ABSTRACT
Introduction: Despite intensive research efforts, there is still no effective prophylactic vaccine available against HIV-1. Currently, substantial efforts are devoted to the development of vaccines aimed at inducing broadly neutralizing antibodies (bNAbs), which are capable of neutralizing most HIV-1 strains. All bNAbs target the HIV-1 envelope glycoprotein (Env), but Env immunizations usually only induce neutralizing antibodies (NAbs) against the sequence-matched virus and not against other strains.
Areas covered: We describe the different strategies that have been explored to improve the breadth and potency of anti-HIV-1 NAb responses. The discussed strategies include the application of engineered Env immunogens, optimization of (bNAb) epitopes, different cocktail and sequential vaccination strategies, nanoparticles and nucleic acid-based vaccines.
Expert opinion: A combination of the strategies described in this review and future approaches are probably needed to develop an effective HIV-1 vaccine that can induce broad, potent and long-lasting NAb responses.
1. Introduction
The last two decades have brought tremendous progress in the fight against HIV-1: the development of effective antiretroviral therapy (cART) has significantly decreased the number of new infections and HIV-related deaths in most parts of the world [Citation1]. However, the pandemic continues to expand in other regions, such as Central Asia and Russia, and still poses one of the biggest threats to global health [Citation2]. Furthermore, cART does not provide a cure for HIV-1 infection and eradication of HIV-1 will likely require an effective and broad-spectrum prophylactic vaccine.
The most important correlate of protection for antiviral vaccines are neutralizing antibodies (NAbs) [Citation3]. Several lines of evidence suggest that a prophylactic HIV-1 vaccine needs to induce NAbs to be effective. First, the immune system cannot clear an established HIV-1 infection and only NAbs could provide the sterilizing protection that would be needed for effective protection against HIV-1. Second, passively administered NAbs protect non-human primates (NHPs) and humanized mice against HIV-1 infection and their protective effect is directly correlated with their neutralizing potency [Citation4,Citation5]. Finally, a recent immunization study demonstrated that an immunogen that is aimed at inducing NAbs could protect NHPs against infection and identified NAbs as the prime correlate of protection [Citation6]. Other immune mechanisms, such as T-cell mediated immunity and antibody-dependent cytotoxicity (ADCC) might also play some role in protection against HIV1 infection, including the weak and short-lived protective effect observed in the RV144 clinical trial [Citation7]. However, these subjects are beyond the scope of this review.
All HIV-1 NAbs target the envelope glycoprotein (Env), which is the only viral protein on the surface of the virus. Env is required at the initial stages of infection for attaching to the host CD4 receptor and CCR5 or CXCR4 co-receptors [Citation8–Citation11]. This glycoprotein consists of a trimer of heterodimers of gp120 and gp41 subunits, which are generated after the cleavage of a gp160 precursor by cellular proteases. The three gp41 subunits anchor the complex to the membrane and the three gp120 are linked by non-covalent interactions to the gp41 anchor. Gp120 is the most exposed moiety and contains five flexible variable loops and five relatively conserved regions.
HIV-1 is a rapidly evolving virus and this has resulted in an enormous sequence diversity between Envs of different isolates [Citation12,Citation13]. Furthermore, the virus has developed strategies to hide essential Env regions from recognition by the immune system [Citation14–Citation16]. These strategies include shielding of conserved epitopes by flexible loops that are highly sequence diverse. Thus, the most conserved epitopes are hidden inside the Env core and are only exposed after CD4 receptor engagement [Citation14]. Moreover, Env has a dense shield of glycans that cover most of the more immunogenic protein epitopes [Citation17].
Despite all of the above, the immune systems of many HIV-1 infected individuals are able to overcome these hurdles and develop NAbs with a significant level of cross-neutralization, called broadly neutralizing antibodies (bNAbs). These bNAb responses fall in a continuum of breadth and potency, and 50% of infected individuals are able to neutralize more than 50% of HIV-1 circulating strains and some individuals develop bNAbs that neutralize even more than 98% of strains [Citation18]. Most bNAbs target exposed but hard-to-reach conserved regions, such as the CD4 binding site, or recognize conserved epitopes in the glycan shield and/or on variable loops. This select group of antibodies are potent and broad enough to neutralize most of the circulating isolates of HIV-1 and have been shown to provide protection against viral challenge when passively transferred to NHPs [Citation4]. Hundreds of bNAbs have now been isolated and characterized and they can target a wide range of Env epitopes. The first identified HIV-1 bNAbs targeted a limited subset of epitopes: the CD4 binding site (CD4bs) (e.g. b12), the glycans on the variable loop 3 (V3) (e.g. 2G12) and the membrane-proximal external region (MPER) (e.g. 2F5 and 4E10) [Citation19,Citation20]. In the last decade, the development of sophisticated single B cell cloning and isolation techniques have greatly expanded the number of bNAbs, which are more broad and potent and target additional epitopes: the glycans on the variable loops 1 and 2 (V1V2) on the trimer apex, the glycans on the variable loop 3 (V3) region, the gp120-gp41 interface, the fusion peptide and the ‘silent’ face of gp120, demonstrating that the whole Env trimer surface could be a potential target for bNAbs [Citation21–Citation23]. The identification of the different bNAbs, their epitopes and binding mechanisms have been extensively reviewed by others [Citation5,Citation24].
Several studies have dissected the virus-antibody co-evolution pathways of several bNAbs. These studies have greatly enhanced our understanding of the often tortuous pathways that B cell lineages undergo before developing into bNAb-producing B cells [Citation21,Citation25–Citation27]. This is mainly due to the uncommon features associated with HIV-1 bNAbs. For instance, many bNAbs contain long heavy complementary determining region 3 (HCDR3) loops, which are needed to penetrate Env’s densely packed glycan shield [Citation28,Citation29]. Other bNAbs (such as the CD4bs-targeting VRC01 bNAb) have undergone high levels of somatic hypermutation [Citation30,Citation31]. Moreover, many (precursors to) bNAbs are reactive to host proteins [Citation32–Citation38] and these B cells are usually depleted during B cell maturation [Citation39–Citation41]. Apart from the fact that germline paratopes are usually related to higher malleability that can result in polyreactivity [Citation42], it has been observed that mutations incorporated during the process of affinity maturation are associated with strong (and sometimes de novo) poly-/auto-reactivity [Citation43–Citation45]. These properties provide major obstacles for an HIV-1 vaccine to induce bNAbs and partially explain why such a vaccine has not been developed yet.
Here, we will discuss the different immunogens and strategies that are being employed to induce bNAb-like responses using vaccination and, in some cases, might improve the durability of these responses. These strategies are schematically represented in .
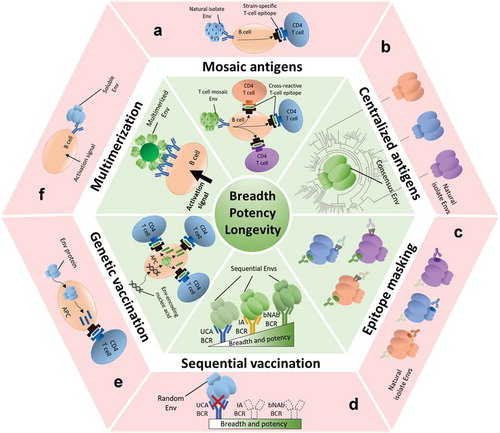
Figure 1. Strategies for increasing the breadth, potency and longevity of NAb responses against HIV-1 Env. The different immunological challenges that HIV-1 poses are indicated in the red outer sectors. The corresponding green sectors show potential strategies to overcome these challenges, as detailed in this review. Env antigens, antibodies and epitopes colored in green are related to broad, potent and/or long-lived neutralizing responses. Envs and epitopes in blue, orange and magenta are related to narrow, weak and/or short-lived responses.
(A) Mosaic antigens: natural HIV-1 isolates present strain-specific T cell epitopes that will attract narrow CD4 T cell help (red), while mosaic-based HIV-1 Envs will attract a broader range of CD4 T cells (green). (B) Centralized Env antigens: consensus sequence-based Envs are more closely related to most Envs (green) than most random natural isolates to each other (red) and thus contain less strain-specific antigenic determinants.(C) Epitope masking: many Envs contain immunodominant strain-specific epitopes that are not suitable for broadening NAb responses (red). By covering or hiding these epitopes (e.g. with glycans), one might focus the responses to more relevant, but subdominant cross-neutralizing epitopes (green).(D) Sequential vaccination strategies: randomly selected Env trimers usually do not engage the unmutated common ancestor (UCA) B cell receptors (BCRs) of bNAb lineages (red). A series of specifically designed or isolated Env immunogens that engage UCA BCRs and recapitulate the antibody maturation process could be administered sequentially to induce bNAb responses (green).(E) Genetic vaccination: protein subunit immunogens are processed by antigen presenting cells (APCs) and presented on major histocompatibility complex (MHC) molecules, but attract only a limited CD4 T cell help (red). However, nucleic acid encoded antigens usually induce longer-lasting responses due to increased CD4 T cell help (green).(F) Multimerization: soluble Envs do not activate cognate B cells efficiently, possibly due to low avidity interactions or lack of BCR cross-linking (red). The higher valency of multimerized Envs increases avidity and BCR crosslinking, thus increasing B cell activation (green).
2. HIV-1 Env immunogen forms
Several types of subunit immunogens have been developed as potential vaccine candidates (extensively reviewed in [Citation46]). The first Env-based immunogen designs were peptides and gp120 monomers. Initially, peptides based on the V3-loop were hailed as promising vaccine candidates because they efficiently induced anti-HIV-1 NAbs [Citation47,Citation48]. However, the initial enthusiasm waned after it became apparent that these NAbs only neutralized lab-adapted and neutralization-sensitive (‘Tier 1’ [Citation49]) viruses. The Tier 1A viruses represent mostly lab-adapted strains and are highly neutralization sensitive, while Tier 1B viruses represent mostly circulating, but neutralization sensitive strains. Tier 1 viruses seem to harbor Envs with a more ‘open’ structure and this might explain why these viruses are efficiently neutralized by patient sera [Citation50,Citation51]. In contrast, most circulating HIV-1 strains harbor a more compact Env and, based on their higher neutralization resistance to patient sera, are categorized as ‘Tier 2’ strains [Citation49,Citation50]. Strains that are highly resistant to serum neutralization are categorized as Tier 3 viruses. A vaccine that is capable of inducing NAbs that neutralize most Tier 2 strains is a major goal for HIV-1 vaccinology. Other peptide immunogens are those based on the Env membrane-proximal external region (MPER), which is located at the base of gp41. Some of the broadest bNAbs, such as 10E8 and DH511, target the MPER [Citation52,Citation53]. This, together with the high sequence conservation of MPER bNAbs epitopes, suggests that MPER peptides are attractive vaccine candidates. However, similarly to V3 peptide immunizations, MPER peptide-based immunogens have mostly induced binding Abs or Tier 1 NAbs only [Citation54,Citation55]. Recently, immunization strategies based on fusion peptides have shown promising results and this will be discussed separately (see section: ‘Epitope-focused vaccination’).
Gp120-based immunogens are being applied in a number of strategies. First, as gp120 monomers, but these immunogens usually do not induce Abs that neutralize Tier 2 strains [Citation56]. This was confirmed when some of the participants in the Phase I Vaxgen AIDSVAX clinical trial became infected despite having developed NAb titres against the lab-adapted strains [Citation56]. The lack of neutralization of Tier 2 strains is possibly because gp120 monomers do not engage the precursors of bNAbs nor does gp120 recapitulate the structure of the complete Env trimer. The gp120 monomer displays epitopes on the non-neutralizing face of Env and the neutralizing epitopes present on gp120, such as the CD4 binding site, can be engaged from many angles of approach [Citation57–Citation59]. On viral Env, the same non-neutralizing epitopes are occluded and the possible angles of approach for the same neutralizing epitopes are much more restricted [Citation58,Citation60,Citation61]. The second gp120-based immunogens are truncated gp120 monomers that lack non-NAb epitopes, such as the inner domains [Citation62–Citation64] or the V3 loop [Citation65]. However, gp120 monomers without these immunogenic non-neutralizing epitopes have only elicited binding Abs or Tier 1 NAbs. Third, gp120 outer domains (eOD) have been engineered that interact with specific precursor B cell receptors [Citation66,Citation67]. These promising immunogens are now being applied in immunization strategies aimed at activating inferred germline versions of bNAbs and are discussed in detail later (see section 7: ‘Targeting the precursors of bNAb producing B cells’).
The Env complex is highly metastable and removing the transmembrane domain causes the complex to fall apart in gp120 and gp41 subunits. Therefore, the first trimeric soluble Env designs consisted of gp140 subunits that included inactivated furin cleavage sites that kept the gp120 associated with the gp41 ectodomain and/or contained heterologous trimerization domains, such as foldon and isoleucine zipper, fused to gp41 to stabilize the trimeric conformation [Citation68,Citation69]. However, these uncleaved Env trimers do not resemble the typical propeller shape of viral Env [Citation70], but instead have a splayed open non-native Env conformation with gp120 monomers loosely tethered to gp41 ectodomains that are mostly in a postfusion conformation [Citation71]. Moreover, these non-native trimers present epitopes in the Env core and the heterologous trimerization domain that are usually hidden (or absent) on neutralization-resistant viruses and that induce potentially distracting and unwanted non-NAbs (see section 4: ‘Epitope masking’) [Citation57,Citation69,Citation72,Citation73].
