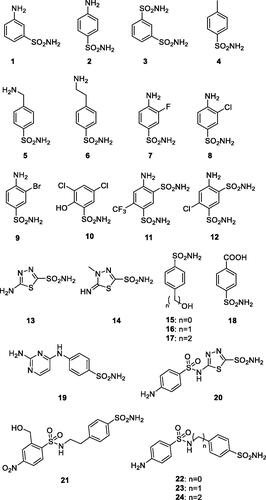
Abstract
Schistosoma mansoni is an intestinal parasite with one β-class carbonic anhydrase, SmaBCA. We report the sequence enhancing, production, catalytic activity, and inhibition results of the recombinant SmaBCA. It showed significant catalytic activity on CO2 hydration in vitro with kcat 1.38 × 105 s−1 and kcat/Km 2.33 × 107 M−1 s−1. Several sulphonamide inhibitors, from which many are clinically used, showed submicromolar or nanomolar inhibitory effects on SmaBCA. The most efficient inhibitor with a KI of 43.8 nM was 4-(2-amino-pyrimidine-4-yl)-benzenesulfonamide. Other effective inhibitors with KIs in the range of 79.4–95.9 nM were benzolamide, brinzolamide, topiramate, dorzolamide, saccharin, epacadostat, celecoxib, and famotidine. The other tested compounds showed at least micromolar range inhibition against SmaBCA. Our results introduce SmaBCA as a novel target for drug development against schistosomiasis, a highly prevalent parasitic disease.
Introduction
Schistosoma mansoni is a parasitic blood fluke affecting ∼200 million people worldwide.Citation1,Citation2 It causes schistosomiasis and has been rated as the second most harmful parasite in the world; only malaria has been stated to cause more mortality.Citation3,Citation4 Schistosoma mansoni is endemic in Africa, South America, the Caribbean, and the Middle East.Citation5 Schistosomiasis is an intestinal infection that produces acute symptoms of diarrhoea,Citation6 abdominal pain,Citation6 and fever.Citation7
Patients are afflicted with S. mansoni as the larvae living in freshwater penetrate through healthy skin and travel via the bloodstream to the recipient’s liver.Citation8 The larvae grow up and reach mesenteric veins to lay eggs.Citation9 Some of the larvae reside in the liver arteriesCitation10 and can live for decades, causing granulomatous infection to the wall of the intestines as the eggs invade the gut wall.Citation2,Citation5 Chronic granulomatous infection occurs in the liver, serving as an incubator for developing larvae and trapping some of the eggs,Citation1 and can lead to irreversible hepatic cirrhosis, especially if the affected person has chronic viral hepatitis, relatively common in the endemic areas of S. mansoni.Citation11 The chronic infection can be cured only via medical intervention as the host’s immune system is unable to overcome the parasite.Citation10
There is no vaccine against schistosomiasisCitation12,Citation13 and the only currently known effective treatment against species of Schistosoma-genus parasitesCitation14 is executed with praziquantel.Citation10 It is used as a preventive medicine and for treating patients,Citation15 but millions of people still get infected yearly.Citation1 Currently, the need for preventive chemotherapy is over two times higher than the number of individuals receiving the required medication (). The pervasive use of praziquantel for schistosomiasis has led to rising resistance against the drug.Citation16,Citation17 Therefore, potential medicines with novel modes of action are urgently needed to fight this devastating disease.
Figure 1. The worldwide requirement of preventive medication compared to the realised medication according to WHO [https://apps.who.int/neglected_diseases/ntddata/sch/sch.html, figure visualisation made by Susanna Haapanen using GraphPad Prism (1992–2020 GraphPad Software, LLC, version 9.0.0)].
![Figure 1. The worldwide requirement of preventive medication compared to the realised medication according to WHO [https://apps.who.int/neglected_diseases/ntddata/sch/sch.html, figure visualisation made by Susanna Haapanen using GraphPad Prism (1992–2020 GraphPad Software, LLC, version 9.0.0)].](/cms/asset/17027dc8-f826-4295-80c6-dfa623c78d8e/ienz_a_2184299_f0001_c.jpg)
In developing new drugs against parasitic diseases, carbonic anhydrases (CAs, EC 4.2.1.1) have recently emerged as potential targets.Citation18–20 CAs catalyse the reversible hydration of CO2, maintaining homeostasis in tissues and fluids of the body. Eight distinct families of CAs have been presented in classifications based on protein structures and the active site. Some of the families have evolved as a result of long divergent evolution, but there are at least four independent ancestors.Citation21–23 The α- and η-CAs share a common protein fold but they contain different catalytic residues in the active sites.Citation22 The group of β-like-CAs includes ε-, θ-, and ζ-CAs (M. Tolvanen, unpublished observations). For γ-CAs and ι-CAs, no structural or evolutionary relatedness to other classes has been reported. On the contrary, δ-CAs show structural resemblance to α-CAs, although they are considered a family of their own.Citation24,Citation25 The earliest discovered enzyme forms are α-CAs found in humans, other animals, and many other pro- and eukaryotic cells.Citation26 β-CAs are not found in vertebrates but are present in most prokaryotes, archaea, plants, fungi, and non-chordate metazoa.Citation26,Citation27 γ-CAs have been identified in some prokaryotes, fungi, plants, and single-cell eukaryotic organisms, such as amoebas.Citation27,Citation28 δ-CAs are widely distributed in marine phytoplankton.Citation29 ζ-CAs are mainly present in diatoms,Citation30 and η-CAs have been discovered in Plasmodium-parasites and in Toxoplasma gondii, thus far.Citation22,Citation31 θ-CAs have been found in marine diatoms, green algae, and bacteria,Citation32 and the most recently found ι-CAs are widely found in bacteria, diatoms, green algae, and cyanobacteria.Citation23,Citation33 S. mansoni has only one β-CA but six α-CAs,Citation34 one of which has been previously produced as a recombinant protein and tested for inhibition properties using various sulphonamide and anion class inhibitors.Citation35 For instance, clorsulon, an antiparasitic sulphonamide, has a good inhibitory activity in vitro.Citation36 This study is focussed on the S. mansoni β-CA, for which we use the abbreviation SmaBCA.
