Abstract
A β-class carbonic anhydrase (CA, EC 4.2.1.1) was cloned from the genome of the Monogenean platyhelminth Gyrodactylus salaris, a parasite of Atlantic salmon. The new enzyme, GsaCAβ has a significant catalytic activity for the physiological reaction, CO2 + H2O ⇋ HCO3− + H+ with a kcat of 1.1 × 105 s−1 and a kcat/Km of 7.58 × 106 M−1 × s−1. This activity was inhibited by acetazolamide (KI of 0.46 µM), a sulphonamide in clinical use, as well as by selected inorganic anions and small molecules. Most tested anions inhibited GsaCAβ at millimolar concentrations, but sulfamide (KI of 81 µM), N,N-diethyldithiocarbamate (KI of 67 µM) and sulphamic acid (KI of 6.2 µM) showed a rather efficient inhibitory action. There are currently very few non-toxic agents effective in combating this parasite. GsaCAβ is subsequently proposed as a new drug target for which effective inhibitors can be designed.
1. Introduction
Gyrodactylus salaris is a flatworm (platyhelminth) parasite belonging to the Monogeneans group, which are hermaphrodite ectoparasites found on the gills, fins, or skin of fishCitation1,Citation2. They do not need an intermediate host for infecting a range of fish species, some of which possess significant commercial status, such as the Atlantic salmon (Salmo salar) and related speciesCitation3,Citation4. The presence of this parasitic pathogen has been reported in 19 countries across Europe and has already produced catastrophic losses of Atlantic salmon mainly in Norway starting in the 1970s and in Russian Karelia since 1992Citation1–4. This small (0.5 mm) parasite attacks the host by attaching its anterior end to the fish through secretions from the cephalic glands, and then releasing a digestive solution rich in proteolytic enzymes which dissolves the fish skin, inducing the formation of large wounds which favour secondary infectionsCitation5. A variety of inorganic and organic compounds, among which are salt (NaCl), hypochlorite, permanganate, aluminium salts, praziquantel, levamisole, mebendazole, toltrazuril, etc., have been tested for efficacy against a broad spectrum of monogenean species, including G. salaris, but only trichlorfon and dichlorvos () showed some efficacyCitation6,Citation7. However, both compounds act as irreversible organophosphoric acetylcholinesterase inhibitors, showing thus a rather high toxicity for all vertebrates, not only for fishCitation8.
Novel potential drug targets present in the proteome of this parasite have been recently explored. This includes excretory/secretory proteins involved in host invasion and colonisationCitation9, with a flatworm host invasion and colonisation, such as a flatworm proteaseCitation9. However, no pharmacological inhibitors were reported so far.
Considering the fact that the metalloenzyme carbonic anhydrase (CA, EC 4.2.1.1) has a fundamental role in many organisms, as it catalyses the hydration of CO2 to bicarbonate and protonsCitation10–17, and also that such enzymes were already investigated in parasitic (Ascaris lumbricoides, Schistosoma spp., etc.)Citation18–24 and non-parasitic (Caenorhabditis elegans) wormsCitation25–27 for their inhibition with various classes of inhibitors, it appeared of interest to investigate whether this enzyme is also present in G. salaris. Here we report the cloning, characterisation and anion inhibition studies of a β-class CA (GsaCAβ) encoded in the genome of G. salaris, which we propose as a potential drug target for the management of this parasitic fish disease.
2. Materials and methods
2.1. Construction of vector for recombinant protein production
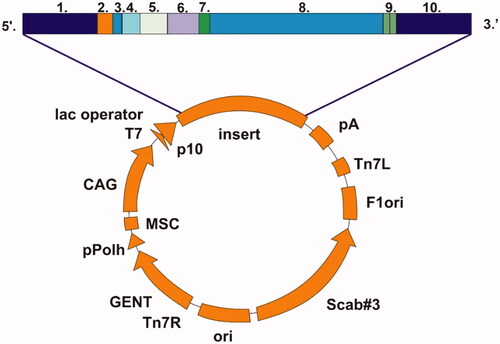
A nearly complete GsaCAβ sequence was obtained from transcriptome data produced at the University of Turku. The open reading frame produced a translation of 229 amino acids, 165 of which were supported by genome data from the University of Oulu. A BLAST search at NCBICitation28 suggested that the sequence is close to full length, and therefore the existing sequence was only extended by an ATG codon at the beginning of the transcript and a stop codons (TAA TAG) at the end. This sequence of the β-CA was inserted in a pBVboost vector construct for the production of recombinant protein (). The construct was obtained by GeneArt (Invitrogen, Regensburg, Germany). The sequence of the β-CA was modified accordingly to produce the protein in bacterial (Escherichia coli) cells.
Figure 2. Schematic presentation of pBVboostFG expression vector designed for production of recombinant protein. The insert contains: 1. attL1, 2. Shine-Dalgarno, 3. Kozak, 4. Met-Ser-Tyr-Tyr, 5. 6 x His, 6. Asp-Tyr-Asp-Ile-Pro-Thr-Thr, 7. Lys-Val, 8. β-CA gene of G. salaris gene of interest, 9. Two stop codons, and 10. AttL2.