3. Native-like Env trimers
A different approach toward generating soluble Env trimers encompasses the so-called SOSIP.664 design, which was iteratively generated over the years. First efforts started almost two decades ago by covalently linking gp120 and gp41 using a disulfide bond (‘SOS’) [Citation74]. Thereafter, an I559P mutation (‘IP’) was introduced in gp41 to stabilize Env in its pre-fusion conformation [Citation75] and gp120/gp41 cleavage was increased by enhancing the furin cleavage site [Citation76]. Lastly, gp140 was truncated at position 664 to prevent unwanted aggregation [Citation77]. Combining this set of ‘SOSIP.664’ mutations with the clade A BG505 viral isolate resulted in the first native-like Env trimer (BG505 SOSIP.664) [Citation78]. The BG505 SOSIP.664 provided the first high-resolution structures of the complete Env trimer [Citation79,Citation80], paving the way for structure-based Env trimer design. Later, it was shown that the SOSIP trimer structure is virtually indistinguishable from that of membrane-anchored HIV-1 Env [Citation81]. Importantly, BG505 and the clade B B41 SOSIP.664 trimers were the first soluble immunogens to induce consistent and moderate to strong autologous NAb responses against a Tier 2 virus in rabbits and macaques [Citation57]. However, these responses did not show neutralizing activity against heterologous Tier 2 viruses [Citation57], as they were primarily targeting BG505- and B41-specific epitopes accessible because of the lack of glycosylation at positions 241 and 289 of the BG505 and B41 envelope protein [Citation82,Citation83].
The developments with SOSIP trimers have encouraged structure-based improvements and alternative native-like Env designs [Citation69,Citation84], such as uncleaved Env trimers that contain a long linker between gp120 and gp41 ('single-chain' or 'native flexibly linked (NFL)' trimers) [Citation85,Citation86] or a redesigned heptad repeat 1 in gp41 as an alternative to the I559P mutation (‘uncleaved prefusion-optimized (UFO)’ design) [Citation87]. Subsequent studies have shown that these designs also present a native-like structure [Citation88,Citation89] and can induce autologous NAbs efficiently [Citation90,Citation91].
During the last years, these Env designs have facilitated the generation of a considerable collection of native-like soluble Env trimers from different isolates and clades [Citation88,Citation90,Citation92–Citation94]. Nevertheless, monovalent immunization regimes with these immunogens have never induced bNAbs in relevant animal models [Citation84], which is not surprising, considering the intricate pathways that bNAbs need to undergo before developing breadth. Thus, more targeted designs and vaccination strategies are needed to optimally exploit the potential of these immunogens for vaccines.
4. Epitope masking
During the co-evolution history of HIV-1 with its hosts, the virus has developed several immune escape mechanisms, which include the incorporation of a glycan shield that covers most of the surface of the Env protein [Citation15,Citation17]. Generally, the most essential and conserved epitopes have evolved into subdominant determinants by glycan masking. On the other hand, strain-specific non-essential determinants are sometimes left exposed and might act as immunodominant ‘epitope-baits’ that hinder the development of broad-spectrum responses. In line with these concepts, it has been found that patients infected by transmitter/founder (T/F) viruses with holes in their glycan shield induce narrower responses than those infected by T/F viruses with intact glycan-shields [Citation95]. Furthermore, detailed characterization of the humoral responses elicited in animals immunized with native-like Env trimers identified several of these immunodominant determinants responsible for the elicitation of strain-specific or narrow-neutralizing antibodies [Citation82,Citation91,Citation94,Citation96,Citation97]. Moreover, trimer immunogens with artificially created glycan holes induce potent neutralization, but these NAbs are usually only reactive with the autologous glycan-deleted virus [Citation98]. Finally, site-specific glycan analyses have shown that glycan occupation of Env is highly heterogeneous [Citation99–Citation101]. Taken together, this suggests that HIV-1 has evolved a dynamic glycan shield that could open to ‘lure’ highly specific strain-specific NAbs that provide little or no protection against other strains.
Furthermore, virtually all engineered soluble HIV-1 Env immunogens present (neo-)epitopes that induce strain-specific or non-neutralizing Abs. First, many soluble Env trimers induce non-neutralizing Abs to the V3-loop and/or the Env core, but these epitopes are hidden on closed viral Env trimers of Tier 2 viruses [Citation51,Citation57]. However, in vitro experiments show that many of the engineered Env trimers do not expose the V3-loop and/or Env core prior to administration, which suggests that these epitopes become exposed due to (partial) degradation or opening up of Env trimers post-administration [Citation102]. Second, heterologous protein scaffolds that are sometimes used to present soluble Env trimers are highly immunogenic [Citation73]. Third, the underside of soluble Env trimers is highly exposed, but this epitope does not exist on membrane-bound viral Env [Citation103]. Polyclonal antibody mapping suggests that a majority of early Ab responses target this non-neutralizing epitope on soluble Env trimer immunogens [Citation104].
These observations open the door to an ‘epitope masking-based design’ strategy in which strain-specific or non-neutralizing epitopes are artificially covered to redirect the responses to other, potentially cross-neutralizing, epitopes [Citation105,Citation106]. Several studies have successfully used stabilizing mutations (reviewed in [Citation84]), interdomain-locking mutations [Citation107] and glycan shielding to decrease the immunogenicity of unwanted non-neutralizing epitopes without compromising the desired Ab responses [Citation73,Citation106–Citation109]. B cell immunology suggests that decreasing the immunogenicity of undesired epitopes should increase the competitive advantage of desired lower affinity bNAb epitopes [Citation110]. However, thus far no in vivo study has directly demonstrated that dampening unwanted Env-responses redirects and increases Ab responses toward the desired epitopes [Citation108,Citation109]. Furthermore, infected individuals that develop bNAbs also induce non-NAbs and this suggests that the induction of bNAbs is not necessarily compromised when non-NAbs are also induced. Still, immunogens lacking most non-neutralizing epitopes will likely be important in cocktail or sequential vaccines in order to subdue responses to immunodominant non-NAb epitopes and to bolster the responses to subdominant cross-neutralizing epitopes. Furthermore, future immunization studies will need to demonstrate the influence of the highly exposed bottom epitope on soluble Env trimers on the induction of NAbs.
5. Centralized and mosaic immunogens
Due to the high diversity of env sequences among HIV-1 strains, Env trimers present strain-specific antigenic determinants that are not likely to induce broad-spectrum responses. One approach to circumvent these strain-specific responses is the use of so-called centralized immunogens [Citation12,Citation111–Citation114], which encompass the variability from several viral strains in a single sequence. In theory, this could result in broader NAb responses by setting the immunological target(s) on the most conserved epitopes or, at least, on epitopes that are shared among a significant percentage of isolates. Two types of centralized sequences are distinguished according to the algorithm used to gather the variability of the viral population: consensus sequences, constructed by the concatenation of the most common amino acid at each position of the protein alignment, and ancestral sequences, which account for the predicted sequence of the common ancestor [Citation12,Citation113].
Despite their artificial origin, several examples of functional immunogens based on centralized env sequences were reported prior to the emergence of soluble native-like Env trimers [Citation115–Citation120]. However, these immunogens were focused on enhancing T-cell responses and only elicited low NAb titres [Citation115–Citation118,Citation120], probably because they were delivered by genetic vaccination or did not present a native-like conformation. Liao et al. compared transmitter/founder (T/F), consensus and chronic Envs from different clades in immunization experiments in guinea pigs. Although all the trimers from the three classes showed a similar antigenic profile, the T/F Env induced the least potent but also the broadest neutralizing responses. The consensus Envs elicited higher NAb titres to both Tier 1 and a subset of Tier 2 viruses (albeit at very low titres) than chronic Envs [Citation121].
Advances in Env trimer protein design have boosted the generation of new consensus sequence-based Env immunogens. For instance, a soluble native-like Env trimer was produced by combining a number of stabilizing mutations with the consensus env sequence of clade C (conC) [Citation122]. Another construct, based on a consensus sequence of all env sequences in group M (conS) [Citation119], which was stabilized by the UFO design [Citation87], showed a favorable antigenic profile and elicited autologous NAb responses in a DNA-protein vaccination experiment in rabbits [Citation90]. The ConM SOSIP, also based on a group M consensus sequence, was structurally indistinguishable from other native-like Env trimers and induced apex-directed antibodies that neutralized the autologous conM virus and related conS virus at relative high NAb titres [Citation123]. These and other consensus-based Env immunogens might be useful additions to NAb-based vaccine strategies, because they should contain less isolate-specific antigenic determinants.
A slightly different approach aimed at maximizing the coverage of an Env-based vaccine is the use of mosaic immunogens, which present as many cross-reactive T- or B- cell epitopes as possible while resembling natural sequences [Citation124,Citation125]. The presence of cross-reactive epitopes representing the whole population of epitope variants should increase the probability of recruitment of more individual lineages of immune cells than those based on single isolates.
T-cell mosaic immunogens have been repeatedly shown to improve the depth and breadth of the CD4 and CD8 cellular immune responses compared to consensus or natural HIV-1 antigens, in DNA- and vectored-based vaccination experiments [Citation126–Citation131]. This is the result of their optimization to include potential T-cell epitopes (PTE), which are nine-amino-acid peptides and thus present the optimal length to be presented on major histocompatibility complex (MHC) molecules [Citation132,Citation133]. There are two types of T-cell epitope mosaic vaccines. First, HIVconsv vaccines that include one or several isolated PTEs corresponding to functionally and structurally conserved subdominant regions of the viral proteins [Citation131,Citation134]. In clinical trials, these immunogens re-focused T cell immune responses to conserved epitopes [Citation135,Citation136]. Other mosaic sequences encode for viral proteins that contain as many PTEs as possible while trying to preserve a native-like conformation. A heterologous gag, pol and env-based mosaic antigen called mosaic M delivered by an adenoviral vector 26 (AdV26) induced higher antigen-specific CD4 T-cell responses than counterparts based on consensus M and clade C natural sequences [Citation126]. Env trimers based on T cell mosaic sequences have been generated, but these are non-native trimers and did not elicit consistent Tier 2 NAb responses [Citation137,Citation138]. On the other hand, the structural complexity of most B cell epitopes [Citation139] has probably hampered the design of B-cell mosaic immunogens, which remains an elusive goal and will probably require intensive protein engineering efforts.
Centralized and mosaic sequence-based immunogens can be useful for suppressing isolate-specific responses and stimulate more universal Env immune responses. However, none of these immunogens have induced bNAbs and a successful vaccination strategy will probably require multiple different Env-based immunogens, as discussed below.
6. Combination vaccines
An obvious approach to increase the breadth of NAb responses is to administer Env antigens from different strains in combination, either simultaneously or sequentially. The rationale behind a combination vaccine is that B cell lineages targeting conserved cross-neutralizing epitopes present in most immunogens of the combination will be selected over the ones that target strain-specific epitopes present only in one or few of the immunogens. This hypothesis is reinforced by observations that show that increased viral diversity in HIV-1 infected patients correlates with neutralization breadth [Citation140–Citation142]. Furthermore, several in silico simulation studies have predicted that combination vaccines would activate cross-neutralizing B cells more efficiently, because these B cells would be able to internalize and present a higher amount of antigen and thus outcompete the strain-specific B cells [Citation143–Citation145].
The first HIV-1 Env-based cocktail or sequential immunization experiments were conducted with DNA vaccines, non-native like trimers or gp120 monomers. In general, these studies showed that such regimens were superior in inducing broader Tier 1 NAb responses compared to monovalent vaccination [Citation36,Citation146–Citation149]. However, consistent NAb responses against Tier 2 viruses were not induced. Two independent studies used native-like Env trimers based on diverse env sequences to assess the potential of sequential and cocktail strategies for eliciting Tier 2 NAbs [Citation83,Citation94]. However, neither simultaneous nor sequential administration of native-like Env trimers elicited bNAbs. Interestingly, both studies found that a clade C Env trimer could cross-boost responses to clade A and B trimers that were administered earlier [Citation83,Citation94]. Furthermore, Tier 1A V3-directed responses were higher in the combination vaccine groups compared to the monovalent vaccine groups [Citation94]. The combination groups also induced less potent autologous NAb responses against the individual Envs in the cocktail than those from the monovalent groups. These results suggested that immune interference might have played a role, since the most immunodominant component in the cocktail, the BG505 SOSIP trimer, seemed to reduce the autologous responses against the other strains [Citation94].