CA inhibitors of the sulphonamide class are already in use as drugs to treat clinical conditions, such as glaucoma,Citation37,Citation38 brain oedema,Citation39 and mountain sickness.Citation40,Citation41 Ongoing research of new drug candidates for targeting different CA families aims at novel treatment of various diseases, such as cancer,Citation38,Citation42 neuropathic pain,Citation43 migraineCitation43 as well as infectious diseases.Citation42,Citation44 The clinically used inhibitors affect more than one CA isoformCitation45 and can contribute to significant adverse side effects.Citation46 The human genome has only α-CAs, and the absence of β-CAs provides an excellent opportunity to design and generate more specific anti-parasitic drugs, explicitly targeting the β-CAs. This study aimed to produce and characterise recombinant SmaBCA and to test the efficacy of selected sulphonamide- and anion-type CA inhibitors against this enzyme. The results could help to determine the best inhibitor candidates for further development as lead compounds of antiparasitic drugs.
Materials and methods
Sequence retrieval
At the start of this study, we identified S. mansoni β-CA sequences by Blast searches at NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and UniProt (https://www.uniprot.org/blast), using β-CA from Drosophila melanogasterCitation47 as a query sequence, finding XP_018647616.1 at NCBI and G4V6B2 at UniProt, identical sequences of 241 aa. When a DNA sequence coding for this sequence could not be produced, we looked at the original sequence more carefully.
Further BlastP searches were carried out against NCBI RefSeq proteins with the S. mansoni β-CA XP_018647616.1 as the query sequence, limiting the search to Platyhelminthes on May 14th, 2018. Selected hits were aligned using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/).Citation48,Citation49 An N-terminally incomplete S. haematobium protein XP_012795040.1 matched well with S. mansoni XP_018647616.1 but had a longer C-terminus, which resembles the C-termini of β-CA sequences of other parasites. This prompted us to revisit the gene model of XP_018647616.1 to see if a sequence coding for a similar C-terminal extension could also be found in the S. mansoni genome. Using the S. haematobium β-CA protein as a query sequence in a Blast search at metazoa.ensembl.org, we identified a sequence coding for a full-length SmBCA, which was used in the successful production of an active enzyme.
Multiple sequence alignments (MSA)
In order to visualise sequence conservation in metazoan β-CAs for this article, we collected a new sequence set on 30 March 2022. S. japonicum KAH8855123.1 was used as a query sequence in BLAST at NCBI, with substitution matrix BLOSUM45, result list size of 5000, and taxonomy filter set to Metazoa. This resulted in 643 hits, filtered (with NCBI Blast result tools) for 85 % query coverage, leaving 520 sequences (https://github.com/MarttiT/S.-mansoni-BCA/blob/main/SmaBCA520%20blast%20hits%20Descriptions.xlsx). Incidentally, at this point, the RefSeq version of S. mansoni β-CA (XP_018647616.1) was too short to pass the coverage filter. The longest 35 sequences (of 998–2153 aa, all from rotifers) were removed. The remaining 485 sequences were aligned preliminarily using the Clustal OmegaCitation48,Citation49 within SeaView 5.0.4.,Citation50 and sequences with unique insertions or clearly mismatching sequences within conserved regions were manually deleted (using SeaView). The remaining 390 sequences were realigned with SeaView, and the resulting MSA (https://github.com/MarttiT/S.-mansoni-BCA/blob/main/SeaView%20MSA%20390.aln) was filtered for sequences with more than 90 % identity against any other sequence in the set. The Decrease redundancy tool at https://web.expasy.org/decrease_redundancy/ was used for this purpose (Cédric Notredame, unpublished), and a final realignment was performed on the obtained set of 162 protein sequences with Clustal Omega at EBI (https://www.ebi.ac.uk/Tools/msa/clustalo/)Citation49 (Supplementary File 1).
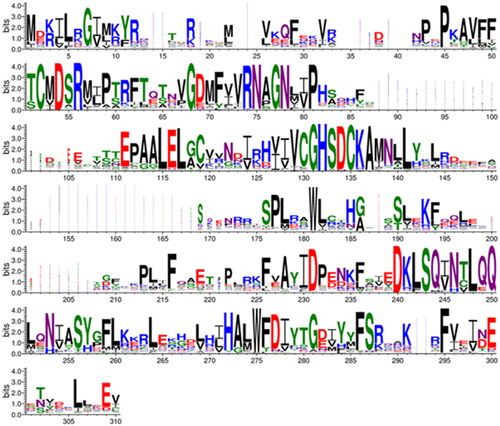
A sequence logo was constructed to visualise this MSA using Berkeley WebLogo version 3.7.12 (https://weblogo.threeplusone.com/)Citation51 with the parameters of 1st pos −112, logo from 1 to 310, no adjustment for composition, no error bars, colour scheme chemistry ().
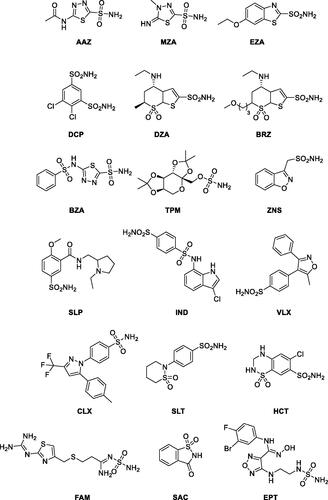
To compare schistosomal β-CA sequences (), we prepared a Clustal Omega alignment of the SmaBCA sequence of this study with S. margrebowiei VDO63334.1, S. haematobium KAH9593836.1, and S. japonicum TNN15731.1. This alignment was visualised using ESPript 3.0 at https://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi.Citation52 A global score cut-off of 0.75 used for colouring the blocks and secondary structure assignments for the top line were taken from the S. mansoni β-CA model retrieved from the AlphaFold protein structure database, https://alphafold.ebi.ac.uk/entry/A0A3Q0KBP5.Citation53,Citation54
Figure 2. Comparison of the β-CA protein sequence of this study (S_mansoni_new) by sequence alignment to homologs from three other Schistosoma species. The conserved residues of the catalytic-site motifs, CXDXR and HXXC, are indicated with black triangles (C: Cys, D: Asp, H: His, R: Arg, X: any residue). Columns with fully conserved residues are shown as red with white letters. Boxed columns denote positions in which at least 75 % of residues are of a similar type, consisting of a total of 244 aa (91.7 %), of which 175 aa are totally conserved (65.8 %). The top line indicates the secondary structures of the AlphaFold model for S. mansoni β-CA. α: α-helices; β: β-strands; η: 310-helices; T: turns.
