentially involved in the cellular effects of ONP. A novel TrxR inhibitor for the first time was isolated from Shangwua and its antitumor activity and action mechanism were reported, which not only revealed the new pharmacological activity of ONP but also provided insights for the development of the medicinal value of the new genus Shangwua.
2. Materials and methods
2.1. Chemistry
2.2.1. Plant materials
The whole plant of Shangwua was collected from Ji-Long County, Tibet, China, in August 2016, and identified by Prof. Wang Yu-Jin of Lanzhou University. A voucher specimen (No. 20170728) was stored at the natural product laboratory of State Key Laboratory of Applied Organic Chemistry, Lanzhou University.
2.2.2. General experimental procedures
200–300 mesh silica gel and 10–40 µm GF 254 silica gel plates (Qingdao Marine Chemical Factory, China), 150–200 mesh RP-C18 silica gel (Merck), and Sephadex LH-20 (Amersham Pharmacia Biotech UK Ltd.) were employed for column chromatography. Semipreparative HPLC with a reversed-phase C18 (150 × 10 mm, 10 µm) column was carried out on the isolation and purification of samples. 1H and 13C NMR spectroscopic data were used to a Varian Mercury-600BB or Bruker Avance III-400 instrument.
2.2.3. Purity analysis
The purity analysis of ONP was performed on Waters pump 1525 and PDA 2998 series HPLC systems with reversed-phase C18 (4.6 × 150 mm, 5 µm) chromatographic columns at room temperature. ONP was dissolved in chromatographic methanol, and the injection volume was 10 µL. The mobile phases are methanol and water, and the flow rate is 1.0 mL/min. The maximum absorbance in the range of 210–400 nm is used as the detection wavelength.
2.2.4. Chemical characterization
The chemical characterization spectrum and HPLC purity analysis diagram of compounds 1-3 were shown in the Supporting Information (Figues S1–S7). The isolated compounds 1-3 were employed in all subsequent activity experiments.
Compound 1 (ONP): yellow oil; 1H NMR (300 MHz, CD3OD, in ppm, J in Hz): 6.29 (1H, d, J = 1.2 Hz, H-4′), 6.10 (1H, br s, H-13), 5.97 (1H, br s, H-4′), 5.81 (1H, d, J = 2.8 Hz, H-13), 5.21 (1H, m, H-6), 5.13 (1H, m, H-8), 5.07 (1H, m, H-1) 4.95 (1H, d, J = Hz, H-5), 4.27 (1H, s, H-3′), 4.25 (1H, m, H-15a) , 4.01 (1H, d, J = 13.2 Hz, H-15b), 3.28 (1H, m, H-7), 2.62 (1H, m, H-3a), 2.55 (1H, m, H-9a) 2.33 (1H, m, H-2a), 2.20 (1H, m, H-2b), 2.01 (1H, m, H-3b), 1.54 (3H, s, H-14); 13C NMR: (75 MHz, CD3OD) 172.1 (C-12), 166.4 (C-1′), 145.6 (C-4), 141.9 (C-2′), 137.5 (C-11), 130.9 (C-1), 129.7 (C-5), 125.8 (C-4′), 125.3 (C-13), 78.6 (C-6), 74.4 (C-8), 61.6 (C-3′), 60.8 (C-15), 54.0 (C-7), 35.2 (C-3), 26.9 (C-2), 17.1 (C-14).
Compound 2 (C2): yellow oil; 1H NMR (300 MHz, CD3OD, in ppm, J in Hz): 6.26 (1H, s, H-4′a), 6.19 (1H, s, H-4′b), 5.25 (1H, m, H-8), 5.09 (1H, m, H-6), 5.04 (1H, m, H-1), 4.86 (1H, m, H-5), 4.30 (1H, s, H-3′), 4.26 (1H, d, J = 13.6 Hz, H-15a), 3.99 (1H, d, J = 13.6 Hz, H-15b), 2.69 (1H, m, H-11), 2.61 (1H, m, H-3a), 2.48 (1H, m, H-9), 2.38 (1H, m, H-7), 2.17 (1H, m, H-2), 2.01 (1H, m, H -2) -3a), 1.52 (1H, s, H-14), 1.36 (1H, d, J = 6.8 Hz, H-13); 13C NMR (75 MHz, CD3OD): 181.0 (C-12), 166.5(C-1′), 144.4(C-4), 142.0(C-2′), 133.7(C-10), 130.7(C-1), 130.3(C-5), 125.5(C-4′), 77.9 (C-6), 74.8 (C-8), 61.7 (C-3′), 60.6 (C-15), 59.1 (C -7), 49.9 (C-9), 35.2 (C- 11), 30.7 (C-3), 26.7 (C-2), 17.3 (C-13), 16.9 (C-14).
Compound 3 (C3): yellow oil; 1H NMR (300 MHz, CD3OD, ppm, J in Hz): 5.19 (1H, m, H-1), 5.02 (1H, d, J = 10.2 Hz, H-5), 4.93 (1H, m, H-6), 4.10 (1H, d, J = 13.8 Hz, H-15a), 3.78 (1H, d, J = 13.8 Hz, H-15 b), 3.32 (1H, d, J = 9.9 Hz, H-15a), 3.11 (1H, m, H-7), 2.95 (1H, d, J = 9.6 Hz, H-15b), 2.59 (1H, m, H-3a), 2.35 (2H, m, H-2), 2.06 (1H, m, H-3b), 1.42 (3H, s, H-14), 1.18 (3H, d, J = 6.6 Hz, H-15a); 13C NMR (75 MHz, CD3OD) 204.0 (C-8), 177.7 (C-12), 145.7 (C-4), 132.5 (C-1), 127.8 (C-5), 126.5 (C-10), 75.6 (C-6), 64.3 (C-7), 59.6 (C-15), 57.5 (C-9), 40.6 (C-11), 33.4 (C-3), 24.8 (C-2), 16.0 (C-14), 13.6 (C-13).
2.2. Enzymes and materials
The recombinant rat TrxR1 was a gift from Prof. Arne Holmgren (Karolinska Institute, Sweden). The recombinant U498C TrxR1 (Sec→Cys) mutant and the Escherichia coli (E. coli) Trx were prepared by our Lab as describedCitation36. Both the plasmids: shTrxR1, shRNA specifically targeting TrxR1, shNT, non-targeting control shRNA, and the cell lines: HEK-TrxR1 cells, HEK cells stably overexpressing TrxR1, HEK-IRES cells, HEK cells stably transfected with a vector were gifts from Prof. Constantinos Koumenis (University of Pennsylvania, USA)Citation40,Citation41. The preparation of HeLa-shNT and HeLa-shTrxR1 cell lines were describedCitation37.