2.2. Transformation of plasmid vector into BL21 cells
The construct of the β-CA sequence from the freeze-dried plasmid supplied by GeneArt was prepared according to the instructions of the manufacturer. The BL21 Star™ (DE3) cells were stored at −80 °C cells (Invitrogen, Carlsbad, CA, USA) and thawed by keeping them on ice. After thawing the competent cells, 25 µl of the cell suspension and 1 µl of the reconstituted plasmid, were transferred into a 1.5 ml centrifuge tube. The suspension was incubated on ice for 30 min. Heat shock was performed by keeping the tube in 42 °C water for 30 s, and transferred immediately on ice for 2 min. 125 µl of SOC Medium (Invitrogen, Carlsbad, CA, USA) was added to the microcentrifuge tube containing the transformed cells, and the tube was incubated at 37 °C for 1 h with shaking (200 rpm). The agar plates containing gentamycin were stored at 37 °C before the transformation. 20 µl or 50 µl of cell suspension described above were spread onto each plate, and the plates were incubated overnight at 37 °C . A volume of 5 ml preculture was prepared by inoculating single colonies from growth plates onto an LB medium with gentamycin (ratio 1:1000), being then incubated overnight at 37 °C with constant shaking of 200 rpm.
2.3. Production of GsaCAβ recombinant protein
Protein production was carried out according to pO-stat fed batch protocol as described earlier with some modificationsCitation29. The fermentation medium contained no glycerol, as the cell line used did not require it. For induction of the culture 1 mM IPTG was used after 12 h of starting the fermentation. The temperature was decreased to 25 °C at the time of the induction. Cell growth was stopped after 12 h of the induction with the OD 34 (A600). The cells were collected by centrifugation and the weight of the cell pellet was recorded. The fermentation was carried out at the Protein Services core facility of Tampere University (https://www.tuni.fi/en/research/protein-services). The cell pellet was suspended in 150 ml of binding buffer containing 50 mM Na2HPO4, 0.5 M NaCl, 50 mM imidazole, and 10% glycerol (pH 8.0). Cell pellet was lysed with sonication (5 min, 30 s off, 20 s on) into the lysing buffer and centrifuged at 20 000×g/15 min. The suspension was homogenised using EmulsiFlex-C3 (AVESTIN, Ottawa, Canada) homogeniser. The lysate was centrifuged at 13 000×g for 15 min at 4 °C, and the clear supernatant was mixed with HisPur™ Ni-NTA Resin (Thermo Fisher Scientific, Waltham, MA, USA) and bound to the resin for 2 h at room temperature on the magnetic stirrer. The obtained resin was washed with the binding buffer and collected onto an empty column with an EMD Millipore™ vacuum filtering flask (Merck, Kenilworth, NJ, USA) and filter paper. The protein from the resin was eluted using 50 mM Na2HPO4, 0.5 M NaCl, 350 mM imidazole, and 10% glycerol (pH 7.0). The protein was re-purified with TALON® Superflow™ cobalt resin (GE Healthcare, Chicago, IL, USA). The eluted protein fractions were diluted with binding buffer (50 mM Na2HPO4, 0.5 M NaCl, and 10% glycerol pH 8.0), so that the imidazole concentration was under 10 mM. The protein binding and elution were performed as described above. The purity of the protein was determined with gel electrophoresis (SDS-PAGE) and visualised with PageBlue Protein staining solution (Thermo Fisher Scientific, Waltham, MA, USA). Protein fractions were pooled and concentrated according to the protocol (https://store.repligen.com/products/floatalyzer) (8–10 kD). Buffer exchange in 50 mM TRIS (pH 7.5) was done using the same centrifugal concentrators. The His-tag was cleaved from the purified protein by Thrombin CleanCleave Kit (Sigma-Aldrich, Saint Louis, MO, USA), according to manufacturer’s manual.
2.4. CA activity and inhibition measurements
An Applied Photophysics stopped-flow instrument has been used for assaying the CA catalysed CO2 hydration activityCitation30. Phenol red at a concentration of 0.2 mM was used as pH indicator, working at the absorbance maximum of 557 nm, with 10 mM TRIS (pH 8.3) as buffer, and in the presence of 10 mM NaClO4 for maintaining constant the ionic strength, following the initial rates of the CA-catalysed CO2 hydration reaction for a period of 10–100 s. The CO2 concentrations ranged from 1.7 to 17 mM for the determination of the kinetic parameters and inhibition constants. For each inhibitor, at least six traces of the initial 5–10% of the reaction have been used for determining the initial velocity. The uncatalyzed rates were determined in the same manner and subtracted from the total observed rates. Stock solutions of inhibitors (10–20 mM) were prepared in distilled-deionized water and dilutions up to 0.01 µM were done thereafter with the assay buffer. Inhibitor and enzyme solutions were preincubated together for 15 min at room temperature prior to assay, in order to allow for the formation of the enzyme-inhibitor complex. The inhibition constantsCitation31–37 represent the mean from at least three different determinations. GsaCAβ concentration in the assay system was 14.3 nM.
2.5. Reagents
Anions and small molecules were commercially sold reagents of the highest available purity from Sigma-Aldrich (Milan, Italy). The purity of tested compounds was higher than 99%.
2.6. Phylogenetic analysis
A BLAST search was performed on the UniPROT webserver (https://www.uniprot.org/blast/) with the novel GsaCAβ sequence as a query and all settings as default. The top 250 closely related sequences, and their annotations (species, phyla), were taken for further analysis.
The 250 β-CAs were clustered to 80% similarity with the "cluster fast" algorithm of the USEARCH tool (version 11.0.667)Citation38 and 111 sequences representing the centroids of clusters resulted. To this list the novel G. salaris β-CA was added, and a custom Python script was then used to further filter out sequences that did not contain both canonical β-CA amino acid motifs (CxDxR and HxxC), resulting in a total of 104 β-CA sequences which were then aligned with Muscle (version 5.1) using all default settingsCitation39.