Dimer modelling
The monomer model retrieved from the AlphaFold protein structure database has a visual resemblance to the pea β-CA model (PDB 1ekj) in that the N-terminal alpha helices are detached from the catalytic domain of the monomer structure. In the full octamer model of 1ekj, we can see that these alpha helices wrap around another monomer to create the basic dimer unit, which is repeated as four copies. This inspired us to create a 3D dimer model for SmaBCA using ChimeraX (daily build 1.4.dev202202030703), developed by the UCSF Resource for Biocomputing, Visualisation, and Informatics (San Francisco, California, USA), supported in part by the National Institutes of Health. The AlphaFold multimer modelling interfaceCitation55 was used to submit the prediction to run at Google Colab, giving two copies of our SmaBCA sequence as input, on 6 April 2022. The dimer model is available at https://github.com/MarttiT/S.-mansoni-BCA/blob/main/SmaBCA%20dimer.pdb.
Vector construction, protein production, and purification
The same production procedure was followed both for the unsuccessful production of the protein corresponding to UniProt G4V6B2 and our amended sequence with a full-length last exon. The amended sequence contains the modifications to C-terminus as described above as well as the silent mutations to prevent Rho-independent termination site forming, on the contrary, to the unsuccessful production of the protein to which the sequence was unmodified.
Since some coding genes may contain termination codes in the middle of mRNA coding sequences and lead to early transcription termination and the consequent production of immature mRNA, a prediction approach was performed using ARNold (http://rssf.i2bc.paris-saclay.fr/toolbox/arnold/) web toolCitation56 to find Rho-independent transcription termination sites on the SmaBCA gene (NCBI gene ID: 8342150). To prevent the formation of immature mRNA for SmaBCA, five silent mutations were introduced to change the nucleotides of Rho-independent termination sites to other nucleotides.
The finalised sequence of the insert was sent to GeneArt (Invitrogen, Regensburg, Germany), where a modified plasmid vector, pBVboost, was constructed.Citation57
The plasmid vector was prepared according to the manufacturer’s instructions and then transformed with heat shock into BL21 StarTM (DE3) cells (Invitrogen, Carlsbad, USA), as described previously.Citation58 The production of recombinant protein was executed manually in LB broth (Sigma-Aldrich, St. Louis, MO, USA) with 1:1000 gentamicin (Sigma-Aldrich) and 1:100 glycerol (VWR International, Radnor, PA, USA)/glucose (Sigma-Aldrich) as proposed in Kopp et al.Citation59 at +37 degrees and shaking with 200 rpm. Both glycerol and glucose proved to be equally effective additives in the growth medium to reduce the number of impurities. The OD (Fisher Scientific Colorimeter Model 45 (WA12173), Fisherbrand, Thermo Fisher Scientific, Waltham, MA, USA) was measured and at the OD 0.4–0.5 1 M isopropyl-β-D-thiogalactopyranoside (IPGT, Thermo Fisher Scientific) was added in relation of 1:1000 to LB medium. Growth continued overnight and was terminated the next day by pelleting the cells by centrifugation at 5000 × g for 20 min resulting in a total incubation and production time of 24 h.
A few alterations were made to the purification protocol compared to Haapanen et al.Citation58 We used 50 mM Na2HPO4 (Sigma-Aldrich), 0.5 M NaCl (VWR International), and 50 mM imidazole (Sigma-Aldrich) (pH 8.0) as a binding buffer and washed the nickel resin (HisPurTM Ni-NTA Resin, Thermo Fisher Scientific) with 15 × 25 ml of 50 mM Na2HPO4, 0.5 M NaCl, 50 mM imidazole, and 20 % glycerol (pH 8.0) with EMD Millipore™ vacuum filtering flask (Merck, Kenilworth, NJ, USA) and filter paper (pore size 2 µm, Whatman™, Maidstone, UK). The glycerol was washed off with 10 × 25 ml of binding buffer with the same vacuum filtering flask. Subsequently, the resin was collected into an empty column (Maxi Columns, G-Biosciences, St. Louis, MO, USA). The protein was eluted from the resin with elution buffer (50 mM Na2HPO4, 0.5 M NaCl, and 350 mM imidazole, pH 8.5). Re-purification was performed twice, similarly to the initial purification, except using 50 mM Na2HPO4, 0.5 M NaCl (pH 8.0) as a binding buffer to make the imidazole concentration under 20 mM. The purity of the collected fractions was verified with 12 % SDS-PAGE (sodium dodecyl sulfate-polyacrylamide gel electrophoresis) under reducing conditions using Precision Plus ProteinTM Standards Dual Colour (Bio-Rad Laboratories, Inc., Hercules, CA, USA) as a standard molecular weight (MW) marker. The MW marker and bands were visualised with PageBlue™ Protein Staining Solution (Thermo Fisher Scientific) on the gel by letting the gel stain for 30 min and then washing with distilled water. The bands were confirmed to be SmaBCA with mass spectrometry (data not shown).