Dimethyl sulfoxide (DMSO), Dulbecco′s modified Eagle’s medium (DMEM), G418, puromycin, 2′, 7′-dichlorfluorescein diacetate (DCFH-DA), Hoechst 33342, dihydroethidium (DHE), insulin (from bovine), DL-dithiothreitol (DTT), N-acetyl-Asp-Glu-Val-Asp-p-nitroanilide (Ac-DEVD-pNA), and 3-[(3-Cholamidopropyl) dimethylammonio]-1-propanesulfonate (CHAPS) were obtained from Sigma-Aldrich (St. Louis, USA). Penicillin, MTT and streptomycin were products of Amresco (Solon, OH). NADPH was purchased from Roche (Mannheim, Germany). Fetal bovine serum (FBS) was a product of Sijiqing (Hangzhou, China). DTNB (5,5′-dithiobis-2-nitrobenzoic acid) was a product of J&K Scientific (Beijing, China). Bovine serum albumin (BSA), Trypan blue, phenylmethylsulfonyl fluoride (PMSF), and Na3VO4 were products of Beyotime (Nantong, China). The propidium iodide (PI) and Annexin V-FITC apoptosis assay kit was a product of Zoman Biotech (Beijing, China). HPLC grade methanol and acetonitrile were products of Merck (Darmstadt, Germany). All other chemicals used were of analytic grade.
2.3. Cell cultures
Different cell lines (HeLa, HepG2, A549, L02, and BEAS-2B) were obtained from the Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, and were cultured under a standard culture condition (5% CO2 atmosphere, 37 °C incubator with a humidified; DMEM supplemented with 10% FBS, 2 mM glutamine, and100 units·mL−1 streptomycin/penicillin). HEK-TrxR1 and HEK-IRES cells were grown in the standard culture conditions supplemented with an additional 0.1 µM sodium selenite and 0.4 mg·mL−1 G418. HeLa-shTrxR1 and HeLa-shNT cells were cultured under the standard culture conditions supplemented with an additional 1 µg·mL−1 puromycin.
2.4. Cell viability analysis
2.4.1. MTT assay
Cells (5 × 103−1 × 104/well) from above different cell lines were incubated with agents and grown in triplicate in a 96-well plate for 24, 48, or 72 h at 37 °C in a final volume of 100 µL. The cells treated with 0.1% DMSO alone were the control group. After incubated for the designed time, MTT reagent (5 µL/well, 5 mg·mL−1) was added and continued culturing for an additional 4 h at 37 °C. Formazan crystals then were solubilized in solvent 100 µL extraction buffer (10% SDS, 0.1% HCl, and 5% iso-butanol). The absorbance was read at 570 nm using a microplate reader (Thermo Scientific Multiskan GO, Finland)) to calculate the cell viability.
2.4.2. Trypan blue exclusion assay
Cells (2 × 105/well) were seeded in a 12-well plate and were cultured overnight. The cells were further treated with indicated concentrations of ONP (20 and 40 µM) for 24 h. The control cells were treated with 0.1% DMSO alone. Then, the cells were stained with trypan blue (0.4%, w/v), and the number of dead (stained) cells and viable (non-stained) cells were counted.
2.5. Apoptosis assays
2.5.1. Hoechst 33342 staining
Hoechst 33342 dye is used to detect the morphological changes of apoptotic cells. Briefly, HeLa cells (2 × 105) were seeded in a 12-well plate and were cultured overnight. The cells subsequently were incubated with a designed concentration of ONP for 24 h. Hoechst 33342 dye (5 µg·mL−1) was added and continued culturing for 30 minutes. Rinsed HeLa cells with the medium to remove the remaining Hoechst 33342. The stained nuclei were photographed under a fluorescent microscope.
2.5.2. Annexin V-FITC/PI staining
HeLa cells (5 × 105) were plated in a 6-well plate and were cultured overnight. The cells were then incubated with different concentrations of ONP for 24 h. Subsequently, the cells were harvested and washed with PBS. According to the Annexin V-FITC/PI double staining apoptosis detection kit (Zoman Biotech, Beijing, China), the apoptotic cells were determined and analyzed by flow cytometry with Cell Quest software (BD Biosciences, USA).
2.5.3. Caspase 3 activity assay
HeLa cells were treated with indicated concentrations of ONP for 24 h. The cells were then collected and lysed with RIPA buffer. Total cellular protein contents were quantified by the Bradford procedure. The extracts containing 60 µg of total proteins were incubated with the protease activity assay mixture (0.2 mM Ac-DEVD-pNA, 0.1% CHAPS, 5% glycerol, 2 mM EDTA and 10 mM DTT in 50 mM Hepes, pH 7.5) at 37 °C for 2 h in a final volume of 100 µL. The caspase 3 activity was determined by measuring the absorbance at 405 nm.
2.6. Measurement of intracellular ROS
DCFH-DA or DHE probes were employed to assess ROS level. HeLa cells (2 × 105) were plated into a 12-well plate and allowed to adhere. HeLa cells were incubated with 0, 20, or 40 µM ONP for 5 h, then removed the medium and subsequently treated with DCFH-DA (10 µM) or DHE (10 µM) in fresh FBS-free medium for 30 min at 37 °C in dark. The images were acquired on a fluorescent microscope (Floid Cell Imaging Station, Thermo Fisher).
2.7. Assay of cellular TrxR activity
2.7.1. TRFS-green-based live-cell imaging TrxR activity assay
TRFS-green is a specific TrxR probe by our previously establishedCitation42. HeLa cells (2 × 105) were plated into a 12-well plate and allowed to adhere. The HeLa cells were incubated with 0, 10, 20, or 40 µM of ONP or 40 µM C2 or C3 for 8 h. Then the cells were removed from the medium and subsequently treated with TRFS-green (10 µM) in a fresh FBS-free medium for 4 h at 37 °C in dark. The cells were washed to remove the residual TRFS-green. The cells were imaged under a fluorescent microscope (Floid Cell Imaging Station, Thermo Fisher).
2.7.2. Fast-TRFS-based cell lysate TrxR activity assay
Fast-TRFS-based cell lysate assay is the rapid detection of TrxR activity in crude protein as the source of TrxR by our previously described methodCitation43. Briefly, HeLa cells were plated in 100-mm culture dishes and were cultured overnight. The cells then were collected and lysed with RIPA buffer. Total cellular protein contents as the source of TrxR were quantified by the Bradford procedure. Subsequently, the HeLa cell lysate (0.3 mg·mL−1) was incubated with NADPH (100 µM) for 5 min at 37 °C. Test drug, ONP (10, 20, or 40 µM each), and blank sample (0.1%DMSO) were then added and the mixture was continued to incubate for 1 h. The probe Fast-TRFS (10 µM) and NADPH (100 µM) were added to initiate the enzymatic reduction of Fast-TRFS. The fluorescence change at 460 nm was recorded (λex = 345 nm) for 10 min on a fluorescent plate reader (Tecan Infinite M200), and the rate of fluorescence increase within the initial 5 min was calculated. The relative TrxR activity was expressed as the percentage of the DMSO-treated sample.