More recently, epitope-focused combination vaccine approaches were described [Citation150,Citation151]. Voss et al. first screened for viral Envs that were neutralized by V2-apex-targeting NAbs. Subsequently, SOSIP trimer versions of these Envs were generated and administered as monovalent or as combination vaccines. Again, the combination groups showed less potent and consistent NAb responses against the individual components, but one rabbit immunized with the cocktail displayed weak heterologous Tier 2 NAb responses, while none of the rabbits in the monovalent groups showed any breadth [Citation151]. Similarly, Bricault and colleagues found that the magnitude and breadth of the NAb responses induced by a cocktail of trimers with optimized V2 epitopes were improved over those of the monovalent vaccines [Citation150]. Together, these studies suggest that combination vaccines can indeed steer responses toward shared epitopes when the appropriate Env trimers are mixed. However, none of the above-mentioned studies induced the level of breadth and potency that would be needed for an effective vaccine and thus more sophisticated strategies are required.
6.1. Sequential vaccines based on natural infection
Longitudinal studies that finely dissected the co-evolutionary pathway between bNAbs and the infecting virus in patients have helped to understand the crucial steps in the development of some bNAbs lineages as well as to elucidate the viral sequences that drove their evolution [Citation27,Citation152–Citation154]. The viral env sequences from these patients are now being turned into soluble immunogens for vaccination strategies that try to recapitulate the virus-antibody co-evolutionary pathways that produced bNAbs. One of the first studies that used viral quasispecies sequences in vaccines was published by Malherbe et al. The authors administred a collection of viral env quasispecies sequences from a SHIV-infected macaque as DNA vaccines to rabbits. These animals developed responses that targeted similar epitopes as those targeted by the macaque [Citation147]. These natural lineage-based vaccination approaches now benefit from the advances in Env protein engineering to design protein immunogens that might be more suitable for guiding bNAb development. The evolutionary pathways of the CD4bs bNAb lineages CH103 and CH235 have been mapped in great detail and viral sequences have provided templates for Env-based sequential vaccines [Citation154,Citation155]. In an effort to raise CH103 or CH235 bNAbs by vaccination, knock-in (KI) mice carrying the unmutated common ancestor of the heavy chain of CH103 (CH103 UCA) and wild-type (WT) NHPs were immunized with immunogens based on Env sequences isolated from the patient who induced CH103 bNAbs [Citation36,Citation156]. Surprisingly, the Tier 2 NAbs induced in both animal models targeted the V1V2 region and not the CD4bs [Citation36,Citation156]. The authors found that the CH103 UCA in the KI mice is polyreactive and that the maturing B cell stemming from this lineage were partially depleted and as a result, other epitope specificities, i.e. V1V2, were being selected by vaccination [Citation36,Citation156]. Together, these and other studies show that inducing certain CD4bs bNAbs with (lineage-based) vaccines can be impeded by tolerance mechanisms [Citation37] and that other epitopes might be more suitable for inducing bNAbs using lineage-based vaccines. As such, studies on the VRC26 and PCT64 bNAb lineages, which target quaternary epitopes on the Env V2-apex, provide interesting avenues for vaccine design [Citation27,Citation152,Citation153,Citation157,Citation158]. Thus far, vaccine studies using immunogens that are based on viral sequences from the individuals who developed the VRC26 and/or PCT64 lineages have been proposed, but their outcomes have not yet been published [Citation159].
7. Targeting the precursors of bNAb producing B cells
A slightly different approach is the use of Env immunogens that are specifically designed to activate specific bNAb precursors, followed by subsequent boosts with tailored immunogens that could shepherd the desired lineage toward breadth [Citation26]. For most bNAbs, the genuine naïve B cell precursors, called unmutated common ancestors (UCA), are not known and therefore these sequences are computationally inferred from the mature bNAb sequence or, ideally, from the early precursor sequences in a bNAb lineage. In theory, these inferred sequences correspond to the naïve (or germline) B cell receptors that were initially engaged by the transmitter/founder virus and later developed into bNAbs. Thus, Env immunogens that bind to the inferred germline bNAb (igl-bNAb) would be potential candidates to prime such bNAb B cell lineages. The observation that most Envs do not efficiently bind to igl-bNAbs [Citation160,Citation161] fueled the design of germline targeting (GT) immunogens, such as engineered outer domain of gp120 (eOD.GT) [Citation66], engineered 426c Env [Citation67] and SOSIP trimers (BG505.SOSIP.GT1 and MD39-11MUTB) [Citation162,Citation163]. These engineered proteins show improved binding to igl-bNAbs that target several epitopes: the CD4 binding site (CD4bs) (e.g. VRC01, 3BNC60) [Citation66,Citation67,Citation162], variable region 2 (V2) apex epitopes (e.g. PG9/16, CH01) [Citation162] and/or the V3 glycan epitope (e.g. PGT121) [Citation163].
Several of these immunogens were tested in knock-in mice that carry the inferred germline versions of 3BNC60, PGT121 or VRC01 [Citation37,Citation66,Citation162,Citation164]. Initially, these studies showed that the germline B cells could be selectively activated and expanded, but none of those mice developed broad neutralization, as expected from these early responses. Follow-up experiments in germline VRC01 and PGT121 KI mice showed that it was possible to shepherd these initial responses with appropriate boosts to induce Abs that neutralize wild-type or near native viruses [Citation165–Citation167]. These studies represent a significant milestone, but these bNAb(-like) responses were elicited in model animals with a controlled and favorable B cell repertoire. How would similar germline-targeting strategies fare when physiologically relevant amounts of germline bNAb B cells are present and have to compete with non-bNAb B cells? Recently, two elegant studies have started to address this question by transferring different amounts of (partly) germline bNAb B cells into WT mice. Subsequently, these mice were immunized with antigens with different affinities for the transferred germline B cells [Citation168,Citation169]. In both studies, the authors found that the activation of the antigen-specific B cells was dependent on B cell frequency and antigen-binding affinity. Moreover, a recent study showed that an engineered variant of the MD39-11MUTB trimer [Citation163] could induce antibodies that resemble the precursors of V3 glycan bNAbs in mice, rabbits and macaques [Citation170]. However, none of the humoral responses induced in the animals of the latter three studies had any neutralizing activity against WT viruses. A next step would be to determine which strategies are needed to guide the responses of these animals toward neutralization potency and breadth.
8. Epitope-focused vaccination
The germline- and lineage-based approaches seem very promising, but they rely on activating the correct B cell lineage and the ability to shepherd these responses toward breadth. An epitope-focused vaccination strategy is B cell agnostic and would potentially be a less complicated strategy for inducing broader NAb responses. However, most conserved neutralizing epitopes are buried or hidden by glycans, or involve conformational epitopes consisting of non-linear peptides. Until recently, it was thought that the linear fusion peptide (FP) was hidden inside the Env protein. Identification of several bNAbs (PGT151, VRC34 and ACS202) that target the N-terminus of the FP [Citation171–Citation174] revealed that this epitope is actually exposed on viral Env. The FP is a promising candidate for epitope-focused vaccination for several reasons: it is essential for the fusion machinery of HIV-1 [Citation175], it is only partially shielded by glycans, it is a linear peptide stretch and its amino acid sequence is highly conserved.
The Kwong research group has used structure-guided design to create several scaffolds that present a FP that is recognizable by FP-specific bNAbs [Citation176]. By immunizing with FP-scaffolds followed by trimer boost, the authors showed that FP-specific NAb responses could be induced. The sera responses were relatively broad: 1-20% of the viruses were neutralized by the polyclonal sera and individual antibodies isolated from immunized mice could even neutralize 31% of a panel of 208 primary isolates [Citation176]. These NAb responses were broader than those achieved before and future studies will reveal whether this strategy will yield similar NAb responses in humans. However, the fusion peptide is highly flexible [Citation177] and not all viruses seem to expose the FP so that it is recognized by NAbs [Citation178]. These features could be potential roadblocks for developing a broadly applicable FP-focused immunization strategy to target HIV-1.
9. Nanoparticle presentation
The immune system has evolved to effectively recognize and mount responses toward bacterial and viral antigens when they are presented in repetitive and continuous arrays on the surface of pathogens [Citation179]. The fact that the only antiviral subunit vaccines licensed to date (hepatitis B, hepatitis E and human papillomavirus) are based on virus-like particle (VLP) formulations signifies the importance of multimerization for immunogenicity [Citation180]. The key immunological concepts underpinning the immunogenicity of multimerized antigens have been extensively reviewed [Citation181,Citation182]. Several of these immunological mechanisms are especially important for vaccines aimed at inducing HIV-1 NAbs. First, multimerization improves B cell activation by increasing the B cell receptor (BCR) cross-linking [Citation181,Citation182]. Second, multimerization increases avidity which could compensate for low affinity antigen-BCR interactions, such as those between naïve precursor B cells and Env antigen [Citation168,Citation183,Citation184]. Third, it has recently been shown that multimerized HIV-1 Env immunogens are more efficiently trafficked to germinal centres by components of the innate immune system [Citation185]. Fourth, presenting soluble HIV-1 Env trimers on nanoparticles hides the non-neutralizing epitopes on the bottom of these trimers [Citation103,Citation104]. Fifth, repetitive display can break immunological tolerance checks that block the maturation of some bNAb lineages [Citation37,Citation186,Citation187].
The physicochemical properties of nanoparticles, such as shape, size and surface chemistry, can affect their effectivity (reviewed in [Citation181]), thus representing important features to be fine-tuned. A number nanoparticle platforms have been used for displaying Env antigens [Citation188,Citation189]. VLPs based on the HIV-1 virion carry native Env spikes, but the intrinsic low density of spikes on the surface of HIV-1 virions negates some of the important immunological benefits of nanoparticle presentation. Furthermore, depending on their manufacturing process, HIV VLPs still present an abundance of misfolded Env, such as gp41 stumps or post-fusion Env, that present non-neutralizing epitopes [Citation190]. An interesting strategy is to enzymatically remove the unwanted Envs, leaving only well-folded Env trimers on the surface [Citation191–Citation193]. These virions still carry a low density of Envs, but they present a potentially more favorable antigenic profile since they display MPER epitopes in a native lipid environment. When immunizing rabbits with these VLPs, two out of eight induced potent Tier 2 autologous NAbs that targeted a hole in the glycan shield near the CD4bs [Citation96].
The use of artificial lipid-based vesicles called liposomes enables controlled immobilization of Env antigens on their surface. However, conventional immobilization procedures are rather stringent and lead to presentation of a percentage of non-native antigenic species. Therefore, His-tagged Env trimers were immobilized on NiNTA-functionalized liposomes for non-covalent, but nondestructive Env-liposome coupling. Such nanoparticles have shown some success in increasing binding [Citation194,Citation195] and neutralizing antibody titres [Citation91,Citation196]. However, it was found that NiNTA-coupled trimers completely dissociate in less than one day [Citation197]. Cobalt coupling and NiNTA-assisted covalent coupling by terminal cysteines [Citation198] have allowed more stable immobilization of Env trimers. These liposomal nanoparticles induced increased Env-specific binding responses in mice [Citation197,Citation198]. Future studies will teach us whether similar approaches can also be used in animals that induce NAbs [Citation199].
Multimerization of Env glycoproteins on viral-resembling particles has also been achieved by their fusion to bacterial self-assembling proteins, like ferritin from Helicobacter pylori, E2 protein from Geobacillus stearothermophilus and lumazine synthase from Aquifex aeolicus [Citation200,Citation201]. BG505.SOSIP antigens presented on ferritin nanoparticles induced stronger neutralizing responses than their soluble trimer counterparts in rabbits [Citation202,Citation203]. The autologous Tier 2 responses were not significantly increased, but the Tier 1A responses were, probably due to the presentation of linear V3 epitopes on non-native Env forms on ferritin particles [Citation202]. However, a next generation of synthetic two-component nanoparticles can be used to present only well-folded Env trimers [Citation204]. Using a two-component system, one could first purify the soluble Env trimers using specific quaternary specific bNAbs followed by addition of a second component to induce nanoparticle formation in vitro. This system has recently been used with success for generating RSV F nanoparticle vaccines and is thus a promising platform for HIV-1 Env trimers as well [Citation204–Citation206].
10. Genetic vaccination
Classical vaccinology has relied on the administration of live-attenuated pathogens or, later, recombinant protein antigens for the induction of immune responses. A more recent approach called genetic vaccination, which was pioneered almost 30 years ago [Citation207], consists of the administration of nucleic acids encoding for one or several target antigens that will be expressed by the cells of the vaccinee to elicit an immune response [Citation208–Citation210]. This approach has been explored to fight various diseases, including tuberculosis [Citation211] and cancer [Citation212]. Recently, a DNA-based vaccine against Zika virus showed promising immunogenicity and safety profiles both in mice and humans [Citation213,Citation214], with 70% of vaccinated volunteers showing 90% inhibition of Zika virus infection in neuronal-cell assays.