CA enzyme activity and inhibition
Before activity measurements, the buffer was changed into 50 mM TRIS (VWR International) (pH 8.5) with 10 kDa Vivaspin® Turbo 15 centrifugal concentrators (Sartorius™, Göttingen, Germany) at 4000 × g at +4°C. The CA-catalysed CO2 hydration activity was measured using an Applied Photophysics stopped-flow instrument.Citation60 The measurement protocol was identical to the previously described in Berrino et al.Citation61
Results
To produce the recombinant protein of SmaBCA, an insert sequence was retrieved from the National Centre of Biotechnology Information (NCBI Reference Sequence: XM_018793067.1). It was included in our construct because it was the only β-CA of S. mansoni available in sequence databases in 2018 (also represented in UniProt protein entry G4V6B2). We did not obtain any protein from recombinant production using this original sequence. Upon closer inspection, the sequence seemed to have an incorrectly predicted splice site in the actual last exon, joined to a superfluous exon, coding only for three residues instead of extending the last exon up to a stop codon. The full-length sequence for the last exon was discovered in the S. mansoni genome by searching with other schistosomal β-CA sequences. The amended last exon coded for 28 additional amino acids (aa), highly similar to the search query sequences. This sequence also led to the successful production of the active enzyme described in this study. A full-length sequence of SmaBCA was added in UniProt after we had already produced our protein (A0A3Q0KBP5, 13 February 2019). This sequence is probably derived from a later genome version than ours (which came from S. mansoni genome assembly ASM23792v2 as of 14 May 2018 at metazoa.ensembl.org), and it differs by one amino acid substitution (N264D) from our sequence. This is a variable site even within Schistosoma species ().
Our analysis also revealed that the SmaBCA gene sequence contains a Rho-independent transcription termination site, a GC-rich area in the sequence causing loop formation and detachment of RNA polymerase, which introduces an early termination of transcription in bacteria and consequently a non-functional messenger RNA (mRNA) (). To enable the recombinant production in Escherichia coli, we chose to create single nucleotide mutations in the coding sequence to disrupt base pairs in the stem (blue in ) without any effect on the translated amino acid sequence.
Figure 3. The coding sequence of the β-CA gene from S. mansoni. The predicted Rho-independent transcription termination site is marked in underlined blue and red text in the sequence, and the corresponding Rho-independent stem-loop is shown on the right.

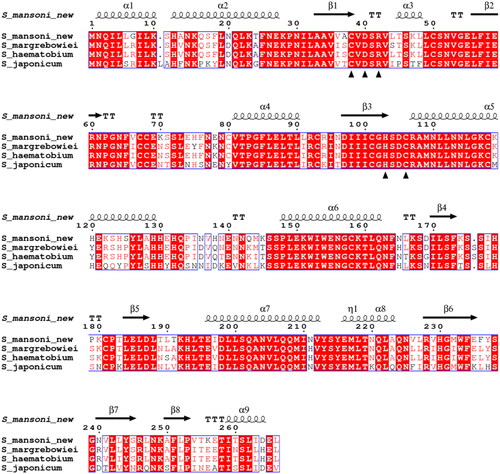
As a result of recombinant protein production, we obtained six protein bands of ∼13, 22, 25, 29, 38, and 75 kDa in size (). All these bands represented different forms of SmaBCA as confirmed by mass spectrometry (data not shown). The calculated molecular weight of the translated coding sequence is 30.4 kDa, not accounting for any post-translational modifications.
Figure 4. SDS-PAGE of β-CA of S. mansoni (SmaBCA) showing six polypeptide bands confirmed to represent SmaBCA by mass-spectrometry (data not shown). Right lane: The most intense bands of 25 and 29 kDa are the main forms of the expressed protein, and the additional polypeptides either are degraded forms (13 and 22 kDa) or oligomers (38 and 75 kDa). Left lane: Standard molecular weight (MW) markers in kDa.
Kinetic analysis of SmaBCA showed that the enzyme (with His-tag) is moderately active with kcat 1.38 × 105 s−1, Km 5.92 mM, and kcat/Km 2.33 × 107 M−1 s−1. Several sulphonamide and anion inhibitorsCitation46,Citation62,Citation63 were tested to evaluate their inhibitory properties against SmaBCA. The most efficient inhibitors showed submicromolar or nanomolar inhibitory effects on SmaBCA ( and ). The most efficient inhibitor with a KI of 43.8 nM was 4-(2-amino-pyrimidine-4-yl)-benzenesulfonamide (compound 19). Other effective inhibitors included several clinically used drugs. Benzolamide (BZA), brinzolamide (BRZ), topiramate (TPM), dorzolamide (DZA), saccharin (SAC), epacadostat (EPT), celecoxib (CLX), and famotidine (FAM) showed KIs in the range of 79.4–95.9 nM. The other tested compounds inhibited SmaBCA at micromolar or millimolar concentrations.
Figure 5. Chemical structures of sulphonamide (1–24) and sulphonamide/sulfamate derivatives (AAZ-EPT) tested as inhibitors against β-CA of Schistosoma mansoni (SmaBCA) in this study.
Table 1. Inhibition data for SmaBCA with sulphonamide analogs 1–24, clinically used compounds, and anions.
Our protein sequence was aligned with β-CA sequences of other Schistosoma species, as shown in . All these sequences are highly similar, with identities to the protein of this study ranging from 75.6 to 86.8 %. The identity percentages are even higher in the protein core and in the active site, which suggests that any drugs developed to inhibit SmaBCA would also be inhibitory for β-CAs of other Schistosoma species.
The sequence logo of extracts information from a multiple sequence alignment (MSA) of 162 metazoan β-CA sequences showing 19 perfectly conserved aa sites and 26 almost conserved sites. Of course, we find highly conserved areas at the motifs, which are part of the active site (CXDXR and HXXC), and additionally, at columns 68–81 and 240–250. The gap region at columns 151–168 in the MSA () is due to an insertion seen only in Schistosoma sequences of our sequence set. The entire MSA is provided as Supplementary File S1. More data is available as noted in the Experimental procedures.
Figure 6. Comparison of metazoan β-CA proteins. This sequence logo represents an MSA of 162 sequences, cropped at both ends to show only the extent of the SmaBCA sequence. The height of each stack of letters represents the conservation (information content) of each column in the alignment. The width of the letters represents the number of non-gap characters in each column (i.e. very narrow letters indicate positions in which only few of the aligned proteins have any sequence).