2.7.3. Trx-mediated endpoint insulin reduction assay
HeLa cells were treated with different concentrations of ONP, C2, and C3 for 12 or 24 h. The cells were washed and harvested. Subsequently, the total cellular proteins were extracted and quantified by the RIPA buffer and the Bradford procedure, respectively. The intracellular TrxR activity was measured by the endpoint insulin reduction assay described as our published proceduresCitation30,Citation37.
2.8. Pure TrxR activity assays
The NADPH-reduced TrxR (170 nM) or U498C TrxR (700 nM) was incubated with 0, 2.5, 5, 10, and 20 µM of ONP or C3 in a 96-well plate at room temperature for 0.5, 1, or 2 h, and the final incubation volume of the mixture was 50 µL. Then added 50 µL TE buffer (50 mM Tris-HCl, 1 mM EDTA, pH 7.5) containing 2 mM DTNB and 200 µM NADPH, recorded the linear increase during the initial 3 min in absorbance at 412 nmCitation30,Citation37. The activity was expressed as the percentage of the control and the same amounts of DMSO were added to the control experiments.
2.9. Molecular docking simulation
The crystal structure of rat TrxR1 (PDB code 3EAN, chain A) was employed in the present docking study as we described previouslyCitation29. The residue Sec498 in chain A was selected and further prepared in the protein preparation wizard module as the reactive residue involved in the Michael addition. In addition, the residue Sec498 was also set as the centroid of the docking pocket. The docking simulation was carried out with the default parameters.
2.10. Statistics
All experiment data are presented as the mean ± S.E. Statistical differences between two groups were assessed by Student’s t-test. p < 0.05 was used as the criterion for statistical significance.
3. Results and discussion
3.1. Chemical isolation and characterization
The dried whole plant (2.375 kg) of Shangwua () was pulverized and extracted with 95% ethanol (3 × 5 L) at room temperature. The mixture was later filtered and concentrated under reduced pressure to provide a crude extract (152.8 g). Subsequently, the crude extract was extracted with ethyl acetate, and the resulting extract (98.9 g) was eluted with ethanol/water (30, 50, 80, 90 and 100%) through macroporous adsorption resin. The 80% ethanol eluted fraction was subjected to silica gel column chromatography and eluted with petroleum ether/acetone to obtain six sub-fractions (Fr. A-F). The Fr. B subfraction was eluted through a reversed-phase silica gel column with a gradient of methanol: water (3:7–0:1). The Fr.B3 part obtained above was separated by Sephadex LH-20 and semi-preparative high-performance liquid chromatography (HPLC) to finally obtain the pure compound 1 (20 mg). Likewise, Fr. E subfraction was directly separated by reversed-phase silica gel column and semi-preparative HPLC (methanol: water = 3:2) to obtain compound 2 (6 mg). Subsequently, the Fr. D subcomponent was eluted by normal phase silica gel (petroleum ether: acetone = 3:1-1:1) to obtain Fr. D2. Then, Fr. D2. was separated and purified by Sephadex LH-20 and semi-preparative HPLC to obtain compound 3 (10 mg).
The structures of compounds 1–3 were characterized by 1H and 13C NMR, and were determined to be onopordopicrin (ONP, )Citation4, 11β,13-dihydro-19-desoxycnicin (C2, )Citation44, and 8-oxo-15-hydroxygermacra-1(10)E, 4Z-dien-11βH-12, 6α-olide (C3, )Citation45, respectively. The purity of ONP is quantified by HPLC (purity >98%, ).
3.2. Inhibiting tumor cell growth
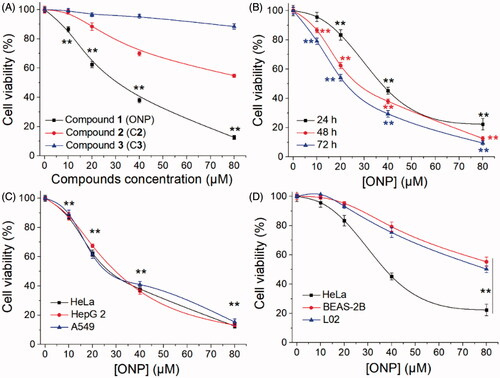
Given that the lack of research on Shangwua and its secondary metabolites, we urgently tested the cytotoxicity of the preceding three isolated compounds on tumor cells. As shown in , the result showed that ONP can significantly inhibit the growth of human cervical cancer HeLa cell line with an IC50 value of 20 µM at 48 h. However, C2 has little cytotoxicity and C3 is almost non-cytotoxic under our experimental conditions towards HeLa cells (). In addition, the cytotoxicity of ONP towards HeLa cells also exhibited a time dependence (). To further confirm the cytotoxicity of ONP to tumor cells, we selected human liver cancer cell line HepG 2 and human lung cancer cell line A549, together with HeLa cells, to assess the growth inhibitory effect of ONP on the tumor cells. Satisfactory results presented that ONP had a remarkable activity of repressing the growth of tumor cells on the above tumor cell lines after 48 h incubation (). More noteworthy is that ONP could selectively kill tumor cells but was less cytotoxicity to normal cells such as human lung epithelial cells BEAS-2B and human liver epithelial cells L02 under the same experimental conditions (). These results indicated that ONP, C2 and C3 isolated from Shangwua have radically different cytotoxicity to HeLa cells due to diversities in their structural fragments. Moreover, ONP with a unique cytotoxic selectivity established by these data deserves a further investigation of its action mechanism for ablating tumor cells.
Figure 2. Inhibiting tumor cell growth by ONP. (A) Cytotoxicity of ONP, C2, and C3 to HeLa cells. After treating HeLa cells with the indicated concentrations of ONP, C2, and C3 for 48 h, the cell viability of HeLa cells was determined by the MTT method. (B) Inhibition of the growth of HeLa cells in a time-dependent manner by ONP. HeLa cells were treated with ONP at designated concentrations for 24, 48, and 72 h, and the cell survival rate was detected by the MTT method. (C) Cytotoxicity of ONP towards multiple tumor cell lines. After different concentrations of ONP were incubated with HeLa, HepG2, and A549 cells for 48 h, the cell viability was detected by the MTT method. (D) Selectivity of ONP for tumor cells. The designated concentrations of ONP acted on HeLa cells, BEAS-2B, and L02 cells for 24 h, and the cell viability was detected by the MTT method. After treatment of HeLa cells, BEAS-2B, and L02 cells with ONP at specified concentrations for 24 h, the cell viability was detected by the MTT method. Data are expressed as mean ± S. E. of three experiments. **p < 0.01 versus the control groups.