The alignment was reduced to a total of 62 conserved amino acid residues which were identified using GBlocks (version 0.91 b)Citation40 with parameters “-t = p -b2 = 6 -b3 = 20 -b4 = 2 -b5 = h -d = y -v = 240”. Model testing was performed to identify the best evolutionary model for analysis of the target sequences using ModelFinderCitation41, with the best model determined to be “LG + I+G4”. A maximum likelihood phylogenetic analysis was performed using the IQTree software (version 2.0.3)Citation42, with parameters set to "-alrt 100000 -bb 100000 -nt AUTO -m LG + I+G4" and all other options run as default. A consensus tree was generated from the 100 000 bootstrap replicates, with a final log-likelihood value of −4717.575. The tree was then visualised using a custom Python script utilising the ETE Toolkit Python libraryCitation43.
The code and workflow used to perform these analyses are provided at https://github.com/thirtysix/Aspatwar.Gsalaris_BCA (Supplemental data).
2.7. Subcellular localizations
TMHMM 2.0Citation44 (https://services.healthtech.dtu.dk/service.php?TMHMM-2.0) was used for the prediction of transmembrane helices. TargetP 2.0Citation45 (https://services.healthtech.dtu.dk/service.php?TargetP-2.0) was used for the prediction of various N-terminal targeting peptides, with parameters for non-plant proteins. Finally, we performed predictions for multiple subcellular localizations with DeepLoc 1.0Citation46 (https://services.healthtech.dtu.dk/service.php?DeepLoc-1.0).
2.8. Multiple sequence alignment
Our protein sequence of GsaCAβ was used as a query in BLAST searchesCitation28 at NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Searches were made limited to metazoa, except vertebrates, in the nr database with wordsize 3 and scoring matrix BLOSUM45, otherwise default parameters. The results were filtered for at least 90% query coverage. Seven homologs were selected based on taxonomical diversity and model organism status from the results with E value cut-off 1e-28 (top 242 hits). Details of all the hits are given in https://bit.ly/3JNRb7i (Supplemental data).
Sequences were aligned with Clustal OmegaCitation47 at EBI (https://www.ebi.ac.uk/Tools/msa/clustalo/) with number of combined iterations = 3, otherwise default parameters. Espript 3Citation48 at https://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi was used in visualising the sequence alignment result. Our AlphaFold model for GsaCAβ (see below) was used for the display of secondary structures above the alignment. The threshold for boxing nearly conserved residues was set to 80%.
2.9. Molecular modelling
All operations with 3 D protein structure models and molecular visualisation were performed using ChimeraX (daily build 1.4.dev202202030703), developed by the UCSF Resource for Biocomputing, Visualisation, and Informatics (San Francisco, California, USA), supported in part by the National Institutes of HealthCitation49.
A 3D model of GsaCAβ was created with AlphaFoldCitation50 using the ChimeraX interface to submit the prediction to run at Google Colab. This model was compared to a crystallographic structure of the pea β-CA, PDB 1EKJCitation51 by 3 D superimposition using the MatchMaker tool of ChimeraX with an iteration cut-off of 1.5 Å for pruning residue pairs in fitting.
3. Results and discussion
3.1. Sequence analysis of GsaCAβ, comparison with other β-CAs, and recombinant protein production
The partial GsaCAβ transcript sequence discovered in this study, of 687 nucleotides, has been submitted to European Nucleotide Archive (ENA) and assigned the accession number OW406882. The translated peptide of 229 amino acids can be found in the CDS/translation feature of the entry at http://www.ebi.ac.uk/ena/data/view/OW406882.
The bioinformatic analyses of G. salaris genomic data revealed the presence of a novel CA sequence that resembled the β-CAs from other metazoan species, as seen in the small MSA of . The sequences in the MSA are detailed in . The GsaCAβ sequence is moderately similar to the β-CAs of other metazoans ( and ), with regions of highest similarity at 32 to 115 and at 176 to 192 (GsaCAβ numbering). The hallmark motifs CxDxR and HxxC of the active site of β-CAs, in which the cysteines and histidine coordinate the catalytic zinc ion, are conserved in GsaCAβ (shown by triangles in ). It is notable that the sequence identity is nearly constant between GsaCAβ and β-CAs from widely different groups of animals, as seen in and . Sequence identity was 29.8% ± 1.4% (mean ± SD) in the whole set of 431 metazoan Blast hits. The MSA of contains 26 fully conserved positions and 59 partially conserved (boxed) positions along the GsaCAβ sequence of 229 residues, in total 85, or 37.1% of the positions.
Figure 3. Alignment of GsaCAβ sequence with β-CA sequences of other metazoans. The conserved hallmark catalytic-site sequences of β-CAs, CXDXR and HXXC, are shown with black triangles (C: Cysteine, D: Aspartic acid, H: Histidine, R: Arginine, X: any residue). Columns with fully conserved residues are shown as red with white letters. Boxed columns denote positions in which at least 80% of residues are of similar type. The top line shows secondary structures derived from our GsaCAβ model. α: α-helices; β: β-strands; η: 310-helices; T: turns.
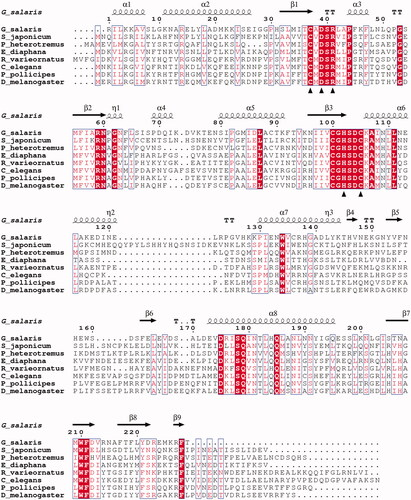
Table 1. Sequences in the multiple sequence alignment of .