We built a computational 3D model of the SmaBCA dimer using AlphaFold multimer. In this model, the alpha-helical segments in the N-terminus are positioned along the side of the other monomer, similar to the dimers of Pisum sativum (pea) β-CA (PDB 1ekj).Citation64 These segments are also present in the AlphaFold monomer model (https://alphafold.ebi.ac.uk/entry/A0A3Q0KBP5), in which their position away from the core of the protein looks odd, but in the dimer model () it makes perfect sense. The orientation between the monomers in our model is nearly identical to pea β-CA dimers (1ekj, e.g. chains C and D, data not shown).
Figure 7. Molecular model of a hypothetical SmBCA dimer, constructed using AlphaFold multimer. Colouring is shown based on the per-residue confidence score (pLDDT), with two different palettes, as shown in the colour keys at the bottom left and right, for the bottom and top monomer, respectively. Metal-binding residues at the catalytic site (Cys 38, His 103, and Cys 106) are shown in balls-and-sticks style. The zinc ion at the catalytic site is displayed in pink.
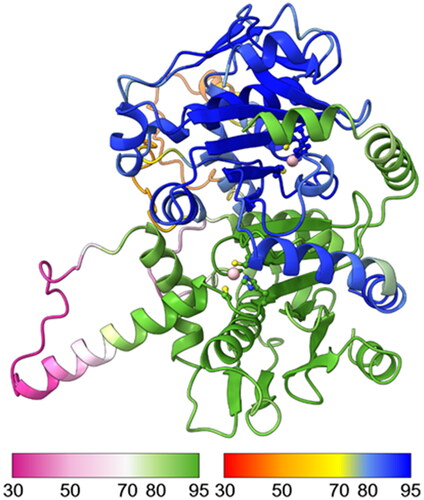
AlphaFold gives a confidence score (predicted local distance difference test, pLDDT) to the position of each residue in the model, pLDDT >90 indicating “very high confidence” and pLDDT >70 “confident”. In our model, two regions (69–79 and 119–145) have pLDDT values <70. They correspond to regions of low conservation and insertions of variable lengths in our MSA of metazoan β-CA sequences (columns 83–108 and 148–174 in ). These two regions are also slightly different between the AlphaFold database monomer model and our dimer model (data not shown).
Discussion
Schistosomiasis causes high morbidity in tropical and subtropical countries, and despite the treatment with praziquantel,Citation65 the infection remains a significant health problem.Citation66 Unfortunately, there are already initial signs of developing resistance against praziquantelCitation67 which emphasises the need for novel medication. In this study, we cloned the SmaBCA and found many already clinically used CA inhibitors with significant inhibitory effects against SmaBCA.
SmaBCA has a similar distribution of bands in SDS-PAGE as β-CAs from Entamoeba histolytica (EhiCA)Citation58 and Trichomonas vaginalisCitation57 before His-tag removal: they all contain the major dual-band at a size which is predicted according to the aa chain composition. However, the dual-band formation disappears from β-CAs of T. vaginalis as the His-tag is removed, contrary to EhiCA, which retains the double band appearance after the cleavage of His-tag.
SmaBCA shows enzyme activity with kcat 1.38 × 105 s−1 and kcat/Km 2.33 × 107 M−1 s−1, within a similar range compared with the β-CAs from T. vaginalis (kcat 4.9 × 105 and kcat/Km 8.0 × 107),Citation57 Leishmania donovani chagasi (kcat 9.35 × 105 s−1 and kcat/Km 5.9 × 107 s−1M−1),Citation68 Ascaris lumbricoides (kcat 6.0 × 105 s−1, kcat/Km 4.3 × 107 M−1 s−1),Citation69 and EhiCA (kcat of 6.7 × 105 s−1 and a kcat/Km of 8.9 × 107 M−1 × s−1),Citation58 demonstrating the possibly crucial role in the metabolism of the organisms, as the magnitude of activity is considered from moderate (SmaBCA) to high (β-CAs from T. vaginalis, Leishmania donovani chagasi, Ascaris lumbricoides, and EhiCA) compared to human CA I (moderate, kcat 2.0 × 105 and kcat/Km 5.0 × 107), for instance.
Schistosomiasis and amoebiasis (intestinal infection caused by E. histolytica) are endemic in the same areas of the worldCitation70 and coinfections are not unusualCitation71,,Citation72 leading to the conclusion of achievable benefits from treating the infections with only one drug: less adverse side effects for the patient, better treatment compliance, and lower costs for the society. SmaBCA and EhiCA are both well-inhibited with many anion and sulphonamide derivatives. The most effective ones for EhiCA are 4-hydroxymethyl/ethyl-benzenesulfonamides (compounds 16 and 17) with KI values of 89 and 36 nM, respectively,Citation63 with good inhibition activity against SmaBCA with KIs of 600 and 346 nM, respectively. They are weak in inhibiting human CA II (KI of 125 nM) and almost inefficient inhibitors of human CA I (KI of 21 µM),Citation63 indicating a slight parasite selectivity. This kind of inhibitors could potentially have minimal side effects on humans. Other nanomolar range inhibitors are 4-(2-aminoethyl)benzenesulfonamide (compound 6), 4-((2-amino-4-pyrimidinyl)amino)benzenesulfonamide (compound 19), and acetazolamide (AAZ) with KIs of 509–798 nM for EhiCA and KIs of 44–286 nM for SmaBCA, from which acetazolamide is already in clinical use.Citation73–76 With low micromolar range are sulfamide and phenylarsonic acid with KIs of 28–38 and 8–20 µM, for EhiCA and SmaBCA, respectively. The many agents with good inhibitory activity demonstrate the possibilities for developing CA inhibitors as anti-parasitic drugs against both enzymes of these parasites.
Aa sequences of SmaBCA and β-CAs of other Schistosoma species are highly similar, as shown by MSA (). In particular, the essential parts (the protein core and the active site) of the β-CA sequences are highly conserved, hence, opening an exciting opportunity to find functioning CA inhibitor-based drugs effective against all the species as praziquantel nowadays is. To our knowledge, only one other Schistosoma species, S. japonicum, has had its only β-CA produced as a recombinant protein previously.Citation77 Cong-Hui et al. produced 38 kDa recombinant protein with CA activity, but they did not make any comparison to β-CAs of other Schistosoma species. S. mansoni and S. japonicum are genetically distinct as they were separated as their own phylogenetic branches ∼14 million years ago,Citation78 and they are endemic in different parts of the world: S. japonicum in South-East Asia and S. mansoni mainly in Africa, South America, the Caribbean, and Middle East, both causing similar intestinal infections.Citation1,Citation5 A new universal anti-schistosomal agent could have clinical value in very large areas covering most of the globe.