3.3. Inducing tumor cell apoptosis
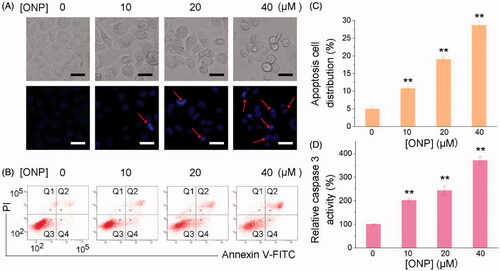
ONP with similar properties as the activities of sesquiterpene lactones reported so far can significantly suppress the growth of tumor cells. Functionally, most of the reported sesquiterpene lactones inhibit tumor cell proliferation via inducing tumor cell apoptosisCitation46–48. Next, we thus sought to confirm whether ONP can induce tumor cell apoptosis. As shown in , we had fully demonstrated that ONP can promote HeLa cell apoptosis through various methods. First, we stained with Hoechst 33342 to observe whether HeLa cells treated with diverse concentrations of ONP can undergo morphological changes of apoptosis. As shown in , our experimental results displayed that after different concentrations of ONP acted on HeLa cells, especially treatment with 40 µM, ONP could significantly cause HeLa cell nuclei to shrink and shine, a typical morphological change of apoptosis. To further confirm the apoptotic morphological changes produced by ONP-treated HeLa cells to eventually induce apoptosis, Annexin V-FITC and propidium iodide (PI) double staining reagents were employed to stain again and detect the number of apoptotic cells by flow cytometry to evaluate the ability of ONP to induce HeLa cell apoptosis. The results in intuitively exhibited that ONP effectively induced HeLa cell apoptosis. The number of apoptotic cells increased with the enhancement of ONP treatment concentration (), indicating directly that ONP could induce HeLa cell apoptosis in a concentration-dependent manner. In addition, our results showed that ONP induced HeLa cell apoptosis in a caspase 3 activity-dependent manner. As shown in , the activity of intracellular caspase 3 activity increased remarkably with the increase of ONP treatment concentration, which further confirmed that ONP kills cells basically in the form of apoptosis. Taken together, ONP can significantly induce caspase 3-dependent apoptosis in HeLa cells in a concentration-dependent manner to contribute to the ability to kill tumor cells.
Figure 3. Killing HeLa cells by ONP primarily via inducing apoptosis. (A) Nuclear morphological changes after ONP treatment. HeLa cells were incubated for 24 h with 0, 10, 20, or 40 μM of ONP treatment, and the nuclei were stained by Hoechst 33342. Nuclear morphology changes then were observed under a fluorescence microscope. The bright filed pictures (top panel) and the fluorescence pictures (bottom panel) were imaged and acquired. Scale bars: 20 μm. Arrows refer to the condensed and irregular nuclei with bright fluorescence stained by Hoechst 33342 in (A). (B) Assessment of apoptosis by Annexin V-FITC/PI double staining assay. HeLa cells were treated with 0, 10, 20, or 40 μM of ONP treatment for 24 h, and the numbers of apoptotic cells were measured by the Annexin V-FITC/PI double staining assay. (C) Quantification of apoptotic cells (Q2 and Q4) from (B) by flow cytometry. (D) Activation of caspase 3 in HeLa cells by ONP. HeLa cells were incubated with 0, 10, 20, or 40 μM of ONP treatment for 24 h, and the caspase 3 activity in the cellular extracts was measured by a colorimetric assay. Data are expressed as mean ± S. E. of three experiments. **p < 0.01 versus the control groups.
3.4. Increasing ROS accumulation
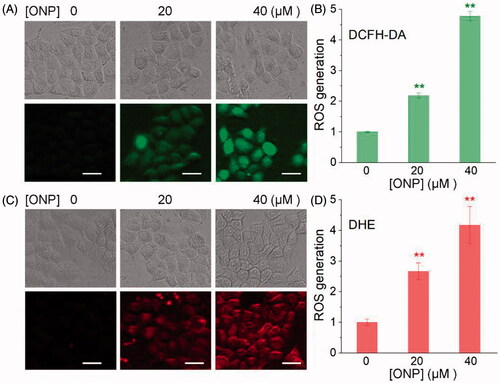
We then explored the mechanism by which ONP induced HeLa apoptosis and showed selective inhibition of tumor cell growth. Considering that one of the biochemical characteristics of carcinoma cells is that they exhibit a higher level of oxidative stress than normal cells, and further stimulation by cytotoxic agents leads to preferentially induce tumor cell apoptosisCitation10,Citation49. We thus observed the changes in ROS levels in HeLa cells treated with ONP to validate the elevated oxidative stress level is the mechanism by which ONP selectively induces tumor cell apoptosis. We administered a fixed concentration of ONP to HeLa cells for different periods and alternately detected the level of intracellular ROS by DCFH-DA probe. Our results clearly showed that the intracellular ROS level reached the highest level after ONP treated HeLa cells for 5 h. As shown in , the green fluorescence enhanced dramatically after treating HeLa cells with 20 and 40 µM ONP for 5 h, suggesting that a large amount of ROS was produced. The effect of 40 µM treatment was much better than that of 20 µM, indicating that excessive accumulation of ROS is also concentration-dependent (). To confirm this result, we used a DHE probe with a different fluorescence wavelength from DCFH-DA to detect again the level of ROS especially superoxide anion in ONP-treated HeLa cells. As shown in , after treatment of HeLa cells with a concentration of 20 µM and 40 µM ONP, intracellular red fluorescence increased remarkably, which once more proved that excessive accumulation of ROS was generated in the HeLa cells treated with ONP. A cascade of ROS generation inevitably broke the upper limit of the cellular adaptive oxidative stress response, and eventually provoked HeLa cell apoptosis.
Figure 4. Induction of ROS accumulation in HeLa cells. Accumulation of ROS in HeLa cells by DCFH-DA staining (A) or DHE staining (C). HeLa cells were treated with 20 or 40 μM of ONP for 24 h, followed by incubation with the ROS probe DCFH-DA (10 μM) or superoxide probe DHE (10 μM) for 30 min. The bright-field (top panel) and the fluorescent-field (bottom panel) pictures from three independent experiments were imaged and acquired by an inverted fluorescence microscope. The fluorescence intensity in individual cells from (A) and (C) was quantified by ImageJ versus the control group and was shown in (B) and (D). **p < 0.01 versus the control groups. Scale bars: 20 μm.