Gene-based vaccines are attractive because, compared to protein immunogens, nucleic acids are easier to design and manufacture at large scale and they require less strict storage conditions due to their higher stability and structure-independent activity. Furthermore, they usually present minimum side effects and in most cases the adjuvant is already contained within the sequence (i.e. unmethylated cytosine-guanine (CpG) motifs) or the adjuvating effect is carried out by the delivery method itself, e.g. in the case of DNA electroporation [Citation215,Citation216]. Nonetheless, the most important benefit of genetic vaccination is its ability to cast both humoral and cellular actors to the immunological play. Protein subunit antigens produced ex vivo elicit mostly antibody-based responses, as they are canonically presented on MHC class II after being processed by host cells. On the other hand, nucleic acid-derived antigens expressed by host antigen presenting cells (APCs) follow both MHC class I and class II pathways, thus stimulating both CD4 helper and CD8 cytotoxic T cell responses [Citation217]. Contrary to protein immunogens, genetic vaccines have also been shown to elicit long-term immune responses derived from the induction of strong effector and memory T follicular helper (Tfh) cells and thus might help to induce more durable NAb responses [Citation218–Citation220]. The expression of the antigen by host cells also allows host-specific modifications important for immunogenicity, such as N-linked glycosylation [Citation221]. Thus, Env antigens expressed by host cells present these modifications exactly as how these would be presented in the context of natural infection. The main drawback of genetic vaccination is that it usually induces lower antibody responses compared to adjuvanted protein subunit vaccines and that there is no control over the quality of the protein antigen being produced within the host.
Genetic vaccination englobes a wide variety of strategies depending on the type of nucleic acid used and the delivery platform. Both DNA and RNA have been widely used as antigen coding molecules. The most common delivery methods include electroporation of the naked nucleic acid, or the use of nanoparticles or viral vectors to deliver them to target cells, as reviewed in [Citation216,Citation222].
10.1. Non-viral vectored genetic vaccines
Current technologies have made it possible to efficiently deliver non-viral-vectored molecules to non-human primates and humans [Citation223–Citation228]. These immunogens present thus comparative benefits compared to viral-vectored ones, as they are not associated with preexisting immunity that can affect viral vector platforms [Citation229]. This is especially important if the approach is to be used as a therapeutic vaccine for seropositive individuals with a pre-mounted immune response [Citation221].
Since the dawn of genetic vaccine development, this approach has shown notable success in the protection of small animals and macaques against diverse pathogens, resulting in the licensure of several DNA vaccines to prevent veterinary infections such as West Nile virus in horses and infectious hematopoietic necrosis factor disease in salmon [Citation230,Citation231]. However, DNA immunogens usually induce suboptimal humoral responses in humans [Citation230]. Thus, DNA vaccines have been mainly explored in DNA prime-protein boost studies, in which DNA kick-starts both humoral and cellular responses that are then boosted by protein-based immunogens to induce higher titre Ab responses [Citation149,Citation232–Citation236]. Other immunization experiments have used DNA and protein co-immunization mixtures to increase the durability of humoral responses [Citation237–Citation239]. Jalah et al. used a SIVmac239 DNA construct and a sequence-matched protein immunogen to compare DNA only (D), DNA prime-protein boost (D-P) and DNA-protein co-immunization (D&P) regimens in rhesus macaques. The DNA only scheme induced no detectable antibody responses after two vaccinations. Both D-P and D&P strategies elicited robust Env-specific antibody responses. However, while the D-P titres showed a decay of 2.4 logs over 6 months, D&P induced Ab titres that showed no decay over a period of 8 months [Citation237]. DNA has also been successfully combined in prime-boost regimens with modified vaccinia Ankara (MVA) for HIV-1 vaccination experiments [Citation240,Citation241].
A relatively recent development are the lipid-nanoparticle-encapsulated nucleoside modified mRNAs (LNP-mRNAs) as vaccine candidates [Citation242,Citation243]. The marginal use of RNA until recently was related to its instability and rejection by anti-RNA innate immune mechanisms. However, LNP-mRNA immunogens include stabilizing groups as well as modified nucleosides that prevent immune sensing, and they are encapsulated for an increased and more durable protein expression. Thus, a single dose of LNP-mRNA encoding for Zika virus glycoproteins induced significantly higher NAb titres than DNA, which provided robust protection of vaccinated NHPs [Citation243]. Further characterization revealed that LNP-mRNA immunogens induced stronger Tfh cell responses, resulting in long-lived and high-affinity neutralizing antibody responses [Citation244]. Obviously, these LNP-mRNA immunogens could be also useful if they are applied to HIV-1 env vaccination. A first proof-of-concept HIV-1 Env LNP-mRNA vaccine candidate induced antibody responses in both rabbits and NHPs. However, the antibodies elicited could not neutralize the autologous Tier 2 virus in most animals [Citation245]. Furthermore, durable persistent expression of gene-based immunogens has been achieved by new delivery platforms, such as self-amplifying RNAs (saRNAs) [Citation246,Citation247]. The ability of these non-viral based molecules to replicate intracellularly allows for the sustained expression of high amounts of protein antigen. For instance, saRNA immunogens have been recently used to elicit long-lived T-cell responses using HIV-1 mosaic vaccines [Citation248].
10.2. Viral-vectored vaccines
Viral-vectored vaccines take advantage of genetically attenuated pathogen-based platforms that can be modified to display an antigen on their surface and/or deliver the genetic material encoding for it. Despite being attenuated to not cause disease, their natural capacity to infect cells of the vaccinee will help to assure that the antigen-encoding gene is delivered to the host cells and is expressed therein [Citation249,Citation250]. Moreover, viral-vectored vaccines are known to induce robust CD4 and CD8 T-cell responses [Citation249,Citation251–Citation254] . The major challenge for the use of these platforms is posed by preexisting immune responses, as humans are common hosts of the viral vectors, which is especially the case for some adenoviruses (AdV) serotypes [Citation255,Citation256]. Anti-Adenovirus 5 (AdV5) preexisting immunity forced the interruption of the STEP clinical trial after it was found that AdV5 seropositive vaccinees had an increased probability of HIV-1 infection [Citation257–Citation259]. However, the mechanism behind this phenomenon has not been completely elucidated yet [Citation259–Citation263].
Nonetheless, heterologous combinations of chimpanzee adenovirus Oxford 1 (ChAdOx1) [Citation264] and modified vaccinia Ankara (MVA) [Citation265] viral vectors have shown a good safety and immunogenicity profile [Citation266–Citation268]. Capucci et al. showed that a sequential regimen of ChAdOx1, MVA and soluble BG505 SOSIP trimer protein induced similar binding-antibody titres to the protein-only regimen, while only the sequential regimen induced cytotoxic T cell responses [Citation269]. These antibody titres also declined faster after immunization for the protein-only group. However, the homologous protein regimen induced more robust Tier 2 NAb titres than the ones that included ChAdOx1 and MVA [Citation269].
Integrase-defective lentiviral vectors (IDLV) are also very safe and have several advantages, such as induction of strong and long-lasting antigen-specific responses related to long DNA persistence in tissues [Citation270]. A single dose of IDLV encoding for HIV-1 transmitter/founder (T/F) 1086c Env induced specific Ab titres that waned by less than four-fold in a one-year period [Citation271]. Another dose after one year notably boosted both cellular and humoral responses [Citation271]. However, NAb titres were not detected.
All in all, genetic vaccination could play an important role in inducing long-lasting immune responses against HIV-1 Env, especially in conjunction with other immunogen forms, such as proteins. However, it is important to note that preexisting immunity against vectors can have detrimental effects. Furthermore, it is obviously not possible to control the quality of the Env antigens in vivo and probably a significant amount of Envs that is produced after genetic vaccination are low-quality non-native Envs that present potentially distracting, non-neutralizing epitopes. The next important challenge would be to generate an immunogen that is expressed as a fully native-like Env trimer after genetic vaccination in vivo.
11. Conclusion: an effective HIV-1 vaccine needs a combinatorial approach
Although the last decade has seen great progress with the discovery of potent bNAbs, there is no vaccine that is able to induce these bNAbs yet. The strategies described in this review could help to overcome the most important hurdles of HIV-1 Env vaccinology. Most conserved Env epitopes are subdominant and consensus, mosaic and epitope-focusing antigens are useful for directing NAb responses to these essential subdominant conserved epitopes. Recently, sequential immunization strategies have been developed that employ bNAb-lineage-based optimized antigens to guide specific naïve B cells toward breadth. Furthermore, multimerization of Env is an effective strategy to increase B cell activation and to stimulate more potent NAb responses. Lastly, genetic vaccination strategies induce stronger T cell responses that could help elicit more durable humoral responses. Obviously, none of these approaches are mutually exclusive and an effective combination is probably needed to elicit broad, potent and durable NAb responses against HIV-1.
Some of the strategies and immunogens aimed at inducing (b)NAbs have made it past laboratory experiments and animal testing and are currently being evaluated in human clinical trials. The immunogens being tested in humans include native-like Env trimers (BG505 and ConM SOSIP), germline-targeting immunogens (eOD-GT8 and BG505.SOSIP.GT1.1) and sequential immunogens (HVTN115 trial). The outcome of these clinical trials will inform the HIV-1 vaccinology field on the effectivity of these approaches and on the directions for the rest of the field.
12. Expert opinion
The generation of the first native-like Env trimers brought optimism to the HIV-1 vaccinology field and boosted structure-based design of different Env trimers that mimic the structure, antigenicity and biochemical properties of viral Env. Moreover, they have been useful for the discovery of a wide panel of bNAbs and the characterization of their developmental pathways. These virus-antibody coevolution studies, together with the repeated failures in inducing broad-spectrum NAb responses, taught us that a sole Env immunogen will probably never induce bNAb responses. A shepherding vaccine approach is therefore needed to guide the right B cells from naïve B cells to bNAb-producing B cells. However, only a small percentage of naïve germline precursor B cells in our repertoire have the potential to develop into mature bNAb-producing B cells and most Envs do not engage these germline B cells. Designed immunogens can effectively stimulate bNAbs B cell precursors in vitro and in knock-in mice that harbor the targeted germline BCR. Recent results of the adoptive transfer experiments and in non-human primates suggest that it is possible to activate the desired germline precursors when these are present in physiologically relevant numbers. The moment of truth has arrived to show that it is possible to steer these germline B cell responses toward breadth. Here, multimerization on nanoparticles is probably essential to provide enough affinity to specifically activate the sought bNAb cell lineages. Moreover, immunization schedules for guiding the B cell evolution to neutralization breadth involve different immunogens and these should not present potentially distracting non-neutralizing epitopes. Consensus sequence-based Env trimers are promising candidates for this, since these immunogens harbor less strain-specific epitopes that are potentially distracting. In this regard, our increased ability to modify and control Env glycosylation will help in focusing the responses to the selected epitope targets, by covering the non-interesting immunodominant native epitopes.
HIV-1 NAb responses wane quickly and this might be solved or mitigated by several of the above-mentioned strategies, such as genetic vaccination. However, the scarcity of studies directly testing effectivity of genetic vaccination is remarkable. Strategies described here, such as DNA-protein co-immunization [Citation237], LNP-mRNA immunogens [Citation244] and IDLV-vectored immunization [Citation271] provide long-lasting Ab responses compared to the ones usually observed after protein immunization. However, none of these studies directly compared the durability of these responses to standard protein immunization schedules. Furthermore, while most immunization studies assesses the breadth and/or potency of NAb responses, only very little attention is given to evaluating the durability of NAb responses. Thus, more studies are needed that systematically assess different strategies for inducing long-lived NAb responses and other elements involved in new candidate vaccines, such as adjuvants. Furthermore, extensive research on the immune mechanisms induced by successful and long-lasting antiviral vaccines (i.e. hepatitis B virus vaccine) [Citation272] might help to design vaccination strategies that provide durable protection against HIV-1.
HIV-1 is an escape artist and it poses one of the greatest challenges in vaccinology. This challenge forces the field to develop new strategies, combine the existing ones or adopt approaches used by other vaccinology fields. These integration efforts can only be beneficial for our future potential to generate new vaccines and will probably be the key to defeat HIV-1. Thus, it is vital that the HIV-1 vaccine field is open to new advances, since the necessary toolbox for an HIV-1 vaccine might not be complete yet. For instance, Kanekiyo et al. performed immunization experiments with ferritin nanoparticles co-displaying the conserved receptor-binding domains (RBDs) from multiple influenza hemagglutinin (HA) molecules on the same nanoparticle [Citation273], effectively combining the cocktail and epitope-focusing strategies with nanoparticle-presentation. The avidity effect of nanoparticle-presentation provided an even stronger advantage to the B cell lineages that targeted cross-neutralizing epitopes on the co-displayed RBDs, which led to broader NAb responses. Other recently developed or future technologies that we have not discussed, such as CRISPR/Cas9 genetic editing to produce bNAb-producing B cells for transplantation [Citation274–Citation277] or the use of osmotic pumps for the slow delivery of Env immunogens [Citation278], might also be key for the development of the long-sought HIV-1 vaccine.
Article highlights
An effective vaccine against HIV-1 should probably induce broadly neutralizing antibodies (bNAbs);
The HIV-1 envelope glycoprotein (Env) is the only target of bNAbs, but thus far no vaccine has been able to induce bNAbs in humans or relevant animal models;
Most Env immunogens induce narrow NAb responses that usually wane quickly after immunization;
Stabilized soluble Envs, epitope masking, artificial consensus sequences and sequential lineage-based vaccines are promising strategies that could improve or even broaden NAb responses;
Nanoparticle presentation and genetic vaccination could be used to increase the potency and durability of the NAb responses;
Approaches that combine these and other novel strategies will probably be needed for an effective HIV-1 vaccine that induces bNAbs.