The β-CA of S. mansoni is a promising target for the development of new anti-schistosomal drugs. In this study, we produced a novel SmaBCA recombinant protein, tested it against different CA inhibitors, leading to the discovery of several well-inhibiting compounds, from which some are already used in treating other conditions. Based on structural and sequence analyses, we also propose that it is feasible to develop one universally functional anti-parasitic drug against several Schistosoma species which could also be effective against other parasites, such as Entamoeba histolytica.
Author contributions
SH, AA, SP, and CTS designed the experiments. SH and AA performed the experiments in the laboratory. MT and RZE planned the modifications to the sequence. MT planned and prepared the bioinformatic analyses. SH drafted this manuscript. SP, MT, and CTS reviewed the drafts and modified the manuscript. All authors read and approved the final manuscript.
Supplemental Material
Download PDF (164 KB)Acknowledgements
We thank Marianne Kuuslahti and Sanna Kavén for their skillful technical assistance; Juha Määttä, PhD, and Niklas Kähkönen for the help with protein purification and Maarit Patrikainen, PhD, for the help with refining the article text. We acknowledge the Tampere Facility of Protein Services (PS) for their service.
Disclosure statement
No potential conflict of interest was reported by all authors except Claudiu T. Supuran. C. T. Supuran is Editor-in-Chief of the Journal of Enzyme Inhibition and Medicinal Chemistry. He was not involved in the assessment, peer review, or decision-making process of this paper. The authors have no relevant affiliations of financial involvement with any organisation or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Data availability statement
The data that support the findings of this study are openly available in MarttiT/S.-mansoni-BCA at https://github.com/MarttiT/S.-mansoni-BCA.
Additional information
Funding
References
- Gryseels B, Polman K, Clerinx J, Kestens L. Human schistosomiasis. Lancet. 2006;368(9541):1106–1118.
- McManus DP, Dunne DW, Sacko M, Utzinger J, Vennervald BJ, Zhou XN. Schistosomiasis. Nat Rev Dis Prim. 2018;4(1).
- Kamau E, Yates A, Maisiba R, Singoei V, Opot B, Adeny R, Arima CO, Otieno V, Sumbi CS, Okoth RO, et al. Epidemiological and clinical implications of asymptomatic malaria and schistosomiasis co-infections in a rural community in western Kenya. BMC Infect Dis. 2021;21(1).
- Wilson S, Vennervald BJ, Dunne DW. Chronic hepatosplenomegaly in African school children: a common but neglected morbidity associated with schistosomiasis and malaria. PLOS Negl Trop Dis. 2011;5(8):e1149.
- Gryseels B. Schistosomiasis. Infect Dis Clin North Am. 2012;26(2):383–397.
- Elbaz T, Esmat G. Hepatic and intestinal schistosomiasis: review. J Adv Res. 2013;4(5):445–452.
- Nelwan ML. Schistosomiasis: life cycle, diagnosis, and control. Curr Ther Res Clin Exp. 2019;91:5–9.
- Colley DG, Bustinduy AL, Secor WE, King CH. Human schistosomiasis. Lancet. 2014;383(9936):2253–2264.
- Costain AH, MacDonald AS, Smits HH. Schistosome egg migration: mechanisms, pathogenesis and host immune responses. Front Immunol. 2018;9.
- Siqueira L. d P, Fontes DAF, Aguilera CSB, Timóteo TRR, Ângelos MA, Silva LCPBB, de Melo CG, Rolim LA, da Silva RMF, Neto PJR, et al. Schistosomiasis: drugs used and treatment strategies. Acta Trop. 2017;176:179–187.
- Gasim GI, Bella A, Adam I. Schistosomiasis, hepatitis B and hepatitis C co-infection. Virol J. 2015;12(1).
- Rahmani A, Baee M, Rostamtabar M, Karkhah A, Alizadeh S, Tourani M, Nouri HR. Development of a conserved chimeric vaccine based on helper T-cell and CTL epitopes for induction of strong immune response against Schistosoma mansoni using immunoinformatics approaches. Int J Biol Macromol. 2019;141:125–136.
- Eyayu T, Zeleke AJ, Worku L. Current status and future prospects of protein vaccine candidates against Schistosoma mansoni infection. Parasite Epidemiol Control. 2020;11:e00176.
- LoVerde PT. Schistosomiasis. Adv Exp Med Biol. 2019;1154:45–70.
- Cioli D, Pica-Mattoccia L, Basso A, Guidi A. Schistosomiasis control: praziquantel forever? Mol Biochem Parasitol. 2014;195(1):23–29.
- Doenhoff MJ, Kusel JR, Coles GC, Cioli D. Resistance of Schistosoma mansoni to praziquantel: is there a problem? Trans R Soc Trop Med Hyg. 2002;96(5):465–469.
- Botros SS, Bennett JL. Praziquantel resistance. Expert Opin Drug Discov. 2007;2(sup1):S35–S40.
- Capasso C, Supuran CT. Anti-infective carbonic anhydrase inhibitors: a patent and literature review. Expert Opin Ther Pat. 2013;23(6):693–704.
- Bonardi A, Vermelho AB, da Silva Cardoso V, de Souza Pereira MC, da Silva Lara L, Selleri S, Gratteri P, Supuran CT, Nocentini A. N-nitrosulfonamides as carbonic anhydrase inhibitors: a promising chemotype for targeting Chagas disease and Leishmaniasis. ACS Med Chem Lett. 2019;10(4):413–418.
- D'Ambrosio K, Supuran CT, De Simone G. Are carbonic anhydrases suitable targets to fight protozoan parasitic diseases? Curr Med Chem. 2018;25(39):5266–5278.
- Liljas A, Laurberg M. A wheel invented three times. The molecular structures of the three carbonic anhydrases. EMBO Rep. 2000;1(1):16–17.