Table 2. Percent identity matrix of the aligned protein sequences of , as computed by Clustal Omega
Analysis of the GsaCAβ sequence with TMHMM 2.0 suggests that no transmembrane helices are present. TargetP 2.0 predicts a cytoplasmic (“other”) localisation, from among the choices of secreted, mitochondrial, or other. DeepLoc 1.0, on the other hand, predicts mitochondrial localisation from among 10 different localizations with a likelihood of 0.77. DeepLoc does not depend solely on N-terminal sequences in its inference, contrary to TargetP 2.0. Furthermore, the authors of TargetP 2.0 state that for non-plant proteins, the most common confusion is between mitochondrial targeting peptides and no targeting peptidesCitation45. The same article also notes that the second residue in the sequence has a markedly strong predictive value for metazoan mitochondrial targeting peptides. Considering all this, and the fact that our protein sequence is incomplete at the N-terminus, we prefer to accept the prediction of DeepLoc and suggest tentatively a mitochondrial localisation for GsaCAβ. This would also be consistent with mitochondrial localisation observed or predicted for many other metazoan β-carbonic anhydrases (BCAs)Citation20,Citation52,Citation53.
The sequence comparisons carried out here confirm that GsaCAβ belongs to β-CA enzyme family and could be a potential target for developing suitable inhibitors for control of this parasite in fish culture farms.
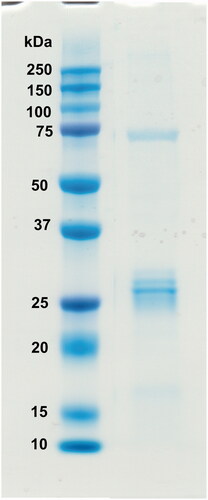
The recombinant GsaCAβ protein was produced in E. coli cells. The purified recombinant protein showed a single band close to the expected size on SDS-PAGE (, measured MW 26.0 kDa).
3.2. Phylogeny of 131 β-CA sequences
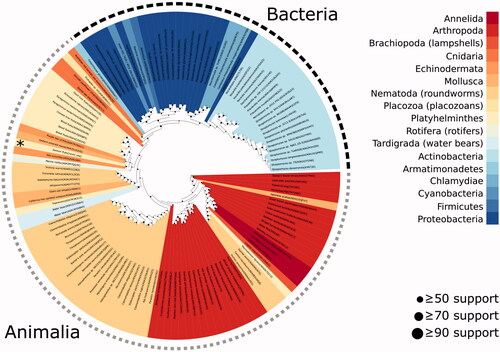
Phylogenetic analysis was performed using the GsaCAβ sequence together with 130 related β-CA sequences identified via BLAST search. Among all sequences included there were 17 distinct phyla represented, 5 of which came from bacteria (46 sequences) and 12 from animalia (85 sequences). Within animalia, the largest two groups were nematoda (29 sequences) and arthropoda (28 sequences). The most closely related sequence to the novel GsaCAβ was from sea cucumber (A0A2G8LGE8), and together these occurred within a larger clade of 13 total sequences, which included 8 other platyhelminthes. shows the final phylogenetic tree.
A total of 130 βCA sequences identified by BLAST search of the UniProt database were combined with the GsaCAβ sequence and used to perform maximum likelihood phylogenetic analysis (IQTree version 2.0.3)Citation42 to identify potential evolutionary relationships. Colours represent the phyla of the species from which each sequence originates. G. salaris is marked with an asterisk. Dot sizes in the internal nodes of the tree indicate bootstrap support level of each node.
3.2. Catalytic activity of GsaCAβ and its inhibition with acetazolamide
The catalytic activity of the new β-CA, GsaCAβ, was investigated for the physiological reaction catalysed by CAs, CO2 hydration to bicarbonate and protons (). The experiments were performed at a pH value of 8.3 where the active site of these enzymes is openCitation54–65. We compared the activity of GsaCAβ with that of other earlier characterised β-CAs from fungi, yeasts and bacteria ().
Table 3. Kinetic parameters for the CO2 hydration reactionCitation30 catalysed by α- and β-class CA enzymes: the human cytosolic isozymes hCA I and II (α-class CAs) at 20 °C and pH 7.5 in 10 mM HEPES buffer, Can2 (from Cryptococcus neoformans), CalCA (from Candida albicans), SceCA (from Saccharomyces cerevisiae), and Cab (from the archaeon Methanobacterium thermoautotrophicum) measured at 20 °C, pH 8.3 in 20 mM TRIS buffer and 10 mM NaClO4.
Data presented in show that GsaCAβ has a low but significant catalytic activity for the physiological reaction, with a kcat of 1.1 × 105 s−1 and kcat/Km of 7.58 × 106 M−1 × s−1 (KM of 14.5 mM), being thus around 12 times less efficient compared to hCA II ()Citation10–13. However, this catalytic activity is relevant, being comparable to that of similar enzymes from well-known bacterial/fungal pathogens ()Citation54–65 where a relevant physiological/pathological role has been demonstrated by using specific inhibitors of enzymatic activityCitation54–65. Indeed, inhibition of CAs has been shown to interfere with the growth and virulence of the pathogensCitation66–68. Also, in the case of GsaCAβ, the enzymatic activity was inhibited with a KI of 460 nM by the classical sulphonamide inhibitor acetazolamide ().
3.3. Anion inhibition studies of GsaCAβ
A panel of anions and small molecules known for interacting with CAsCitation69 were chosen to be tested as inhibitors of GsaCAβ ().
As seen from the data in , where the inhibition of the human α-class isoforms hCA I and II was also included for comparison, many inorganic/organic anions and small molecules, such as sulfamide and sulphamic acid, inhibited GsaCAβ. The action of inhibition is presumably through coordination of the molecule to the metal ion in the active site, as with other CAs previously investigated for their interaction with this type of inhibitorCitation13–17,Citation54,Citation55,Citation70,Citation71. The following should be noted regarding the inhibition data of :
Table 4. Anion inhibition data of the β-CA from G. salaris and human isoforms hCA I and hCA II as determined by stopped-flow CO2 hydrase assay.