Declaration of interest
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Reviewer disclosures
Peer reviewers on this manuscript have no relevant financial or other relationships to disclose
Acknowledgments
We would like to thank Prof. Rogier W. Sanders for reviewing and providing constructive criticism of this manuscript.
Additional information
Funding
References
- World Health Organization. WHO HIV update: global epidemic, progress in scale up and policy uptake. 2018.
- Beyrer C, Wirtz AL, O’Hara G, et al. The expanding epidemic of HIV-1 in the Russian federation. PLoS Med. 2017;14:e1002462.
- Plotkin SA. Correlates of protection induced by vaccination. Clin Vaccine Immunol. 2010;17:1055–1065.
- van Gils MJ, Sanders RW. In vivo protection by broadly neutralizing HIV antibodies. Trends Microbiol. 2014;22:550–551.
- Sok D, Burton DR. Recent progress in broadly neutralizing antibodies to HIV. Nat Immunol. 2018;19:1179–1188.
- Pauthner MG, Nkolola JP, Havenar-Daughton C, et al. Vaccine-induced protection from homologous tier 2 SHIV challenge in nonhuman primates depends on serum-neutralizing antibody titers. Immunity. 2019;50:241–252.e6.
- Corey L, Gilbert PB, Tomaras GD, et al. Immune correlates of vaccine protection against HIV-1 acquisition. Sci Transl Med. 2015;7:310rv7.
- Feng Y, Broder CC, Kennedy PE, et al. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872.
- Maddon PJ, Dalgleish AG, McDougal JS, et al. The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell. 1986;47:333–348.
- McDougal JS, Kennedy MS, Sligh JM, et al. Binding of HTLV-III/LAV to T4+ T cells by a complex of the 110K viral protein and the T4 molecule. Science. 1986;231:382–385.
- Moore JP, Trkola A, Dragic T. Co-receptors for HIV-1 entry. Curr Opin Immunol. 1997;9:551–562.
- Gaschen B, Taylor J, Yusim K, et al. Diversity considerations in HIV-1 vaccine selection. Science. 2002;296:2354–2360.
- Korber B, Gaschen B, Yusim K, et al. Evolutionary and immunological implications of contemporary HIV-1 variation. Br Med Bull. 2001;58:19–42.
- Kwong PD, Doyle ML, Casper DJ, et al. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature. 2002;420:678.
- Wei X, Decker JM, Wang S, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307.
- West AP Jr, Scharf L, Scheid JF, et al. Structural insights on the role of antibodies in HIV-1 vaccine and therapy. Cell. 2014;156:633–648.
- Seabright GE, Doores KJ, Burton DR, et al. Protein and glycan mimicry in HIV vaccine design. J Mol Biol. 2019;431:2223–2247.
- Hraber P, Seaman MS, Bailer RT, et al. Prevalence of broadly neutralizing antibody responses during chronic HIV-1 infection. AIDS Lond Engl. 2014;28:163–169.
- Stiegler G, Kunert R, Purtscher M, et al. A potent cross-clade neutralizing human monoclonal antibody against a novel epitope on gp41 of human immunodeficiency virus type 1. AIDS Res Hum Retroviruses. 2001;17:1757–1765.
- Burton DR, Pyati J, Koduri R, et al. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266:1024–1027.
- Kwong PD, Mascola JR. HIV-1 vaccines based on antibody identification, B cell ontogeny, and epitope structure. Immunity. 2018;48:855–871.
- McCoy LE. The expanding array of HIV broadly neutralizing antibodies. Retrovirology. 2018;15:70.
- Derking R, Ozorowski G, Sliepen K, et al. Comprehensive antigenic map of a cleaved soluble HIV-1 envelope trimer. PLOS Pathog. 2015;11:1–22.
- McCoy LE, Burton DR. Identification and specificity of broadly neutralizing antibodies against HIV. Immunol Rev. 2017;275:11–20.
- Dashti A, DeVico AL, Lewis GK, et al. Broadly neutralizing antibodies against HIV: back to blood. Trends Mol Med. 2019;25:228–240.
- Haynes BF, Kelsoe G, Harrison SC, et al. B-cell-lineage immunogen design in vaccine development with HIV-1 as a case study. Nat Biotechnol. 2013;30:423–433.
- Rantalainen K, Berndsen ZT, Murrell S, et al. Co-evolution of HIV envelope and apex-targeting neutralizing antibody lineage provides benchmarks for vaccine design. Cell Rep. 2018;23:3249–3261.
- Crispin M, Bowden TA. Antibodies expose multiple weaknesses in the glycan shield of HIV. Nat Struct Amp Mol Biol. 2013;20:771.
- Lee JH, Andrabi R, Su C-Y, et al. A broadly neutralizing antibody targets the dynamic HIV envelope trimer apex via a long, rigidified, and anionic β-hairpin structure. Immunity. 2017;46:690–702.
- Huang J, Kang BH, Ishida E, et al. Identification of a CD4-binding-site antibody to HIV that evolved near-pan neutralization breadth. Immunity. 2016;45:1108–1121.
- Wu X, Yang Z-Y, Li Y, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861.
- Haynes BF, Fleming J, St. Clair EW, et al. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science. 2005;308:1906.
- Kelsoe G, Haynes BF. Host controls of HIV broadly neutralizing antibody development. Immunol Rev. 2017;275:79–88.
- Liu M, Yang G, Wiehe K, et al. Polyreactivity and autoreactivity among HIV-1 antibodies. Silvestri G, editor. J Virol. 2015;89:784–798.
- Moody MA, Pedroza-Pacheco I, Vandergrift NA, et al. Immune perturbations in HIV-1–infected individuals who make broadly neutralizing antibodies. Sci Immunol. 2016;1:aag0851.
- Williams WB, Zhang J, Jiang C, et al. Initiation of HIV neutralizing B cell lineages with sequential envelope immunizations. Nat Commun. 2017;8:1732.
- McGuire AT, Gray MD, Dosenovic P, et al. Specifically modified Env immunogens activate B-cell precursors of broadly neutralizing HIV-1 antibodies in transgenic mice. Nat Commun. 2016;7:10618.
- Finney J, Kelsoe G. Poly- and autoreactivity of HIV-1 bNAbs: implications for vaccine design. Retrovirology. 2018;15:53.
- Zhang R, Verkoczy L, Wiehe K, et al. Initiation of immune tolerance-controlled HIV gp41 neutralizing B cell lineages. Sci Transl Med. 2016;8:336ra62.
- Verkoczy L, Diaz M, Holl TM, et al. Autoreactivity in an HIV-1 broadly reactive neutralizing antibody variable region heavy chain induces immunologic tolerance. Proc Natl Acad Sci U S A. 2010;107:181–186.
- Doyle-Cooper C, Hudson KE, Cooper AB, et al. Immune tolerance negatively regulates B cells in knock-in mice expressing broadly neutralizing HIV antibody 4E10. J Immunol. 2013;191:3186–3191.
- Khan T, Salunke DM. Adjustable locks and flexible keys: plasticity of epitope-paratope interactions in germline antibodies. J Immunol. 2014;192:5398–5405.
- Rudicell RS, Kwon YD, Ko S-Y, et al. Enhanced potency of a broadly neutralizing HIV-1 antibody in vitro improves protection against lentiviral infection in vivo. J Virol. 2014;88:12669–12682.
- Prigent J, Jarossay A, Planchais C, et al. Conformational plasticity in broadly neutralizing HIV-1 antibodies triggers polyreactivity. Cell Rep. 2018;23:2568–2581.
- Kaur H, Salunke DM. Antibody promiscuity: understanding the paradigm shift in antigen recognition. IUBMB Life. 2015;67:498–505.
- Sliepen K, Sanders RW. HIV-1 envelope glycoprotein immunogens to induce broadly neutralizing antibodies. Expert Rev Vaccines. 2016;15:349–365.
- Hart MK, Palker TJ, Matthews TJ, et al. Synthetic peptides containing T and B cell epitopes from human immunodeficiency virus envelope gp120 induce anti-HIV proliferative responses and high titers of neutralizing antibodies in rhesus monkeys. J Immunol. 1990;145:2677.
- Javaherian K, Langlois A, LaRosa G, et al. Broadly neutralizing antibodies elicited by the hypervariable neutralizing determinant of HIV-1. Science. 1990;250:1590.
- Montefiori DC, Roederer M, Morris L, et al. Neutralization tiers of HIV-1. Curr Opin HIV AIDS. 2018;13:128–136.
- Seaman MS, Janes H, Hawkins N, et al. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J Virol. 2010;84:1439–1452.
- Munro JB, Gorman J, Ma X, et al. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science. 2014;346:759.
- Huang J, Ofek G, Laub L, et al. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature. 2012;491:406–412.
- Williams LD, Ofek G, Schätzle S, et al. Potent and broad HIV-neutralizing antibodies in memory B cells and plasma. Sci Immunol. 2017;2(7):eaal2200.
- Matyas GR, Wieczorek L, Beck Z, et al. Neutralizing antibodies induced by liposomal HIV-1 glycoprotein 41 peptide simultaneously bind to both the 2F5 or 4E10 epitope and lipid epitopes. AIDS Lond Engl. 2009;23:2069–2077.
- Watson DS, Szoka FCJ. Role of lipid structure in the humoral immune response in mice to covalent lipid-peptides from the membrane proximal region of HIV-1 gp41. Vaccine. 2009;27:4672–4683.
- Mascola JR, Snyder SW, Weislow OS, et al. Immunization with envelope subunit vaccine products elicits neutralizing antibodies against laboratory-adapted but not primary isolates of human immunodeficiency virus type 1. J Infect Dis. 1996;173:340–348.
- Sanders RW, van Gils MJ, Derking R, et al. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science. 2015;349:aac4223.
- Wyatt R, Kwong PD, Desjardins E, et al. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393:705–711.
- Kovacs JM, Nkolola JP, Peng H, et al. HIV-1 envelope trimer elicits more potent neutralizing antibody responses than monomeric gp120. Proc Natl Acad Sci U S A. 2012;109:12111–12116.
- Yasmeen A, Ringe R, Derking R, et al. Differential binding of neutralizing and non-neutralizing antibodies to native-like soluble HIV-1 Env trimers, uncleaved Env proteins, and monomeric subunits. Retrovirology. 2014;11:41.
- Tran K, Poulsen C, Guenaga J, et al. Vaccine-elicited primate antibodies use a distinct approach to the HIV-1 primary receptor binding site informing vaccine redesign. Proc Natl Acad Sci U S A. 2014;111:E738–747.
- Chen H, Xu X, Jones IM. Immunogenicity of the outer domain of a HIV-1 clade C gp120. Retrovirology. 2007;4:33.
- Bhattacharyya S, Rajan RE, Swarupa Y, et al. Design of a non-glycosylated outer domain-derived HIV-1 gp120 immunogen that binds to CD4 and induces neutralizing antibodies. J Biol Chem. 2010;285:27100–27110.
- Qin Y, Banasik M, Kim S, et al. Eliciting neutralizing antibodies with gp120 outer domain constructs based on M-group consensus sequence. Virology. 2014;462–463:363–376.
- Joyce MG, Kanekiyo M, Xu L, et al. Outer domain of HIV-1 gp120: antigenic optimization, structural malleability, and crystal structure with antibody VRC-PG04. J Virol. 2013;87:2294–2306.
- Jardine J, Julien J-P, Menis S, et al. Rational HIV immunogen design to target specific germline B cell receptors. Science. 2013;340:711–716.
- McGuire AT, Hoot S, Dreyer AM, et al. Engineering HIV envelope protein to activate germline B cell receptors of broadly neutralizing anti-CD4 binding site antibodies. J Exp Med. 2013;210:655–663.
- Forsell MNE, Schief WR, Wyatt RT. Immunogenicity of HIV-1 envelope glycoprotein oligomers. Curr Opin HIV AIDS. 2009;4:380–387.
- Sanders RW, Moore JP. Native-like Env trimers as a platform for HIV-1 vaccine design. Immunol Rev. 2017;275:161–182.
- Liu J, Bartesaghi A, Borgnia MJ, et al. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455:109–113.
- Ringe RP, Sanders RW, Yasmeen A, et al. Cleavage strongly influences whether soluble HIV-1 envelope glycoprotein trimers adopt a native-like conformation. Proc Natl Acad Sci. 2013;110:18256.
- Du SX, Idiart RJ, Mariano EB, et al. Effect of trimerization motifs on quaternary structure, antigenicity, and immunogenicity of a noncleavable HIV-1 gp140 envelope glycoprotein. Virology. 2009;395:33–44.
- Sliepen K, van Montfort T, Melchers M, et al. Immunosilencing a highly immunogenic protein trimerization domain. J Biol Chem. 2015;290:7436–7442.