3.5. Inhibiting cellular TrxR activity
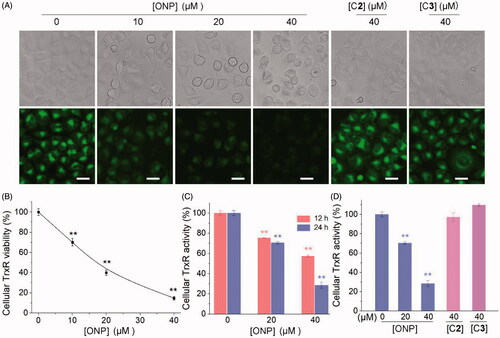
As one of the essential antioxidant enzymes in cells is TrxR. If the TrxR activity is inhibited, the intracellular antioxidant capacity for tumor cells is partially lessened, thereby causing the above-mentioned substantial accumulation of ROSCitation39,Citation50. We thus examined the effect of ONP on cellular TrxR activity. Firstly, the TRFS-green probe established by our groupCitation42 was invested to detect the effects of the ONP, C1, and C2 on TrxR activity in HeLa cells. As shown in , ONP harbored an effective ability to suppress intracellular TrxR activity concentration-dependently from the qualitative results of fluorescence microscopy imaging. While compounds C2 and C3 remained almost silent towards TrxR activity in cells at our observed concentrations (). Likewise, the fluorescence quantification results were also demonstrated by Fast-green, another detection probe for TrxR ()Citation43. To further validate the qualitative and quantitative results of the probes cell imaging, we employed the classic Trx-mediated insulin reduction method to assess the ability of ONP, C2, and C3 to repress TrxR activity in HeLa cells (). Consistent with expectations, ONP remarkably suppressed intracellular TrxR activity in a concentration-and time-dependent manner (). Whereas there was no significant difference in the inhibitory effects of C2 and C3 on intracellular TrxR activity compared with the control group consistent with the probe test results (). By comparing the structures of ONP, C2, and C3, the molecular skeleton bearing α, β unsaturated ketone fragments seems to be highly crucial for their abilities to restrain intracellular TrxR activity, which requires further validation. Collectively, our data strongly confirmed that ONP can excellently inhibit TrxR activity in HeLa cells.
Figure 5. Inhibiting cellular TrxR activity by ONP. (A) TRFS-green-based live-cell imaging TrxR activity assay. HeLa cells were incubated with 0, 10, 20, or 40 μM of ONP or 40 μM C2 or C3 for 12 h. Cellular TrxR activity was assessed by TRFS-green. The fluorescence images were acquired by inverted fluorescence microscopy. (B) Fast-TRFS-based cell lysate TrxR activity assay. HeLa cell lysate (0.3 mg·mL−1) was incubated with NADPH (100 µM) for 5 min at 37 °C. ONP (0, 10, 20, or 40 µM) and blank sample (0.1% DMSO) were added to and the mixture was continued to incubate for 1 h. The probe Fast-TRFS (10 µM) and NADPH (100 µM) were added to initiate the enzymatic reduction of Fast-TRFS. The fluorescence change at 460 nm was recorded (λex = 345 nm) for 10 min on a fluorescent plate reader (Tecan Infinite M200), and the rate of fluorescence increase within the initial 5 min was calculated. The relative TrxR activity was expressed as the percentage of the DMSO-treated sample. Inhibition of intracellular TrxR activity by ONP in a concentration and time-dependent manner (C), and the difference of inhibition of cellular TrxR activity by ONP, C2, and C3 (D) were assayed by the Trx-mediated endpoint insulin reduction. After the HeLa cells were treated with varying concentrations of ONP, C2, and C3 for 12 or 24 h, the enzyme activity of TrxR in HeLa cells was determined by the Trx-mediated endpoint insulin reduction assay. Scale bars: 20 μm. Data are expressed as mean ± S. E. of three experiments. ** p < 0.01 versus the control group.
3.6. Confirmation of the interaction site
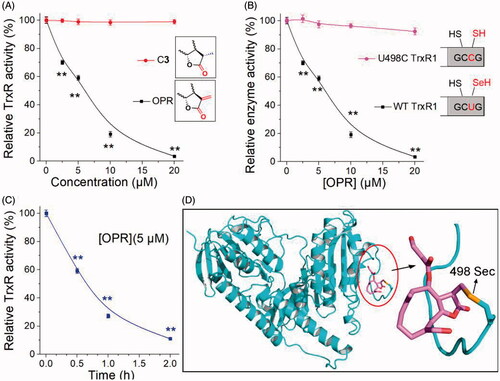
To confirm the key performance of α, β-unsaturated ketone fragments on the lactone ring in the interaction between ONP and TrxR, we explored the interaction of ONP and C3 with pure TrxR. The results were shown in . Compared with C3, ONP could substantially inhibit the pure TrxR activity in a concentration-dependent manner. The vital structural difference between C3 and ONP was that the α, β-unsaturated ketone fragment on the lactone ring (the inset of ). ONP bearing this key fragment had remarkable enzyme inhibitory activity while C3 was almost silenced, heavily validating the importance of α, β-unsaturated ketone fragment in weakening TrxR activity consistent with the literatureCitation51. In addition, ONP at a concentration of 5 µM also had the ability to inhibit pure TrxR in a time-dependent manner (), and the inhibition ability was nearly saturated after incubation for 1 h. Existing evidence highly confirms that the redox-active Sec residue at the C-terminus of TrxR plays a cardinal role in the inhibition of TrxR activity by small moleculesCitation52. The reason is that compared with the homologous cysteine (Cys), Sec has stronger nucleophilicity and stronger resistance to irreversible oxidationCitation53. The high nucleophilicity of Sec residues and exposure to the outer surface of the enzyme render TrxR assailable to be modified by electrophilesCitation11,Citation13. To validate whether Sec residues also play a key role in the interaction between ONP and TrxR, we constructed a control protein for the WT TrxR1, which mutated the Sec at position 498 of the C-terminus to Cys to obtain the U498C TrxR1 mutant enzyme (the inset of ). The effect of ONP on wild-type TrxR1 and recombinant U498C TrxR1 was shown in . ONP hardly inhibited U498C TrxR1, suggesting that the site where ONP interacted with pure TrxR is Sec residue at position 498. We then docked the receptor rat TrxR1 with ONP. The result was shown in . The β-position carbon atom of the lactone ring carbonyl in the ONP can covalently bond well with the Sec at position 498 in the protein. This also proved once again that the interaction sites between ONP and pure TrxR were the β-position carbon atom on the lactone ring and the Sec at position 498 of the enzyme. In short, our data fully confirms that ONP with α, β-unsaturated ketone structure can well capture Sec residues to heavily inhibit TrxR activity, which provides strong evidence for explaining the above-mentioned essential intracellular processes.