Anions and small molecules which were non-inhibitory (up to 50 mM) against GsaCAβ included iodide, azide, nitrate, bicarbonate, carbonate, sulphate, phenylarsonic acid, phenylboronic acid, perchlorate, selenate, pyrophosphate, tetraborate, hexafluorophosphate and triflate. Many of these anions/small molecules poorly interact with metal ions in solution or within the metalloenzymes’ active sites (e.g. perchlorate, hexafluorophosphate and triflate)Citation69, whereas others (e.g. azide, iodide, phenylarsonic acid, phenylboronic acid, etc.) inhibit with various efficacy CAs belonging to other classesCitation13,Citation69–71. Azide, for example, is a rather efficient hCA I inhibitor, with a KI of 1.2 µM, and has thoroughly been characterised by diverse techniques, including X-ray crystallographyCitation14–17.
Most of the investigated anions showed a millimolar affinity for GsaCAβ, which is the typical inhibitory profile for this type of compounds. Indeed, KI-s in the range of 1.9–91 mM were measured for the following anions: fluoride, chloride, bromide, cyanate, thiocyanate, nitrite, bisulphite, bisulphide, tellurate, perosmate, divanadate, perrhenate, perruthenate, selenocyanate, imidosulfonate, and trithiocarbonate. Many of these anions are known for their propensity to complex with metal ions and they also act as inhibitors of other CAs, including hCA I and II ()Citation13–17,Citation69–71.
Sub-millimolar inhibition of GsaCAβ was observed for cyanide, stannate, peroxydisulfate, and fluorosulfonate, which showed KI-s in the range of 0.77–0.92 mM.
The most effective GsaCAβ inhibitors were sulfamide, sulphamic acid (presumably acting as sulfamateCitation72) and N,N-diethyldithiocarbamate, which showed KI-s in the range of 6.2–81 µM. These inhibitors incorporate two well-known zinc-binding groups (ZBGs) present in many efficient CA inhibitors: the sulfamoyl moiety (present in sulfamide and sulphamic acidCitation72, which has been demonstrated using crystallography to coordinate the zinc ion from the CA active siteCitation72. The same inhibition mechanism was thereafter observed for dithiocarbamates and their derivativesCitation73–75, which incorporate the CS2− ZBG. The fact that simple derivatives possessing no organic scaffolds (sulfamide and sulphamic acid) or a very small and compact scaffold (as N,N-diethyldithiocarbamate) do inhibit GsaCAβ quite efficiently, prompts us to hypothesise that it might be possible to develop more efficient and selective inhibitors for this enzyme, with potential use as antiparasitic agents.
3.3. Molecular model of GsaCAβ
We created a predicted structural model of GsaCAβ using AlphaFoldCitation50. The model is highly similar to crystallographic structures of other β-CAs. AlphaFold evaluates the per-residue confidence score (pLDDT, between 0 and 100) to be higher than 90 (“very high confidence”) for 65.1% of the residues and higher than 70 (“confident”) for an additional 26.2% of the residues. The N-terminal region 1–30 contains only residues with pLDDT <90. Counting from H31 (just before β-strand β1) to the end of the sequence, 75.3% of the residues are of very high confidence and 19.2% are confident. The only regions with low-confidence residues (50 < pLDDT < 70) are the N-terminus (8 of the first 10 residues) and the irregular helix at 67 to 78. All pLDDT values in our model can be found at https://bit.ly/3qIMIv6 (Supplemental data).
In our model (yellow) is superimposed with pea β-CA (sky blue), with excellent fit over the core secondary structures and the loops of the catalytic site. The models of were superimposed with a strict iteration cut-off of 1.5 Å. There were 97 residue pairs left within the cut-off distance, with an RMSD of 0.858 Å, indicating excellent fit. Note that this RMSD is not a proper measure of the overall similarity of the two models, just between the subsets of the best-fitting residues. These structurally constant residues cover the parallel β-sheet formed of β-strands β2, β1, β3 and β7; α-helices α5 and α8 (on top of the β-sheet in ); the α-helix α6 extending after the HxxC motif; and the loops at the tips of β-strands β1 and β3 which form the catalytic site. Refer to for the numbering and locations of secondary structure elements.
Figure 6. Molecular model of GsaCAβ. Our model constructed using AlphaFold, yellow, superimposed with pea β-CA (PDB 1EKJ, chain C), sky blue. The zinc ion of the catalytic site is shown in red.
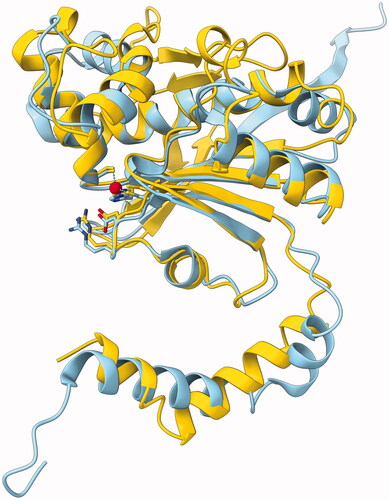
The region 150–168 represents an insertion relative to the pea β-CA sequence. This region is seen near the top of , predicted to form a β-hairpin-like structure containing β-strands β5 and β6. Despite the lack of similar loops in β-CA models in PDB, this region is mainly evaluated to be of high confidence.
To our knowledge, this is the second 3 D model for any platyhelmith β-CA to be made freely accessible, the first one being β-CA of Schistosoma mansoni in the AlphaFold database. Our model is available as a PDB file at https://bit.ly/3tPaNT2.