- Binley JM, Sanders RW, Clas B, et al. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J Virol. 2000;74:627.
- Sanders RW, Vesanen M, Schuelke N, et al. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J Virol. 2002;76:8875–8889.
- Binley JM, Sanders RW, Master A, et al. Enhancing the proteolytic maturation of human immunodeficiency virus type 1 envelope glycoproteins. J Virol. 2002;76:2606.
- Klasse PJ, Depetris RS, Pejchal R, et al. Influences on trimerization and aggregation of soluble, cleaved HIV-1 SOSIP envelope glycoprotein. J Virol. 2013;87:9873.
- Sanders RW, Derking R, Cupo A, et al. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLOS Pathog. 2013;9:1–20.
- Julien J-P, Cupo A, Sok D, et al. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1477.
- Lyumkis D, Julien J-P, de Val N, et al. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1484.
- Lee JH, Ozorowski G, Ward AB. Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science. 2016;351:1043.
- McCoy LE, van Gils MJ, Ozorowski G, et al. Holes in the glycan shield of the native HIV envelope are a target of trimer-elicited neutralizing antibodies. Cell Rep. 2016;16:2327–2338.
- Klasse PJ, LaBranche CC, Ketas TJ, et al. Sequential and simultaneous immunization of rabbits with HIV-1 envelope glycoprotein SOSIP.664 trimers from clades A, B and C. PLOS Pathog. 2016;12:1–31.
- Torrents de la Peña A, Sanders RW. Stabilizing HIV-1 envelope glycoprotein trimers to induce neutralizing antibodies. Retrovirology. 2018;15:63.
- Georgiev IS, Joyce MG, Yang Y, et al. Single-chain soluble BG505.SOSIP gp140 trimers as structural and antigenic mimics of mature closed HIV-1 Env. J Virol. 2015;89:5318.
- Sharma SK, de Val N, Bale S, et al. Cleavage-independent HIV-1 Env trimers engineered as soluble native spike mimetics for vaccine design. Cell Rep. 2015;11:539–550.
- Kong L, He L, De Val N, et al. Uncleaved prefusion-optimized gp140 trimers derived from analysis of HIV-1 envelope metastability. Nat Commun. 2016;7:1–15.
- Guenaga J, Garces F, de Val N, et al. Glycine substitution at helix-to-coil transitions facilitates the structural determination of a stabilized subtype C HIV envelope glycoprotein. Immunity. 2017;46:792–803.e3.
- Sarkar A, Bale S, Behrens A-J, et al. Structure of a cleavage-independent HIV Env recapitulates the glycoprotein architecture of the native cleaved trimer. Nat Commun. 2018;9:1956.
- Aldon Y, McKay PF, Allen J, et al. Rational design of DNA-expressed stabilized native-like HIV-1 envelope trimers. Cell Rep. 2018;24:3324–3338.e5.
- Martinez-Murillo P, Tran K, Guenaga J, et al. Particulate array of well-ordered HIV clade C Env trimers elicits neutralizing antibodies that display a unique V2 cap approach. Immunity. 2017;46:804–817.e7.
- Julien J-P, Lee JH, Ozorowski G, et al. Design and structure of two HIV-1 clade C SOSIP.664 trimers that increase the arsenal of native-like Env immunogens. Proc Natl Acad Sci. 2015;112:11947.
- Pugach P, Ozorowski G, Cupo A, et al. A native-like SOSIP.664 trimer based on an HIV-1 subtype B env gene. J Virol. 2015;89:3380–3395.
- Torrents de la Peña A, de Taeye SW, Sliepen K, et al. Immunogenicity in rabbits of HIV-1 SOSIP trimers from clades A, B, and C, given individually, sequentially, or in combination. J Virol. 2018;92:1–15.
- Wagh K, Kreider EF, Li Y, et al. Completeness of HIV-1 envelope glycan shield at transmission determines neutralization breadth. Cell Rep. 2018;25:893–908.e7.
- Crooks ET, Tong T, Chakrabarti B, et al. Vaccine-elicited tier 2 HIV-1 neutralizing antibodies bind to quaternary epitopes involving glycan-deficient patches proximal to the CD4 binding site. PLOS Pathog. 2015;11:1–34.
- Klasse PJ, Ketas TJ, Cottrell CA, et al. Epitopes for neutralizing antibodies induced by HIV-1 envelope glycoprotein BG505 SOSIP trimers in rabbits and macaques. PLOS Pathog. 2018;14:1–20.
- Zhou T, Doria-Rose NA, Cheng C, et al. Quantification of the impact of the HIV-1-glycan shield on antibody elicitation. Cell Rep. 2017;19:719–732.
- Behrens A-J, Vasiljevic S, Pritchard LK, et al. Composition and antigenic effects of individual glycan sites of a trimeric HIV-1 envelope glycoprotein. Cell Rep. 2016;14:2695–2706.
- Cao L, Pauthner M, Andrabi R, et al. Differential processing of HIV envelope glycans on the virus and soluble recombinant trimer. Nat Commun. 2018;9:3693.
- Struwe WB, Chertova E, Allen JD, et al. Site-specific glycosylation of virion-derived HIV-1 Env is mimicked by a soluble trimeric immunogen. Cell Rep. 2018;24:1958–1966.e5.
- Torrents de la Peña A, Julien J-P, de Taeye SW, et al. Improving the immunogenicity of native-like HIV-1 envelope trimers by hyperstabilization. Cell Rep. 2017;20:1805–1817.
- Hu JK, Crampton JC, Cupo A, et al. Murine antibody responses to cleaved soluble HIV-1 envelope trimers are highly restricted in specificity. Silvestri G, editor. J Virol. 2015;89:10383.
- Bianchi M, Turner HL, Nogal B, et al. Electron-microscopy-based epitope mapping defines specificities of polyclonal antibodies elicited during HIV-1 BG505 envelope trimer immunization. Immunity. 2018;49:288–300.e8.
- Kulp DW, Steichen JM, Pauthner M, et al. Structure-based design of native-like HIV-1 envelope trimers to silence non-neutralizing epitopes and eliminate CD4 binding. Nat Commun. 2017;8:1655.
- Ringe RP, Pugach P, Cottrell CA, et al. Closing and opening holes in the glycan shield of HIV-1 envelope glycoprotein SOSIP trimers can redirect the neutralizing antibody response to the newly unmasked epitopes. J Virol. 2019;93:e01656-18.
- Zhang P, Gorman J, Geng H, et al. Interdomain stabilization impairs CD4 binding and improves immunogenicity of the. Cell Host Microbe. 2018;23:832–844.e6.
- De Taeye SW, Ozorowski G, Torrents De La Peña A, et al. Immunogenicity of stabilized HIV-1 envelope trimers with reduced exposure of non-neutralizing epitopes. Cell. 2015;163:1702–1715.
- Duan H, Chen X, Boyington JC, et al. Glycan masking focuses immune responses to the HIV-1 CD4-binding site and enhances elicitation of VRC01-class precursor antibodies. Immunity. 2018;49:301–311.e5.
- Silva M, Nguyen TH, Philbrook P, et al. Targeted elimination of immunodominant B cells drives the germinal center reaction toward subdominant epitopes. Cell Rep. 2017;21:3672–3680.
- Ellenberger DL, Li B, Lupo LD, et al. Generation of a consensus sequence from prevalent and incident HIV-1 infections in West Africa to guide AIDS vaccine development. Virology. 2002;302:155–163.
- Mullins JI, Nickle DC, Heath L, et al. Immunogen sequence: the fourth tier of AIDS vaccine design. Expert Rev Vaccines. 2004;3:S151–159.
- Nickle DC, Jensen MA, Gottlieb GS, et al. Consensus and ancestral state HIV vaccines. Science. 2003;299:1515–1518.
- Novitsky V, Smith UR, Gilbert P, et al. Human immunodeficiency virus type 1 subtype C molecular phylogeny : consensus sequence for an AIDS vaccine design? J Virol. 2002;76:5435–5451.
- Doria-Rose NA, Learn GH, Rodrigo AG, et al. Human Immunodeficiency virus type 1 subtype B ancestral envelope protein is functional and elicits neutralizing antibodies in rabbits similar to those elicited by a circulating subtype B envelope. J Virol. 2005;79:11214–11224.
- Gao F, Weaver EA, Lu Z, et al. Antigenicity and immunogenicity of a synthetic human immunodeficiency virus type 1 group M consensus envelope glycoprotein. J Virol. 2005;79:1154–1163.
- Kothe DL, Li Y, Decker JM, et al. Ancestral and consensus envelope immunogens for HIV-1 subtype C. Virology. 2006;352:438–449.
- Kothe DL, Decker JM, Li Y, et al. Antigenicity and immunogenicity of HIV-1 consensus subtype B envelope glycoproteins. Virology. 2007;360:218–234.
- Liao H, Sutherland LL, Xia S-M, et al. A group M consensus envelope glycoprotein induces antibodies that neutralize subsets of subtype B and C HIV-1 primary viruses. Virology. 2006;353:268–282.
- Weaver EA, Lu Z, Camacho ZT, et al. Cross-subtype T-cell immune responses induced by a human immunodeficiency virus type 1 group M consensus Env immunogen. J Virol. 2006;80:6745–6756.
- Liao H-X, Tsao C-Y, Alam SM, et al. Antigenicity and immunogenicity of transmitted/ founder,consensus, and chronic envelope glycoproteins of human immunodeficiency virus type 1. J Virol. 2013;87:4185–4201.
- Rutten L, Lai YT, Blokland S, et al. A universal approach to optimize the folding and stability of prefusion-closed HIV-1 envelope trimers. Cell Rep. 2018;23:584–595.
- Sliepen K, Han BW, Bontjer I, et al. Structure and immunogenicity of a stabilized HIV-1 envelope trimer based on a group-M consensus sequence. Nat Commun. 2019;10:2355.
- Fischer W, Perkins S, Theiler J, et al. Polyvalent vaccines for optimal coverage of potential T-cell epitopes in global HIV-1 variants. Nat Med. 2007;13:100–106.
- Palesch D, Kirchhoff F. First steps toward a globally effective HIV/AIDS vaccine. Cell. 2013;155:495–497.
- Barouch DH, Brien KLO, Simmons NL, et al. Mosaic HIV-1 vaccines expand the breadth and depth of cellular immune responses in rhesus monkeys. Nat Med. 2010;16:319–323.
- Barouch DH, Stephenson KE, Borducchi EN, et al. Protective efficacy of a global HIV-1 mosaic vaccine against heterologous SHIV challenges in rhesus monkeys. Cell. 2013;155:531–539.
- Kong W, Wu L, Wallstrom TC, et al. Expanded breadth of the T-cell response to mosaic human immunodeficiency virus type 1 envelope DNA vaccination. J Virol. 2009;83:2201–2215.
- Santra S, Liao H, Zhang R, et al. Mosaic vaccines elicit CD8 + T lymphocyte responses that confer enhanced immune coverage of diverse HIV strains in monkeys. Nat Med. 2010;16:324–328.
- Santra S, Muldoon M, Watson S, et al. Breadth of cellular and humoral immune responses elicited in rhesus monkeys by multi-valent mosaic and consensus immunogens. Virology. 2012;428:121–127.
- Wee EG, Ondondo B, Berglund P, et al. HIV-1 conserved mosaics delivered by regimens with integration-deficient DC-targeting lentiviral vector induce robust T cells. Mol Ther. 2017;25:494–503.
- Korber BT, Letvin NL, Haynes BF. T-cell vaccine strategies for human immunodeficiency virus, the virus with a thousand faces. J Virol. 2009;83:8300–8314.
- Li F, Malhotra U, Gilbert PB, et al. Peptide selection for human immunodeficiency virus type 1 CTL-based vaccine evaluation. Vaccine. 2006;24:6893–6904.
- Ondondo B, Murakoshi H, Clutton G, et al. Novel conserved-region T-cell mosaic vaccine with high global HIV-1 coverage is recognized by protective responses in untreated infection. Mol Ther. 2016;24:832–842.
- Ahmed T, Borthwick NJ, Gilmour J, et al. Control of HIV-1 replication in vitro by vaccine-induced human CD8(+) T cells through conserved subdominant Pol epitopes. Vaccine. 2016;34:1215–1224.
- Borthwick N, Ahmed T, Ondondo B, et al. Vaccine-elicited human T cells recognizing conserved protein regions inhibit HIV-1. Mol Ther J Am Soc Gene Ther. 2014;22:464–475.
- Hulot SL, Korber B, Giorgi EE, et al. Comparison of Immunogenicity in rhesus macaques of transmitted- founder, HIV-1 group M consensus, and trivalent mosaic envelope vaccines formulated as a DNA prime, NYVAC, and envelope protein boost. J Virol. 2015;89:6462–6480.
- Nkolola JP, Bricault CA, Cheung A, et al. Characterization and immunogenicity of a novel mosaic M HIV-1 gp140 trimer. J Virol. 2014;88:9538.
- Korber B, Gnanakaran S. The implications of patterns in HIV diversity for neutralizing antibody induction and susceptibility. Curr Opin HIV AIDS. 2009;4:408–417.