Figure 6. Evidence for the interaction site between ONP and TrxR. (A) ONP and C3 selectively inhibited pure TrxR. (The illustration is the structural difference between ONP and C3). (B) Varying concentrations of ONP showed the difference in activity inhibition of WT TrxR and U498C TrxR1. (The illustration exhibited the difference between WT TrxR and U498C TrxR1). (C) ONP at a concentration of 5 μM inhibited the activity of pure TrxR in a time-dependent manner. The enzyme activities in (A), (B), and (C) are all detected by the DTNB method. The NADPH pre-reduced enzyme was incubated with different concentrations of ONP or C3 for 1 h, and then the enzyme activity in the system was detected by the DTNB method. (D) Covalent docking for ONP with the C-terminal site 498 of the chain A of the mouse TrxR1. The docking experiment was conducted using the covalent docking protocol in the SchrödingerSuite 2015-1 program. The monomer of TrxR1 is represented by a cyan cartoon. The interacting residues in the ONP and active site are shown in orange and purple sticks. Data are expressed as mean ± S. E. of three experiments. **p < 0.01 versus the control groups.
3.7. Contribution of targeting TrxR to ONP cytotoxicity
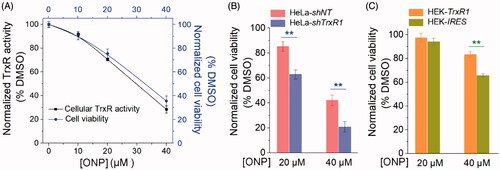
Since we have well verified the interaction site of ONP and TrxR, and this interaction highly caused ONP to suppress both pure TrxR and intracellular TrxR activity. In particular, ONP inhibited TrxR activity in cells, inevitably limiting the function of cellular TrxR. Therefore, when the tumor cells were further stressed, the cellular anti-oxidation ability was partially weakened. As a consequence, the upper limit of tumor cell vulnerability to ROS toxicity may be broken, finally leading to oxidative stress-mediated carcinoma cell apoptosis. The biological behavior of ONP targeting TrxR reasonably contribute to the cytotoxicity of ONP towards tumor cells. As shown in , the survival rate of ONP on HeLa cells we determined previously had a certain extent correlation with its inhibitory rate on intracellular TrxR activity. To validate this correlation, we employed the HeLa-shTrxR1 that had previously knocked down TrxR expression in HeLa cells and the control cell line HeLa-shNT generated by transfecting non-targeted shRNA plasmidsCitation54. After ONP at a concentration of 20 and 40 µM acted respectively on HeLa-shTrxR1 with low TrxR expression and HeLa-shNT with empty plasmid cells, the results showed that the cytotoxicity of ONP at the same concentration to HeLa-shTrxR1 cells was remarkably increased compared with HeLa-shNT cells (), which means TrxR participated in the physiological process of ONP inhibiting the growth of HeLa cells. Correspondingly, overexpression of the functional TrxR protein in HEK-TrxR1 cells rendered the cytotoxicity of ONP to decrease when ONP at a concentration of 40 µM was used to act on the HEK-TrxR1 overexpressing TrxR and the control cell line HEK-IRES (). Attenuation of cytotoxicity by ONP towards the HEK-TrxR1 cell line illustrated further that TrxR was involved in the intracellular process of ONP. Taken together, our data supported that targeting TrxR strongly contributes to the cytotoxicity of ONP.
Figure 7. Contribution of targeting TrxR to ONP cytotoxicity. (A) The positive correlation between inhibition of HeLa cell viability by ONP and inhibition of intracellular TrxR activity by ONP. Different concentrations of ONP were incubated with HeLa cells for 24 h, and the intracellular TrxR activity was detected by the Trx-mediated endpoint insulin reduction assay. The HeLa cell survival rate under the same administration conditions was detected by the trypan blue exclusion staining method. (B) The difference in cytotoxicity of ONP towards the HeLa cell line knocked down TrxR1. ONP (20 and 40 μM) acted on HeLa-shNT and HeLa-shTrxR1 cells for 24 h, respectively, and the cell viability was detected by the trypan blue exclusion staining method (average of three independent experiments). (C) The difference in cytotoxicity of ONP to HEK 293T cell line overexpressing TrxR1. The survival rate of HEK-IRES and HEK-TrxR1 cells under the same experimental conditions as (B) was detected by the trypan blue exclusion staining method (average of three independent experiments). Data are expressed as mean ± S. E. of three experiments. **p < 0.01 versus the control groups.
3.8. Significance of ONP as a new TrxR inhibitor with anticancer activity
Isolation and identification of bioactive molecules in natural products and uncovering their action mechanisms are of great significance to the discovery and development of medicinal value for secondary metabolites and natural plants. As the first major family of dicots, Asteraceae contains numerous genera and species, and most of their medicinal value and secondary metabolites have been systematically studiedCitation55. However, as a new genus of Asteraceae discovered in recent yearsCitation3, the secondary metabolites from Shangwua and their bioactivities have not been reported yet. Since Shangwua grows in a high-altitude hypoxic environmentCitation3, research on its secondary metabolites and medicinal value would inevitably receive attention.
We herein isolated ONP, C2 and C3 from Shangwua for the first time, indicating that sesquiterpene lactones may be one of its primary secondary components (). Intriguingly, these three compounds, e.g. ONP with significant cytotoxicity, C2 with weaker cytotoxicity, and C3 with almost no cytotoxicity, harbor significant differences in cytotoxicity towards tumor cells (). We revealed the reasons for the difference in their antitumor activity from the perspective of compound functional groups and described the antitumor mechanism of ONP targeting TrxR ( and ). Mechanistically, our results revealed that ONP selectively acts on the alteration of redox homeostasis of carcinoma cells, thereby selectively killing tumor cells via apoptosis ( and ). The ability of ONP to vary the redox homeostasis of carcinoma cells was connected with targeting intracellular main antioxidant enzyme TrxR (). Importantly, we not only demonstrated the interaction site of ONP and TrxR but also confirmed that TrxR is involved in the cytotoxicity of ONP. Our data supported that α, β unsaturated ketone in the ONP and the Sec residues with stronger nucleophilicity of the enzyme are the sites of action of both. Selective inhibition of TrxR activity impaired TrxR function in cells, resulting in a series of changes in intracellular redox levels to contribute to the antitumor activity of ONP.
Figure 8. Revealing the mechanism of ONP isolated from the new genus Shangwua for targeting TrxR to induce oxidative stress-mediated apoptosis.