4. Conclusions
We report here the cloning and characterisation of a β-class CA identified in the genome of the Monogenean platyhelminth Gyrodactylus salaris, a parasite of Atlantic salmon and other economically relevant aquaculture fish species. Sequence analysis and successful modelling of the protein confirm its membership in the β-CA class. Sequence comparisons and the observed enzymatic activity also suggest that the N- and C-terminal sequences missing from our incomplete sequence are insignificant. This new enzyme, GsaCAβ, showed a low but significant catalytic activity for the physiological CO2 hydration reaction, with a kcat of 1.1 × 105 s−1 and a kcat/Km of 7.58 × 106 M−1 × s−1. This activity was inhibited by acetazolamide (KI of 0.46 µM), a sulphonamide in clinical use, as well as by some inorganic anions and small molecules. Most investigated anions (fluoride, chloride, bromide, cyanate, thiocyanate, nitrite, bisulphite, bisulphide, tellurate, perosmate, divanadate, perrhenate, perruthenate, selenocyanate, imidosulfonate, and trithiocarbonate) were millimolar GsaCAβ inhibitors. Cyanide, stannate, peroxydisulfate, and fluorosulfonate, showed submillimolar range KI-s of 0.77–0.92 mM. Sulfamide (KI of 81 µM), N,N-diethyldithiocarbamate (KI of 67 µM) and sulphamic acid (KI of 6.2 µM) were the most efficient GsaCAβ inhibitors. Correlated to the fact that there are very few non-toxic agents effective in combating this parasite, GsaCAβ is proposed as a new antiparasitic drug target for which effective inhibitors could be designed.
Disclosure statement
CT Supuran is Editor-in-Chief of the Journal of Enzyme Inhibition and Medicinal Chemistry. He was not involved in the assessment, peer review, or decision-making process of this paper. The authors have no relevant affiliations of financial involvement with any organisation or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Data availability statement
Data files pertaining to this study, including the 3 D protein model, are available at https://github.com/MarttiT/G.-salaris-BCA. Individual supplementary files in this repository are indicated as bit.ly links in the text. Further data files and code are stored at https://github.com/thirtysix/Aspatwar.Gsalaris_BCA.
Additional information
Funding
References
- Hahn C, Fromm B, Bachmann L. Comparative genomics of flatworms (platyhelminthes) reveals shared genomic features of ecto- and endoparastic neodermata. Genome Biol Evol 2014;6:1577–17.
- Zueva KJ, Lumme J, Veselov AE, et al. Genomic signatures of parasite-driven natural selection in north European Atlantic salmon (Salmo salar). Mar Genomics 2018;39:26–38.
- Hansen H, Cojocaru CD, Mo TA. Infections with Gyrodactylus spp. (Monogenea) in Romanian fish farms: Gyrodactylus salaris Malmberg, 1957 extends its range. Parasit Vectors 2016;9:444.
- Ramírez R, Bakke TA, Harris PD. Same barcode, different biology: differential patterns of infectivity, specificity and pathogenicity in two almost identical parasite strains. Int J Parasitol 2014;44:543–9.
- Hopkins C, Introduced marine organisms in Norwegian waters, including Svalbard. Parasites and diseases. In: Leppäkoski E, Gollasch S, Olenin S, editors. Invasive aquatic species of Europe. Distribution, impacts and management. Dordrecht: Springer Netherlands; 2002.
- Schmahl G. The chemotherapy of monogeneans which parasitize fish: a review. Folia Parasitol 1991;38:97–106.
- Soleng A, Poléo AB, Alstad NE, Bakke TA. Aqueous aluminium eliminates Gyrodactylus salaris (Platyhelminthes, Monogenea) infections in Atlantic salmon. Parasitology 1999;119: 19–25.
- Harder A. Chemotherapeutic approaches to trematodes (except schistosomes) and cestodes: current level of knowledge and outlook. Parasitol Res 2002;88:587–7.
- Caña-Bozada V, Chapa-López M, Díaz-Martín RD, et al. In silico identification of excretory/secretory proteins and drug targets in monogenean parasites. Infect Genet Evol 2021;93:104931.
- Supuran CT. Structure and function of carbonic anhydrases. Biochem J 2016;473:2023–32.
- Supuran CT. Emerging role of carbonic anhydrase inhibitors. Clin Sci 2021;135:1233–49.
- Mishra CB, Tiwari M, Supuran CT. Progress in the development of human carbonic anhydrase inhibitors and their pharmacological applications: where are we today? Med Res Rev 2020;40:2485–565.
- Supuran CT. Exploring the multiple binding modes of inhibitors to carbonic anhydrases for novel drug discovery. Expert Opin Drug Discov 2020;15:671–86.
- Supuran CT. Carbonic anhydrases: novel therapeutic applications for inhibitors and activators. Nat Rev Drug Discov 2008;7:168–81.
- Supuran CT. Carbonic anhydrase inhibitors as emerging agents for the treatment and imaging of hypoxic tumors. Expert Opin Investig Drugs 2018;27:963–70.
- Supuran CT. Carbonic anhydrases and metabolism. Metabolites 2018;8:25.
- Nocentini A, Angeli A, Carta F, et al. Reconsidering anion inhibitors in the general context of drug design studies of modulators of activity of the classical enzyme carbonic anhydrase. J Enzyme Inhib Med Chem 2021;36:561–80.
- Zolfaghari Emameh R, Kuuslahti M, Vullo D, et al. Ascaris lumbricoides β carbonic anhydrase: a potential target enzyme for treatment of ascariasis. Parasit Vectors 2015;8:479.