- Euler Z, van Gils MJ, Boeser-Nunnink BD, et al. Genome-wide association study on the development of cross-reactive neutralizing antibodies in HIV-1 infected individuals. Plos One. 2013;8:1–7.
- Piantadosi A, Panteleeff D, Blish CA, et al. Breadth of neutralizing antibody response to human immunodeficiency virus type 1 is affected by factors early in infection but does not influence disease progression. J Virol. 2009;83:10269–10274.
- Rademeyer C, Moore PL, Taylor N, et al. Genetic characteristics of HIV-1 subtype C envelopes inducing cross-neutralizing antibodies. Virology. 2007;368:172–181.
- Chaudhury S, Reifman J, Wallqvist A. Simulation of B cell affinity maturation explains enhanced antibody cross-reactivity induced by the polyvalent malaria vaccine AMA1. J Immunol. 2014;193:2073–2086.
- Shaffer JS, Moore PL, Kardar M, et al. Optimal immunization cocktails can promote induction of broadly neutralizing Abs against highly mutable pathogens. Proc Natl Acad Sci U S A. 2016;113:E7039–E7048.
- Wang S, Mata-fink J, Kardar M, et al. Manipulating the selection forces during affinity maturation to generate cross-reactive HIV antibodies theory manipulating the selection forces during affinity maturation to generate cross-reactive HIV antibodies. Cell. 2015;160:785–797.
- Bricault CA, Kovacs JM, Nkolola JP, et al. A multivalent clade C HIV-1 Env trimer cocktail elicits a higher magnitude of neutralizing antibodies than any individual component. J Virol. 2015;89:2507–2519.
- Malherbe DC, Doria-Rose NA, Misher L, et al. Sequential immunization with a subtype B HIV-1 envelope quasispecies partially mimics the in vivo development of neutralizing antibodies. J Virol. 2011;85:5262–5274.
- Seaman MS, Xu L, Beaudry K, et al. Multiclade human immunodeficiency virus type 1 envelope immunogens elicit broad cellular and humoral immunity in rhesus monkeys. J Virol. 2005;79:2956–2963.
- Vaine M, Wang S, Hackett A, et al. Antibody responses elicited through homologous or heterologous prime-boost DNA and protein vaccinations differ in functional activity and avidity. Vaccine. 2010 February 17;28:2999–3007.
- Bricault CA, Yusim K, Seaman MS, et al. HIV-1 neutralizing antibody signatures and application to epitope-targeted vaccine design. Cell Host Microbe. 2019;25:59–72.e8.
- Voss JE, Andrabi R, McCoy LE, et al. Elicitation of neutralizing antibodies targeting the V2 Apex of the HIV envelope trimer in a wild-type animal model. Cell Rep. 2017;21:222–235.
- Bhiman JN, Anthony C, Doria-Rose NA, et al. Viral variants that initiate and drive maturation of V1V2-directed HIV-1 broadly neutralizing antibodies. Nat Med. 2015;21:1332–1336.
- Doria-Rose NA, Schramm CA, Gorman J, et al. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature. 2014;509:55–62.
- Liao H-X, Lynch R, Zhou T, et al. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 2013;496:469–476.
- Gao F, Bonsignori M, Liao H-X, et al. Cooperation of B cell lineages in induction of HIV-1-broadly neutralizing antibodies. Cell. 2014;158:481–491.
- Saunders KO, Verkoczy LK, Jiang C, et al. Vaccine induction of heterologous tier 2 HIV-1 neutralizing antibodies in animal models. Cell Rep. 2017;21:3681–3690.
- Doria-Rose NA, Bhiman JN, Roark RS, et al. New member of the V1V2-directed CAP256-VRC26 lineage that shows increased breadth and exceptional potency. J Virol. 2015;90:76–91.
- Landais E, Murrell B, Briney B, et al. HIV envelope glycoform heterogeneity and localized diversity govern the initiation and maturation of a V2 apex broadly neutralizing antibody lineage. Immunity. 2017;47:990–1003.e9.
- Moore PL, Gorman J, Doria-Rose NA, et al. Ontogeny-based immunogens for the induction of V2-directed HIV broadly neutralizing antibodies. Immunol Rev. 2017;275:217–229.
- Hoot S, McGuire AT, Cohen KW, et al. Recombinant HIV envelope proteins fail to engage germline versions of anti-CD4bs bNAbs. PLOS Pathog. 2013;9:1–14.
- Sliepen K, Medina-Ramírez M, Yasmeen A, et al. Binding of inferred germline precursors of broadly neutralizing HIV-1 antibodies to native-like envelope trimers. Virology. 2015;486:116–120.
- Medina-Ramírez M, Garces F, Escolano A, et al. Design and crystal structure of a native-like HIV-1 envelope trimer that engages multiple broadly neutralizing antibody precursors in vivo. J Exp Med. 2017;214:2573–2590.
- Steichen JM, Kulp DW, Tokatlian T, et al. HIV vaccine design to target germline precursors of glycan-dependent broadly neutralizing antibodies. Immunity. 2016;45:483–496.
- Dosenovic P, von Boehmer L, Escolano A, et al. Immunization for HIV-1 broadly neutralizing antibodies in human Ig knockin mice. Cell. 2015;161:1505–1515.
- Briney B, Sok D, Jardine JG, et al. Tailored immunogens direct affinity maturation toward HIV neutralizing antibodies article tailored immunogens direct affinity maturation toward HIV neutralizing antibodies. Cell. 2016;166:1459–1464.e11.
- Escolano A, Steichen JM, Dosenovic P, et al. Sequential immunization elicits broadly neutralizing anti-HIV-1 antibodies in Ig knockin mice. Cell. 2016;166:1445–1458.e12.
- Tian M, Cheng C, Chen X, et al. Induction of HIV neutralizing antibody lineages in mice with diverse precursor repertoires. Cell. 2016;166:1471–1484.e18.
- Abbott RK, Lee JH, Menis S, et al. Precursor frequency and affinity determine B cell competitive fitness in germinal centers, tested with germline-targeting HIV vaccine immunogens. Immunity. 2018;48:133–146.e6.
- Dosenovic P, Kara EE, Pettersson A-K, et al. Anti–HIV-1 B cell responses are dependent on B cell precursor frequency and antigen-binding affinity. Proc Natl Acad Sci. 2018;115:4743.
- Escolano A, Gristick HB, Abernathy ME, et al. Immunization expands B cells specific to HIV-1 V3 glycan in mice and macaques. Nature. 2019;570:468–473.
- Blattner C, Lee JH, Sliepen K, et al. Structural delineation of a quaternary, cleavage-dependent epitope at the gp41-gp120 interface on intact HIV-1 Env trimers. Immunity. 2014;40:669–680.
- van Gils MJ, van Den Kerkhof TLGM, Ozorowski G, et al. An HIV-1 antibody from an elite neutralizer implicates the fusion peptide as a site of vulnerability. Nat Microbiol. 2016;2:16199.
- Kong R, Xu K, Zhou T, et al. Fusion peptide of HIV-1 as a site of vulnerability to neutralizing antibody. Science. 2016;352:828.
- Lee JH, Leaman DP, Kim AS, et al. Antibodies to a conformational epitope on gp41 neutralize HIV-1 by destabilizing the Env spike. Nat Commun. 2015;6:8167.
- Dimitrov AS, Rawat SS, Jiang S, et al. Role of the fusion peptide and membrane-proximal domain in HIV-1 envelope glycoprotein-mediated membrane fusion. Biochemistry. 2003;42:14150–14158.
- Xu K, Acharya P, Kong R, et al. Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1. Nat Med. 2018;24:857–867.
- Kumar S, Sarkar A, Pugach P, et al. Capturing the inherent structural dynamics of the HIV-1 envelope glycoprotein fusion peptide. Nat Commun. 2019;10:763.
- Ananthaswamy N, Fang Q, AlSalmi W, et al. A sequestered fusion peptide in the structure of an HIV-1 transmitted founder envelope trimer. Nat Commun. 2019;10:873.
- Bachmann MF, Zinkernagel RM. The influence of virus structure on antibody responses and virus serotype formation. Immunol Today. 1996;17:553–558.
- Zhao Q, Li S, Yu H, et al. Virus-like particle-based human vaccines: quality assessment based on structural and functional properties. Trends Biotechnol. 2013;31:654–663.
- Bachmann MF, Jennings GT. Vaccine delivery: a matter of size, geometry, kinetics and molecular patterns. Nat Rev Immunol. 2010;10:787.
- Kelly HG, Kent SJ, Wheatley AK. Immunological basis for enhanced immunity of nanoparticle vaccines. Expert Rev Vaccines. 2019;18:269–280.
- Jardine JG, Ota T, Sok D, et al. Priming a broadly neutralizing antibody response to HIV-1 using a germline-targeting immunogen. Science. 2015;349:156.
- Temchura VV, Kozlova D, Sokolova V, et al. Targeting and activation of antigen-specific B-cells by calcium phosphate nanoparticles loaded with protein antigen. Biomaterials. 2014;35:6098–6105.
- Tokatlian T, Read BJ, Jones CA, et al. Innate immune recognition of glycans targets HIV nanoparticle immunogens to germinal centers. Science. 2018;9120:eaat9120.
- Schiller J, Chackerian B. Why HIV virions have low numbers of envelope spikes: implications for vaccine development. PLOS Pathog. 2014;10:1–4.
- Verkoczy L, Kelsoe G, Haynes BF. HIV-1 envelope gp41 broadly neutralizing antibodies: hurdles for vaccine development. PLOS Pathog. 2014;10:1–4.
- Aikins ME, Bazzill J, Moon JJ. Vaccine nanoparticles for protection against HIV infection. Nanomed. 2017;12:673–682.
- Gao Y, Wijewardhana C, Mann JFS. Virus-like particle, liposome, and polymeric particle-based vaccines against HIV-1. Front Immunol. 2018;9:345.
- Crooks ET, Moore PL, Franti M, et al. A comparative immunogenicity study of HIV-1 virus-like particles bearing various forms of envelope proteins, particles bearing no envelope and soluble monomeric gp120. Virology. 2007;366:245–262.
- Cale EM, Gorman J, Radakovich NA, et al. Virus-like particles identify an HIV V1V2 apex-binding neutralizing antibody that lacks a protruding loop. Immunity. 2017;46:777–791.e10.
- Crooks ET, Tong T, Osawa K, et al. Enzyme digests eliminate nonfunctional Env from HIV-1 particle surfaces, leaving native Env trimers intact and viral infectivity unaffected. J Virol. 2011;85:5825–5839.
- Tong T, Crooks ET, Osawa K, et al. HIV-1 virus-like particles bearing pure env trimers expose neutralizing epitopes but occlude nonneutralizing epitopes. J Virol. 2012;86:3574–3587.
- Leaman DP, Lee JH, Ward AB, et al. Immunogenic display of purified chemically cross-linked HIV-1 spikes. J Virol. 2015;89:6725–6745.
- Pejawar-Gaddy S, Kovacs JM, Barouch DH, et al. Design of lipid nanocapsule delivery vehicles for multivalent display of recombinant Env trimers in HIV vaccination. Bioconjug Chem. 2014;25:1470–1478.
- Ingale J, Stano A, Guenaga J, et al. High-density array of well-ordered HIV-1 spikes on synthetic liposomal nanoparticles efficiently activate B cells. Cell Rep. 2016;15:1986–1999.
- Tokatlian T, Kulp DW, Mutafyan AA, et al. Enhancing humoral responses against HIV envelope trimers via nanoparticle delivery with stabilized synthetic liposomes. Sci Rep. 2018;8:16527.
- Bale S, Goebrecht G, Stano A, et al. Covalent linkage of HIV-1 trimers to synthetic liposomes elicits improved B cell and antibody responses. J Virol. 2017;91:e00443-17.
- Pauthner M, Havenar-Daughton C, Sok D, et al. Elicitation of robust tier 2 neutralizing antibody responses in nonhuman primates by HIV envelope trimer immunization using optimized approaches. Immunity. 2017;46:1073–1088.e6.
- He L, Val ND, Morris CD, et al. Presenting native-like trimeric HIV-1 antigens with self-assembling nanoparticles. Nat Commun. 2016;7:1–15.
- López-sagaseta J, Malito E, Rappuoli R, et al. Self-assembling protein nanoparticles in the design of vaccines. CSBJ. 2016;14:58–68.
- Sliepen K, Ozorowski G, Burger JA, et al. Presenting native-like HIV-1 envelope trimers on ferritin nanoparticles improves their immunogenicity. Retrovirology. 2015;12:82.
- Brinkkemper M, Sliepen K. Nanoparticle vaccines for inducing HIV-1 neutralizing antibodies. Vaccines. 2019;7:76.
- Bale JB, Gonen S, Liu Y, et al. Accurate design of megadalton-scale two-component icosahedral protein complexes. Science. 2016;353:389.
- Marcandalli J, Fiala B, Ols S, et al. Induction of potent neutralizing antibody responses by a designed protein nanoparticle vaccine for respiratory syncytial virus. Cell. 2019;176:1420–1431.e17.