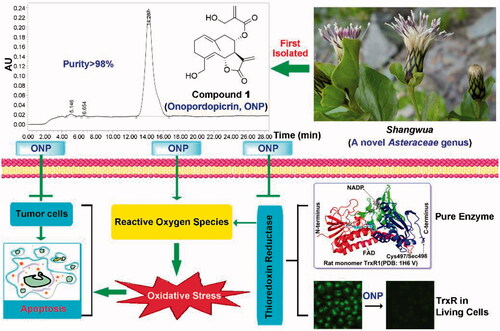
We first studied the secondary metabolites of the new genus Shangwua and discovered a TrxR inhibitor with antitumor activity from it. Our results provided a basis not only for the development of bioactive components of the genus Shangwua but also for the establishment of the medicinal value of the Shangwua. For the first time, we revealed the antitumor activity of the bioactive ingredient ONP and its antitumor mechanism targeting TrxR, inspiring the development of new pharmacological activities of ONP and providing support for the contribution of the target TrxR to the antitumor effect. Therefore, the discovery of ONP from Shangwua and the report of its antitumor activity are of great significance to the establishment of new genus resource components and medicinal value.
4. Conclusions
We isolated and characterized compounds ONP, C2, and C3 from a new genus Shangwua, and evaluated the pharmacological function of ONP with interfering with cellular redox signaling. Further experiments demonstrated that ONP robustly inhibits TrxR, causing perturbation of cellular redox homeostasis with the favor of oxidative stress. Knockdown of the TrxR expression sensitizes cells to the ONP treatment while overexpression of the enzyme reduces the potency of ONP, underpinning the correlation of TrxR inhibition to the observed cytotoxicity of ONP. Our results for the first time revealed the bioactive ingredients and antitumor activity from the new genus Shangwua and served as the discovery of the secondary metabolites and the validation of medicinal value for Shangwua.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
Reference
- Newman DJ, Cragg GM. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J Nat Prod 2020;83(3):770–803.
- Wu QX, Shi YP, Jia ZJ. Eudesmane sesquiterpenoids from the Asteraceae family. Nat Prod Rep 2006;23(5):699–734.
- Wang YJ, von Raab-Straube E, Susanna A, Liu JQ. Shangwua (Compositae), a new genus from the Qinghai-Tibetan Plateau and Himalayas. Taxon 2013;62(5):984–96.
- Lonergan G, Routsi E, Georgiadis T, et al. Isolation, NMR studies, and biological activities of onopordopicrin from Centaurea sonchifolia. J Nat Prod 1992;55(2):225–8.
- Moricz AM, Kruzselyi D, Alberti A, et al. Layer chromatography-bioassays directed screening and identification of antibacterial compounds from Scotch thistle. J Chromatogr A 2017;1524:266–72.
- Bordignon A, Frederich M, Ledoux A, et al. In vitro antiplasmodial and cytotoxic activities of sesquiterpene lactones from Vernonia fimbrillifera Less. (Asteraceae). Nat Prod Res 2018;32(12):1463–6.
- Suzuki M, Iwasaki A, Suenaga K, Kato-Noguchi H. Phytotoxic activity of crop residues from Burdock and an active substance. J Environ Sci Health B 2019;54(11):877–82.
- de Almeida AB, Luiz-Ferreira A, Cola M, et al. Anti-ulcerogenic mechanisms of the sesquiterpene lactone onopordopicrin-enriched fraction from Arctium lappa L. (Asteraceae): role of somatostatin, gastrin, and endogenous sulfhydryls and nitric oxide. J Med Food 2012;15(4):378–83.
- de Almeida AB, Sanchez-Hidalgo M, Martin AR, et al. Anti-inflammatory intestinal activity of Arctium lappa L. (Asteraceae) in TNBS colitis model. J Ethnopharmacol 2013;146(1):300–10.
- Zhang J, Duan D, Song ZL, Liu T, Hou Y, Fang J. Small molecules regulating reactive oxygen species homeostasis for cancer therapy. Med Res Rev 2021;41(1):342–94.
- Zhang J, Li X, Han X, Liu R, Fang J. Targeting the thioredoxin system for cancer therapy. Trends Pharmacol Sci 2017;38(9):794–808.
- Arnér ESJ. Chapter 31 – perspectives of TrxR1-based cancer therapies. In: Sies H, ed. Oxidative Stress. Academic Press; 2020, p. 639–67.
- Bian M, Fan R, Zhao S, Liu W. Targeting the thioredoxin system as a strategy for cancer therapy. J Med Chem 2019;62(16):7309–21.
- Zhang J, Zhang B, Li X, Han X, Liu R, Fang J. Small molecule inhibitors of mammalian thioredoxin reductase as potential anticancer agents: an update. Med Res Rev 2019;39(1):5–39.
- Scalcon V, Bindoli A, Rigobello MP. Significance of the mitochondrial thioredoxin reductase in cancer cells: An update on role, targets and inhibitors. Free Radic Biol Med 2018;127:62–79.
- Jastrzab A, Skrzydlewska E. Thioredoxin-dependent system. Application of inhibitors. J Enzyme Inhib Med Chem 2021;36(1):362–71.
- Zhong L, Arner ES, Holmgren A. Structure and mechanism of mammalian thioredoxin reductase: the active site is a redox-active selenolthiol/selenenylsulfide formed from the conserved cysteine-selenocysteine sequence. Proc Natl Acad Sci USA 2000;97(11):5854–9.
- Lee SR, Bar-Noy S, Kwon J, Levine RL, Stadtman TC, Rhee SG. Mammalian thioredoxin reductase: oxidation of the C-terminal cysteine/selenocysteine active site forms a thioselenide, and replacement of selenium with sulfur markedly reduces catalytic activity. Proc Natl Acad Sci USA 2000;97(6):2521–6.
- Cai W, Zhang L, Song Y, et al. Small molecule inhibitors of mammalian thioredoxin reductase. Free Radic Biol Med 2012;52(2):257–65.
- Dagnell M, Schmidt EE, Arner ESJ. The A to Z of modulated cell patterning by mammalian thioredoxin reductases. Free Radic Biol Med 2018;115:484–96.
- Arner ES. Focus on mammalian thioredoxin reductases – important selenoproteins with versatile functions. Biochim Biophys Acta 2009;1790(6):495–526.
- Lu J, Holmgren A. Thioredoxin system in cell death progression. Antioxid Redox Signal 2012;17(12):1738–47.
- Arner ES, Holmgren A. Physiological functions of thioredoxin and thioredoxin reductase. Eur J Biochem 2000;267(20):6102–6109.
- Arner ESJ. Targeting the selenoprotein thioredoxin reductase 1 for anticancer therapy. Adv Cancer Res 2017;136:139–51.
- Schmidt C, Albrecht L, Balasupramaniam S, et al. A gold(i) biscarbene complex with improved activity as a TrxR inhibitor and cytotoxic drug: comparative studies with different gold metallodrugs. Metallomics 2019;11(3):533–45.