- De Simone G, Di Fiore A, Capasso C, Supuran CT. The zinc coordination pattern in the η-carbonic anhydrase from Plasmodium falciparum is different from all other carbonic anhydrase genetic families. Bioorg Med Chem Lett. 2015;25(7):1385–1389.
- Jensen EL, Clement R, Kosta A, Maberly SC, Gontero B. A new widespread subclass of carbonic anhydrase in marine phytoplankton. ISME J. 2019;13(8):2094–2106.
- Cox EH, McLendon GL, Morel FM, Lane TW, Prince RC, Pickering IJ, George GN. The active site structure of Thalassiosira weissflogii carbonic anhydrase 1. Biochemistry. 2000;39(40):12128–12130.
- Del Prete S, Vullo D, De Luca V, Supuran CT, Capasso C. Biochemical characterization of the δ-carbonic anhydrase from the marine diatom Thalassiosira weissflogii, TweCA. J Enzyme Inhib Med Chem. 2014;29(6):906–911.
- Capasso C, Supuran CT. An overview of the alpha-, beta- and gamma- carbonic anhydrases from bacteria: can bacterial carbonic anhydrases shed new light on evolution of bacteria? J Enzyme Inhib Med Chem. 2015;30(2):325–332.
- Zolfaghari Emameh R, Kuuslahti M, Nosrati H, Lohi H, Parkkila S. Assessment of databases to determine the validity of β-A nd γ-carbonic anhydrase sequences from vertebrates. BMC Genomics. 2020;21(1):352.
- Ferry JG. The γ class of carbonic anhydrases. Biochim Biophys Acta Proteins Proteomics. 2010;1804(2):374–381.
- Alterio V, Langella E, Viparelli F, Vullo D, Ascione G, Dathan NA, Morel FMM, Supuran CT, De Simone G, Monti SM. Structural and inhibition insights into carbonic anhydrase CDCA1 from the marine diatom Thalassiosira weissflogii. Biochimie. 2012;94(5):1232–1241.
- Park H, Song B, Morel FMM. Diversity of the cadmium-containing carbonic anhydrase in marine diatoms and natural waters. Environ Microbiol. 2007;9(2):403–413.
- Chasen NM, Asady B, Lemgruber L, Vommaro RC, Kissinger JC, Coppens I, Moreno SNJ. A glycosylphosphatidylinositol-anchored carbonic anhydrase-related protein of Toxoplasma gondii is important for rhoptry biogenesis and virulence. mSphere. 2017;2(3).
- Kikutani S, Nakajima K, Nagasato C, Tsuji Y, Miyatake A, Matsuda Y. Thylakoid luminal Θ-carbonic anhydrase critical for growth and photosynthesis in the marine diatom Phaeodactylum tricornutum. Proc Natl Acad Sci USA. 2016;113(35):9828–9833.
- Angeli A, Berrino E, Carradori S, Supuran CT, Cirri M, Carta F, Costantino G. Amine- and amino acid-based compounds as carbonic anhydrase activators. Molecules. 2021;26(23):7331.
- Berriman M, Haas BJ, LoVerde PT, Wilson RA, Dillon GP, Cerqueira GC, Mashiyama ST, Al-Lazikani B, Andrade LF, Ashton PD, et al. The genome of the blood fluke Schistosoma mansoni. Nature. 2009;460(7253):352–358.
- Da’dara AA, Angeli A, Ferraroni M, Supuran CT, Skelly PJ. Crystal structure and chemical inhibition of essential schistosome host-interactive virulence factor carbonic anhydrase SmCA. Commun Biol. 2019;2(1).
- Ferraroni M, Angeli A, Carradori S, Supuran CT. Inhibition of Schistosoma mansoni carbonic anhydrase by the antiparasitic drug clorsulon: X-ray crystallographic and in vitro studies. Acta Crystallogr D Struct Biol. 2022;78(Pt 3):321–327.
- Supuran CT, Altamimi ASA, Carta F. Carbonic anhydrase inhibition and the management of glaucoma: a literature and patent review 2013–2019. Expert Opin Ther Pat. 2019;29(10):781–792.
- Lomelino C, McKenna R. Carbonic anhydrase inhibitors: a review on the progress of patent literature (2011–2016). Expert Opin Ther Pat. 2016;26(8):947–956.
- Li Y, Zhang Y, Zhang Y. Research advances in pathogenesis and prophylactic measures of acute high altitude illness. Respir Med. 2018;145:145–152.
- Imray C, Wright A, Subudhi A, Roach R. Acute mountain sickness: pathophysiology, prevention, and treatment. Prog Cardiovasc Dis. 2010;52(6):467–484.
- Davis C, Hackett P. Advances in the prevention and treatment of high altitude illness. Emerg Med Clin North Am. 2017;35(2):241–260.
- Supuran CT. Advances in structure-based drug discovery of carbonic anhydrase inhibitors. Expert Opin Drug Discov. 2017;12(1):61–88.
- Supuran CT. Carbonic anhydrase inhibitors and their potential in a range of therapeutic areas. Expert Opin Ther Pat. 2018;28(10):709–712.
- McKenna R, Supuran CT. Carbonic anhydrase inhibitors drug design. Subcell Biochem. 2014;75:291–323.
- Supuran CT. Novel carbonic anhydrase inhibitors. Future Med Chem. 2021;13(22):1935–1937.
- Supuran CT. Carbonic anhydrases: novel therapeutic applications for inhibitors and activators. Nat Rev Drug Discov. 2008;7(2):168–181.
- Emameh RZ, Syrjänen L, Barker H, Supuran CT, Parkkila S. Drosophila melanogaster: a model organism for controlling Dipteran vectors and pests. J Enzyme Inhib Med Chem. 2015;30(3):505–513.
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using clustal omega. Mol Syst Biol. 2011;7(1):539.
- Madeira F, Pearce M, Tivey ARN, Basutkar P, Lee J, Edbali O, Madhusoodanan N, Kolesnikov A, Lopez R. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 2022;50(W1):W276–W279.
- Gouy M, Guindon S, Gascuel O. Sea view version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27(2):221–224.