- Zolfaghari Emameh R, Barker H, Hytönen VP, et al. Beta carbonic anhydrases: novel targets for pesticides and anti-parasitic agents in agriculture and livestock husbandry. Parasit Vectors 2014;7:403.
- Zolfaghari Emameh R, Barker HR, Syrjänen L, et al. Identification and inhibition of carbonic anhydrases from nematodes. J Enzyme Inhib Med Chem 2016;31:176–84.
- Da'dara AA, Angeli A, Ferraroni M, Supuran CT, et al. Crystal structure and chemical inhibition of essential schistosome host-interactive virulence factor carbonic anhydrase SmCA. Commun Biol 2019;2:333.
- Angeli A, Pinteala M, Maier SS, et al. Sulfonamide inhibition studies of an α-carbonic anhydrase from Schistosoma mansoni, a platyhelminth parasite responsible for Schistosomiasis. Int J Mol Sci 2020;21:1842.
- Angeli A, Ferraroni M, Da'dara AA, et al. Structural insights into Schistosoma mansoni carbonic anhydrase (SmCA) inhibition by selenoureido-substituted benzenesulfonamides. J Med Chem 2021;64:10418–28.
- Ferraroni M, Angeli A, Carradori S, Supuran CT. Inhibition of Schistosoma mansoni carbonic anhydrase by the antiparasitic drug clorsulon: X-ray crystallographic and in vitro studies. Acta Crystallogr D Struct Biol 2022;78:321–7.
- Hall RA, Vullo D, Innocenti A, et al. External pH influences the transcriptional profile of the carbonic anhydrase, CAH-4b in Caenorhabditis elegans. Mol Biochem Parasitol 2008;161:140–9.
- Crocetti L, Maresca A, Temperini C, et al. A thiabendazole sulfonamide shows potent inhibitory activity against mammalian and nematode alpha-carbonic anhydrases. Bioorg Med Chem Lett 2009;19:1371–5.
- Güzel O, Innocenti A, Hall RA, et al. Carbonic anhydrase inhibitors. The nematode alpha-carbonic anhydrase of Caenorhabditis elegans CAH-4b is highly inhibited by 2-(hydrazinocarbonyl)-3-substituted-phenyl-1H-indole-5-sulfonamides. Bioorg Med Chem 2009;17:3212–5.
- Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool. J Mol Biol 1990;215:403–10.
- Määttä JA, Eisenberg-Domovich Y, Nordlund HR, et al. Chimeric avidin shows stability against harsh chemical conditions-biochemical analysis and 3D structure. Biotechnol Bioeng 2011;108:481–90.
- Khalifah RG. The carbon dioxide hydration activity of carbonic anhydrase. I. Stop-flow kinetic studies on the native human isoenzymes B and C. J Biol Chem 1971;246:2561–73.
- Angeli A, Pinteala M, Maier SS, et al. Inhibition of α-, β-, γ-, δ-, ζ- and η-class carbonic anhydrases from bacteria, fungi, algae, diatoms and protozoans with famotidine. J Enzyme Inhib Med Chem 2019;34:644–50.
- Urbański LJ, Di Fiore A, Azizi L, et al. Biochemical and structural characterisation of a protozoan beta-carbonic anhydrase from Trichomonas vaginalis. J Enzyme Inhib Med Chem 2020;35:1292–9.
- Petreni A, De Luca V, Scaloni A, et al. Anion inhibition studies of the Zn(II)-bound ι-carbonic anhydrase from the Gram-negative bacterium Burkholderia territorii. J Enzyme Inhib Med Chem 2021;36:372–6.
- Winum JY, Temperini C, El Cheikh K, et al. Carbonic anhydrase inhibitors: clash with Ala65 as a means for designing inhibitors with low affinity for the ubiquitous isozyme II, exemplified by the crystal structure of the topiramate sulfamide analogue. J Med Chem 2006;49:7024–31.
- Pastorekova S, Casini A, Scozzafava A, et al. Carbonic anhydrase inhibitors: the first selective, membrane-impermeant inhibitors targeting the tumor-associated isozyme IX. Bioorg Med Chem Lett 2004;14:869–73.
- Bonardi A, Nocentini A, Bua S, et al. Sulfonamide inhibitors of human carbonic anhydrases designed through a three-tails approach: improving ligand/isoform matching and selectivity of action. J Med Chem 2020;63:7422–44.
- Briganti F, Pierattelli R, Scozzafava A, Supuran CT. Carbonic anhydrase inhibitors. Part 37. Novel classes of carbonic anhydrase inhibitors and their interaction with the native and cobalt-substituted enzyme: kinetic and spectroscopic investigations. Eur J Med Chem 1996;31:1001–10.
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010;26:2460–1.
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 2004;5:113.
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 2000;17:540–52.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, et al. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 2017;14:587–9.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 2015;32:268–74.
- Huerta-Cepas J, Serra F, Bork P. ETE 3: reconstruction, analysis, and visualization of phylogenomic data. Mol Biol Evol 2016;33:1635–8.
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 2001;305:567–80.
- Almagro Armenteros JJ, Salvatore M, Emanuelsson O. Detecting sequence signals in targeting peptides using deep learning. Life Sci Alliance 2019;2:e201900429.
- Almagro Armenteros JJ, Sønderby CK, Sønderby SK, et al. DeepLoc: prediction of protein subcellular localization using deep learning. Bioinformatics 2017;33:3387–95.
- Sievers F, Wilm A, Dineen D, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 2011;7:539.
- Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 2014;42:W320–324.
- Pettersen EF, Goddard TD, Huang CC, et al. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci 2021;30:70–82.
- Jumper J, Evans R, Pritzel A, et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021;596:583–9.