- Jovanovic M, Zhukovsky D, Podolski-Renic A, et al. Further exploration of DVD-445 as a lead thioredoxin reductase (TrxR) inhibitor for cancer therapy: optimization of potency and evaluation of anticancer potential. Eur J Med Chem 2020;191:112119.
- Krasavin M, Zalubovskis R, Grandane A, Domraceva I, Zhmurov P, Supuran CT. Sulfocoumarins as dual inhibitors of human carbonic anhydrase isoforms IX/XII and of human thioredoxin reductase. J Enzyme Inhib Med Chem 2020;35(1):506–10.
- Li XM, Hou YN, Zhao JT, Li J, Wang S, Fang JG. Combination of chemotherapy and oxidative stress to enhance cancer cell apoptosis. Chem Sci 2020;11(12):3215–22.
- Liu R, Shi D, Zhang J, et al. Virtual screening-guided discovery of thioredoxin reductase inhibitors. Toxicol Appl Pharmacol 2019;370:106–16.
- Liu T, Zhang J, Han X, Xu J, Wu Y, Fang J. Promotion of HeLa cells apoptosis by cynaropicrin involving inhibition of thioredoxin reductase and induction of oxidative stress. Free Radic Biol Med 2019;135:216–26.
- Li X, Hou Y, Meng X, et al. Selective activation of a prodrug by thioredoxin reductase providing a strategy to target cancer cells. Angew Chem Int Ed Engl 2018;57(21):6141–5.
- Zhang J, Liu Y, Shi D, et al. Synthesis of naphthazarin derivatives and identification of novel thioredoxin reductase inhibitor as potential anticancer agent. Eur J Med Chem 2017;140:435–47.
- Zhang J, Li Y, Duan D, Yao J, Gao K, Fang J. Inhibition of thioredoxin reductase by alantolactone prompts oxidative stress-mediated apoptosis of HeLa cells. Biochem Pharmacol 2016;102:34–44.
- Duan D, Zhang J, Yao J, Liu Y, Fang J. Targeting thioredoxin reductase by parthenolide contributes to inducing apoptosis of HeLa cells. J Biol Chem 2016;291(19):10021–31.
- Zhang B, Duan D, Ge C, et al. Synthesis of xanthohumol analogues and discovery of potent thioredoxin reductase inhibitor as potential anticancer agent. J Med Chem 2015;58(4):1795–805.
- Liu Y, Duan D, Yao J, et al. Dithiaarsanes induce oxidative stress-mediated apoptosis in HL-60 cells by selectively targeting thioredoxin reductase. J Med Chem 2014;57(12):5203–11.
- Duan D, Zhang B, Yao J, Liu Y, Fang J. Shikonin targets cytosolic thioredoxin reductase to induce ROS-mediated apoptosis in human promyelocytic leukemia HL-60 cells. Free Radic Biol Med 2014;70:182–193.
- Zhang J, Yao J, Peng S, Li X, Fang J. Securinine disturbs redox homeostasis and elicits oxidative stress-mediated apoptosis via targeting thioredoxin reductase. Biochim Biophys Acta Mol Basis Dis 2017;1863(1):129–38.
- Zhang J, Duan D, Osama A, Fang J. Natural molecules targeting thioredoxin system and their therapeutic potential. Antioxid Redox Signal 2020.doi:https://doi.org/10.1089/ars.2020.8213.
- Nalvarte I, Damdimopoulos AE, Nystom C, et al. Overexpression of enzymatically active human cytosolic and mitochondrial thioredoxin reductase in HEK-293 cells. Effect on cell growth and differentiation. J Biol Chem 2004;279(52):54510–17.
- Javvadi P, Hertan L, Kosoff R, et al. Thioredoxin reductase-1 mediates curcumin-induced radiosensitization of squamous carcinoma cells. Cancer Res 2010;70(5):1941–50.
- Zhang L, Duan D, Liu Y, et al. Highly selective off-on fluorescent probe for imaging thioredoxin reductase in living cells. J Am Chem Soc 2014;136(1):226–33.
- Li X, Zhang B, Yan C, et al. A fast and specific fluorescent probe for thioredoxin reductase that works via disulphide bond cleavage. Nat Commun 2019;10(1):2745.
- Djeddi S, Karioti A, Sokovic M, Stojkovic D, Seridi R, Skaltsa H. Minor sesquiterpene lactones from Centaurea pullata and their antimicrobial activity. J Nat Prod 2007;70(11):1796–9.
- Marco JA, Sanz-Cervera JF, Yuste A, Sancenon F, Carda M. Sesquiterpenes from Centaurea aspera. Phytochemistry 2005;66(14):1644–50.
- Ding Y, Gao H, Zhang Y, et al. Alantolactone selectively ablates acute myeloid leukemia stem and progenitor cells. J Hematol Oncol 2016;9(1):93.
- Suvannasankha A, Crean CD, Shanmugam R, et al. Antimyeloma effects of a sesquiterpene lactone parthenolide. Clin Cancer Res 2008;14(6):1814–22.
- Abu-Izneid T, Rauf A, Shariati MA, et al. Sesquiterpenes and their derivatives-natural anticancer compounds: an update. Pharmacol Res 2020;161:105165.
- Sies H, Berndt C, Jones DP. Oxidative stress. Annu Rev Biochem 2017;86:715–48.
- Lu J, Holmgren A. The thioredoxin antioxidant system. Free Radic Biol Med 2014;66:75–87.
- Gan FF, Kaminska KK, Yang H, et al. Identification of Michael acceptor-centric pharmacophores with substituents that yield strong thioredoxin reductase inhibitory character correlated to antiproliferative activity. Antioxid Redox Signal 2013;19(11):1149–65.
- Seki H, Xue S, Pellett S, Silhar P, Johnson EA, Janda KD. Cellular protection of SNAP-25 against botulinum neurotoxin/A: inhibition of thioredoxin reductase through a suicide substrate mechanism. J Am Chem Soc 2016;138(17):5568–75.
- Maroney MJ, Hondal RJ. Selenium versus sulfur: reversibility of chemical reactions and resistance to permanent oxidation in proteins and nucleic acids. Free Radic Biol Med 2018;127:228–37.
- Zhang J, Duan D, Xu J, Fang J. Redox-dependent copper carrier promotes cellular copper uptake and oxidative stress-mediated apoptosis of cancer cells. Acs Appl Mater Inter 2018;10(39):33010–21.
- Heinrich M, Robles M, West JE, Ortiz de Montellano BR, Rodriguez E. Ethnopharmacology of Mexican asteraceae (Compositae). Annu Rev Pharmacol Toxicol 1998;38:539–65.