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14(6):1188–1190.
- Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42(W1):W320–W324.
- Varadi M, Anyango S, Deshpande M, Nair S, Natassia C, Yordanova G, Yuan D, Stroe O, Wood G, Laydon A, et al. AlphaFold protein structure database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022;50(D1):D439–D444.
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):583–589.
- Evans R, O’Neill M, Pritzel A, et al. Protein complex prediction with AlphaFold-Multimer. bioRxiv. 2021:2021.10.04.463034.
- Naville M, Ghuillot-Gaudeffroy A, Marchais A, Gautheret D. ARNold: a web tool for the prediction of rho-independent transcription terminators. RNA Biol. 2011;8(1):11–13.
- Urbański LJ, Di Fiore A, Azizi L, Hytönen VP, Kuuslahti M, Buonanno M, Monti SM, Angeli A, Zolfaghari Emameh R, Supuran CT, et al. Biochemical and structural characterisation of a protozoan beta-carbonic anhydrase from Trichomonas vaginalis. J Enzyme Inhib Med Chem. 2020;35(1):1292–1299.
- Haapanen S, Bua S, Kuuslahti M, Parkkila S, Supuran CT. Cloning, characterization and anion inhibition studies of a β-carbonic anhydrase from the pathogenic protozoan Entamoeba histolytica. Molecules. 2018;23(12):3112.
- Kopp J, Slouka C, Ulonska S, Kager J, Fricke J, Spadiut O, Herwig C. Impact of glycerol as carbon source onto specific sugar and inducer uptake rates and inclusion body productivity in E. coli BL21(DE3). Bioengineering. 2017;5(1):1.
- Khalifah RG. The carbon dioxide hydration activity of carbonic anhydrase. I. Stop-flow kinetic studies on the native human isoenzymes B and C. J Biol Chem. 1971;246(8):2561–2573.
- Berrino E, Bua S, Mori M, Botta M, Murthy VS, Vijayakumar V, Tamboli Y, Bartolucci G, Mugelli A, Cerbai E, et al. Novel sulfamide-containing compounds as selective carbonic anhydrase I inhibitors. Molecules. 2017;22(7):1049.
- Urbanski LJ, Bua S, Angeli A, Kuuslahti M, Hytönen VP, Supuran CT, Parkkila S. Sulphonamide inhibition profile of Staphylococcus aureus β-carbonic anhydrase. J Enzyme Inhib Med Chem. 2020;35(1):1834–1839.
- Bua S, Haapanen S, Kuuslahti M, Parkkila S, Supuran CT. Sulfonamide inhibition studies of a new β-carbonic anhydrase from the pathogenic protozoan entamoeba histolytica. Int J Mol Sci. 2018;19(12):3946.
- Kimber MS, Pai EF. The active site architecture of Pisum sativum beta-carbonic anhydrase is a mirror image of that of alpha-carbonic anhydrases. EMBO J. 2000;19(7):1407–1418.
- Vale N, Gouveia MJ, Rinaldi G, Brindley PJ, Gärtner F, Da Costa JMC. Praziquantel for schistosomiasis: single-drug metabolism revisited, mode of action, and resistance. Antimicrob Agents Chemother. 2017;61(5).
- Li EY, Gurarie D, Lo NC, Zhu X, King CH. Improving public health control of schistosomiasis with a modified WHO strategy: a model-based comparison study. Lancet Glob Health. 2019;7(10):e1414–e1422.
- Wang W, Wang L, Liang YS. Susceptibility or resistance of praziquantel in human schistosomiasis: a review. Parasitol Res. 2012;111(5):1871–1877.
- Syrjänen L, Vermelho AB, Rodrigues I. d A, Corte-Real S, Salonen T, Pan P, Vullo D, Parkkila S, Capasso C, Supuran CT. Cloning, characterization, and inhibition studies of a β-carbonic anhydrase from Leishmania donovani chagasi, the protozoan parasite responsible for leishmaniasis. J Med Chem. 2013;56(18):7372–7381.
- Zolfaghari Emameh R, Kuuslahti M, Vullo D, Barker HR, Supuran CT, Parkkila S. Ascaris lumbricoides β carbonic anhydrase: a potential target enzyme for treatment of ascariasis. Parasites Vectors. 2015;8(1).
- Khubchandani IT, Bub DS. Parasitic infections. Clin Colon Rectal Surg. 2019;32(5):364–371.
- Mansour NS, Youssef FG, Mikhail EM, Mohareb EW. Amebiasis in schistosomiasis endemic and non-endemic areas in Egypt. J Egypt Soc Parasitol. 1997;27(3):617–628.
- Dolabella SS, Coelho PMZ, Borçari IT, Mello NAST, Andrade ZDA, Silva EF. Morbidity due to Schistosoma mansoni – Entamoeba histolytica coinfection in hamsters (Mesocricetus auratus). Rev Soc Bras Med Trop. 2007;40(2):170–174.
- Gulati S, Aref AA. Oral acetazolamide for intraocular pressure lowering: balancing efficacy and safety in ophthalmic practice. Expert Rev Clin Pharmacol. 2021;14(8):955–961.
- Leaf DE, Goldfarb DS. Mechanisms of action of acetazolamide in the prophylaxis and treatment of acute mountain sickness. J Appl Physiol. 2007;102(4):1313–1322.
- Adamson R, Swenson ER. Acetazolamide use in severe chronic obstructive pulmonary disease pros and cons. Ann Am Thorac Soc. 2017;14(7):1086–1093.
- Van Berkel MA, Elefritz JL. Evaluating off-label uses of acetazolamide. Am J Health Syst Pharm. 2018;75(8):524–531.
- Cong-Hui Z, Huai-Min Z. Cloning and characterization of β-carbonic anhydrase, a potential drug target of Schistosoma japonicum. Zhongguo Xue Xi Chong Bing Fang Zhi Za Zhi. 2016;28(2):161–166.
- Llanwarne F, Helmby H. Granuloma formation and tissue pathology in Schistosoma japonicum versus Schistosoma mansoni infections. Parasite Immunol. 2021;43(2):e12778.