- Kimber MS, Pai EF. The active site architecture of Pisum sativum beta-carbonic anhydrase is a mirror image of that of alpha-carbonic anhydrases. Embo j 2000;19:1407–18.
- Syrjanen L, Tolvanen M, Hilvo M, et al. Characterization of the first beta-class carbonic anhydrase from an arthropod (Drosophila melanogaster) and phylogenetic analysis of beta-class carbonic anhydrases in invertebrates. BMC Biochem 2010;11:28.
- Zolfaghari Emameh R, Barker H, Tolvanen ME, et al. Bioinformatic analysis of beta carbonic anhydrase sequences from protozoans and metazoans. Parasit Vectors 2014;7:38.
- Schlicker C, Hall RA, Vullo D, et al. Structure and inhibition of the CO2-sensing carbonic anhydrase Can2 from the pathogenic fungus Cryptococcus neoformans. J Mol Biol 2009;385:1207–20.
- Innocenti A, Muhlschlegel FA, Hall RA, et al. Carbonic anhydrase inhibitors: inhibition of the beta-class enzymes from the fungal pathogens Candida albicans and Cryptococcus neoformans with simple anions. Bioorg Med Chem Lett 2008;18:5066–70.
- Innocenti A, Hall RA, Schlicker C, et al. Carbonic anhydrase inhibitors. Inhibition of the beta-class enzymes from the fungal pathogens Candida albicans and Cryptococcus neoformans with aliphatic and aromatic carboxylates. Bioorg Med Chem 2009;17:2654–7.
- Innocenti A, Hall RA, Schlicker C, et al. Carbonic anhydrase inhibitors. Inhibition and homology modeling studies of the fungal beta-carbonic anhydrase from Candida albicans with sulfonamides. Bioorg Med Chem 2009;17:4503–9.
- Innocenti A, Hall RA, Scozzafava A, et al. Carbonic anhydrase activators: activation of the beta-carbonic anhydrases from the pathogenic fungi Candida albicans and Cryptococcus neoformans with amines and amino acids. Bioorg Med Chem 2010;18:1034–7.
- Isik S, Guler OO, Kockar F, et al. Saccharomyces cerevisiae β-carbonic anhydrase: inhibition and activation studies. Curr Pharm Des 2010;16:3327–36.
- Isik S, Kockar F, Aydin M, et al. Carbonic anhydrase inhibitors: inhibition of the beta-class enzyme from the yeast Saccharomyces cerevisiae with sulfonamides and sulfamates. Bioorg Med Chem 2009;17:1158–63.
- Innocenti A, Zimmerman S, Ferry JG, et al. Carbonic anhydrase inhibitors. Inhibition of the beta-class enzyme from the methanoarchaeon Methanobacterium thermoautotrophicum (Cab) with anions. Bioorg Med Chem Lett 2004;14:4563–7.
- Zimmerman SA, Ferry JG, Supuran CT. Inhibition of the archaeal beta-class (Cab) and gamma-class (Cam) carbonic anhydrases. Curr Top Med Chem 2007;7:901–8.
- Del Prete S, De Luca V, Nocentini A, et al. Anion inhibition studies of the beta-carbonic anhydrase from Escherichia coli. Molecules 2020;25:2564.
- Del Prete S, De Luca V, Bua S, et al. The effect of substituted benzene-sulfonamides and clinically licensed drugs on the catalytic activity of CynT2, a carbonic anhydrase crucial for Escherichia coli life cycle. Int J Mol Sci 2020;21:4175.
- Nocentini A, Del Prete S, Mastrolorenzo MD, et al. Activation studies of the β-carbonic anhydrases from Escherichia coli with amino acids and amines. J Enzyme Inhib Med Chem 2020;35:1379–86.
- Supuran CT, Capasso C. A highlight on the inhibition of fungal carbonic anhydrases as drug targets for the antifungal armamentarium. Int J Mol Sci 2021;22:4324.
- Campestre C, De Luca V, Carradori S, et al. Carbonic anhydrases: new perspectives on protein functional role and inhibition in Helicobacter pylori. Front Microbiol 2021;12:629163.
- Grande R, Carradori S, Puca V, et al. Selective inhibition of helicobacter pylori carbonic anhydrases by carvacrol and thymol could impair biofilm production and the release of outer membrane vesicles. Int J Mol Sci 2021;22:11583.
- De Simone G, Supuran CT. (In)organic anions as carbonic anhydrase inhibitors. J Inorg Biochem 2012;111:117–29.
- Supuran CT. Carbon- versus sulphur-based zinc binding groups for carbonic anhydrase inhibitors? J Enzyme Inhib Med Chem 2018;33:485–95.
- Supuran CT. How many carbonic anhydrase inhibition mechanisms exist? J Enzyme Inhib Med Chem 2016;31:345–60.
- Abbate F, Supuran CT, Scozzafava A, et al. Nonaromatic sulfonamide group as an ideal anchor for potent human carbonic anhydrase inhibitors: role of hydrogen-bonding networks in ligand binding and drug design. J Med Chem 2002;45:3583–7.
- Aspatwar A, Hammaren M, Koskinen S, et al. β-CA-specific inhibitor dithiocarbamate Fc14-584B: a novel antimycobacterial agent with potential to treat drug-resistant tuberculosis. J Enzyme Inhib Med Chem 2017;32:832–40.
- Carta F, Aggarwal M, Maresca A, et al. Dithiocarbamates strongly inhibit carbonic anhydrases and show antiglaucoma action in vivo. J Med Chem 2012;55:1721–30.
- Carta F, Aggarwal M, Maresca A, et al. Dithiocarbamates: a new class of carbonic anhydrase inhibitors. Crystallographic and kinetic investigations. Chem Commun (Camb) 2012;48:1868–70.