- Brouwer PJM, Sanders RW. Presentation of HIV-1 envelope glycoprotein trimers on diverse nanoparticle platforms. Curr Opin HIV AIDS. 2019;14:302–308.
- Wolff J, Malone R, Williams P, et al. Direct gene transfer into mouse muscle in vivo. Science. 1990;247:1465.
- Tang D, DeVit M, Johnston SA. Genetic immunization is a simple method for eliciting an immune response. Nature. 1992;356:152–154.
- Ulmer J, Donnelly J, Parker S, et al. Heterologous protection against influenza by injection of DNA encoding a viral protein. Science. 1993;259:1745.
- Wang B, Ugen KE, Srikantan V, et al. Gene inoculation generates immune responses against human immunodeficiency virus type 1. Proc Natl Acad Sci. 1993;90:4156.
- Bruffaerts N, Huygen K, Romano M. DNA vaccines against tuberculosis. Expert Opin Biol Ther. 2014;14:1801–1813.
- Yang B, Jeang J, Yang A, et al. DNA vaccine for cancer immunotherapy. Hum Vaccines Immunother. 2014;10:3153–3164.
- Gaudinski MR, Houser KV, Morabito KM, et al. Safety, tolerability, and immunogenicity of two Zika virus DNA vaccine candidates in healthy adults: randomised, open-label, phase 1 clinical trials. Lancet. 2018;391:552–562.
- Tebas P, Roberts CC, Muthumani K, et al. Safety and immunogenicity of an anti-zika virus DNA vaccine - preliminary report. N Engl J Med. 2017;NEJMoa1708120.
- Flingai S, Czerwonko M, Goodman J, et al. Synthetic DNA vaccines: improved vaccine potency by electroporation and co-delivered genetic adjuvants. Front Immunol. 2013;4:354.
- Saade F, Petrovski N. Technologies for enhanced efficacy of DNA vaccines. Expert Rev Vaccines. 2012;11:189–209.
- Gurunathan S, Stobie L, Prussin C, et al. Requirements for the maintenance of Th1 immunity in vivo following DNA vaccination: a potential immunoregulatory role for CD8+ T cells. J Immunol. 2000;165:915.
- Gurunathan S, Wu CY, Freidag BL, et al. DNA vaccines: a key for inducing long-term cellular immunity. Curr Opin Immunol. 2000;12:442–447.
- Méndez S, Gurunathan S, Kamhawi S, et al. The potency and durability of DNA- and protein-based vaccines against leishmania major evaluated using low-dose, intradermal challenge. J Immunol. 2001;166:5122–5128.
- Patel V, Valentin A, Kulkarni V, et al. Long-lasting humoral and cellular immune responses and mucosal dissemination after intramuscular DNA immunization. Vaccine. 2010;28:4827–4836.
- Weiner DB. RNA-based vaccination: sending a strong message. Mol Ther. 2013;21:506–508.
- Jorritsma SHT, Gowans EJ, Grubor-Bauk B, et al. Delivery methods to increase cellular uptake and immunogenicity of DNA vaccines. Vaccine. 2016;34:5488–5494.
- Felber BK, Valentin A, Rosati M, et al. HIV DNA vaccine: stepwise improvements make a difference. Vaccines. 2014;2:354–379.
- Kowalski PS, Rudra A, Miao L, et al. Delivering the messenger: advances in technologies for therapeutic mRNA delivery. Mol Ther. 2019;27:710–728.
- Pardi N, Hogan MJ, Porter FW, et al. mRNA vaccines - a new era in vaccinology. Nat Rev Drug Discov. 2018;17:261–279.
- Sahin U, Karikó K, Türeci Ö. mRNA-based therapeutics–developing a new class of drugs. Nat Rev Drug Discov. 2014;13:759–780.
- Weissman D. mRNA transcript therapy. Expert Rev Vaccines. 2015;14:265–281.
- Williams JA. Vector design for improved DNA vaccine efficacy, safety and production. Vaccines (Basel). 2013;1:225–249.
- MacGregor RR, Weiner DB, Boyer JD, et al. Safety and immune responses to a DNA-based human immunodeficiency virus (HIV) type I Env/Rev Vaccine in HIV-infected recipients: follow-up data. J Infect Dis. 2000;181:406.
- Klinman DM, Klaschik S, Tross D, et al. FDA guidance on prophylactic DNA vaccines: analysis and recommendations. Vaccine. 2010;28:2801–2805.
- Kutzler MA, Weiner DB. DNA vaccines: ready for prime time? Nat Rev Genet. 2008;9:776–788.
- Letvin NL, Montefiori DC, Yasutomi Y, et al. Potent, protective anti-HIV immune responses generated by bimodal HIV envelope DNA plus protein vaccination. Proc Natl Acad Sci. 1997;94:9378.
- Pal R, Wang S, Kalyanaraman VS, et al. Polyvalent DNA prime and envelope protein boost HIV-1 vaccine elicits humoral and cellular responses and controls plasma viremia in rhesus macaques following rectal challenge with an R5 SHIV isolate. J Med Primatol. 2005;34:226–236.
- Wang S, Pal R, Mascola JR, et al. Polyvalent HIV-1 Env vaccine formulations delivered by the DNA priming plus protein boosting approach are effective in generating neutralizing antibodies against primary human immunodeficiency virus type 1 isolates from subtypes A, B, C, D and E. Virology. 2006;350:34–47.
- Wise MC, Hutnick NA, Pollara J, et al. An enhanced synthetic multiclade DNA prime induces improved cross-clade-reactive functional antibodies when combined with an adjuvanted protein boost in nonhuman primates. J Virol. 2015;89:9154–9166.
- Zhang M, Zhang L, Zhang C, et al. DNA prime-protein boost using subtype consensus Env was effective in eliciting neutralizing antibody responses against subtype BC HIV-1 viruses circulating in China. Hum Vaccines Immunother. 2012;8:1630–1637.
- Jalah R, Kulkarni V, Patel V, et al. DNA and protein co-immunization improves the magnitude and longevity of humoral immune responses in macaques. Plos One. 2014;9:e91550.
- Pissani F, Malherbe DC, Schuman JT, et al. Improvement of antibody responses by HIV envelope DNA and protein co-immunization. Vaccine. 2014;32:507–513.
- Singh S, Ramírez-Salazar EG, Doueiri R, et al. Control of heterologous simian immunodeficiency virus SIVsmE660 infection by DNA and protein coimmunization regimens combined with different toll-like-receptor-4-based adjuvants in macaques. Kirchhoff F, editor. J Virol. 2018;92:e00281–18.
- Chea LS, Amara RR. Immunogenicity and efficacy of DNA/MVA HIV vaccines in rhesus macaque models. Expert Rev Vaccines. 2017;16:973–985.
- Shen X, Basu R, Sawant S, et al. HIV-1 gp120 and Modified Vaccinia Virus Ankara (MVA) gp140 boost immunogens increase immunogenicity of a DNA/MVA HIV-1 vaccine. Silvestri G, editor. J Virol. 2017;91:e01077–17.
- Cullis PR, Hope MJ. Lipid nanoparticle systems for enabling gene therapies. Mol Ther J Am Soc Gene Ther. 2017;25:1467–1475.
- Pardi N, Hogan MJ, Pelc RS, et al. Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination. Nature. 2017;543:248–251.
- Pardi N, Hogan MJ, Naradikian MS, et al. Nucleoside-modified mRNA vaccines induce potent T follicular helper and germinal center B cell responses. J Exp Med. 2018;215:1571–1588.
- Pardi N, LaBranche CC, Ferrari G, et al. Characterization of HIV-1 nucleoside-modified mRNA vaccines in rabbits and rhesus macaques. Mol Ther Nucleic Acids. 2019;15:36–47.
- Geall AJ, Verma A, Otten GR, et al. Nonviral delivery of self-amplifying RNA vaccines. Proc Natl Acad Sci. 2012;109:14604.
- Vogel AB, Lambert L, Kinnear E, et al. Self-amplifying RNA vaccines give equivalent protection against influenza to mRNA vaccines but at much lower doses. Mol Ther. 2018;26:446–455.
- Moyo N, Vogel AB, Buus S, et al. Efficient induction of T cells against conserved HIV-1 regions by mosaic vaccines delivered as self-amplifying mRNA. Mol Ther - Methods Clin Dev. 2019;12:32–46.
- Nayak S, Herzog RW. Progress and prospects: immune responses to viral vectors. Gene Ther. 2010;17:295–304.
- Ura T, Okuda K, Shimada M. Developments in viral vector-based vaccines. Vaccines (Basel). 2014;2:624–641.
- Harari A, Bart P-A, Stöhr W, et al. An HIV-1 clade C DNA prime, NYVAC boost vaccine regimen induces reliable, polyfunctional, and long-lasting T cell responses. J Exp Med. 2008;205:63–77.
- Rowe HM, Lopes L, Ikeda Y, et al. Immunization with a lentiviral vector stimulates both CD4 and CD8 T cell responses to an ovalbumin transgene. Mol Ther. 2006;13:310–319.
- Smaill F, Jeyanathan M, Smieja M, et al. A human type 5 adenovirus–based tuberculosis vaccine induces robust T cell responses in humans despite preexisting anti-adenovirus immunity. Sci Transl Med. 2013;5:205ra134.
- Webster DP, Dunachie S, Vuola JM, et al. Enhanced T cell-mediated protection against malaria in human challenges by using the recombinant poxviruses FP9 and modified vaccinia virus Ankara. Proc Natl Acad Sci U S A. 2005;102:4836–4841.
- Barouch DH, Kik SV, Weverling GJ, et al. International seroepidemiology of adenovirus serotypes 5, 26, 35, and 48 in pediatric and adult populations. Vaccine. 2011;29:5203–5209.
- Trojnar Z, Ciepiela O, Demkow UA. The prevalence of IgG and IgA against adenoviruses in serum of children aged 11–26 months, hospitalised in the clinical paediatric Hospital in Warsaw, Poland. Cent-Eur J Immunol. 2014;39:91–95.
- Buchbinder SP, Mehrotra DV, Duerr A, et al. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet. 2008;372:1881–1893.
- Duerr A, Huang Y, Buchbinder S, et al. Extended follow-up confirms early vaccine-enhanced risk of HIV acquisition and demonstrates waning effect over time among participants in a randomized trial of recombinant adenovirus HIV vaccine (Step Study). J Infect Dis. 2012;206:258–266.
- Gray G, Buchbinder S, Duerr A. Overview of STEP and Phambili trial results: two phase IIb test-of-concept studies investigating the efficacy of MRK adenovirus type 5 gag/pol/nef subtype B HIV vaccine. Curr Opin HIV AIDS. 2010;5:357–361.
- Benlahrech A, Harris J, Meiser A, et al. Adenovirus vector vaccination induces expansion of memory CD4 T cells with a mucosal homing phenotype that are readily susceptible to HIV-1. Proc Natl Acad Sci. 2009;106:19940.
- Curlin ME, Cassis-Ghavami F, Magaret AS, et al. Serological immunity to adenovirus serotype 5 is not associated with risk of HIV infection: a case-control study. AIDS Lond Engl. 2011;25:153–158.
- Day TA, Kublin JG. Lessons learned from HIV vaccine clinical efficacy trials. Curr HIV Res. 2013;11:441–449.
- Hutnick NA, Carnathan DG, Dubey SA, et al. Baseline Ad5 serostatus does not predict Ad5 HIV vaccine-induced expansion of adenovirus-specific CD4+ T cells. Nat Med. 2009;15:876–878.
- Dicks MDJ, Spencer AJ, Edwards NJ, et al. A novel chimpanzee adenovirus vector with low human seroprevalence: improved systems for vector derivation and comparative immunogenicity. Plos One. 2012;7:e40385.
- Drexler I, Staib C, Sutter G. Modified vaccinia virus Ankara as antigen delivery system: how can we best use its potential? Curr Opin Biotechnol. 2004;15:506–512.
- Brandler S, Lepelley A, Desdouits M, et al. Preclinical studies of a modified vaccinia virus ankara-based HIV candidate vaccine: antigen presentation and antiviral effect. J Virol. 2010;84:5314.
- Keefer MC, Frey SE, Elizaga M, et al. A phase I trial of preventive HIV vaccination with heterologous poxviral-vectors containing matching HIV-1 inserts in healthy HIV-uninfected subjects. Vaccine. 2011;29:1948–1958.
- Verheust C, Goossens M, Pauwels K, et al. Biosafety aspects of modified vaccinia virus Ankara (MVA)-based vectors used for gene therapy or vaccination. Vaccine. 2012;30:2623–2632.
- Capucci S, Wee EG, Schiffner T, et al. HIV-1-neutralizing antibody induced by simian adenovirus- and poxvirus MVA-vectored BG505 native-like envelope trimers. Plos One. 2017;12:1–18.
- Blasi M, Negri D, LaBranche C, et al. IDLV-HIV-1 Env vaccination in non-human primates induces affinity maturation of antigen-specific memory B cells. Commun Biol. 2018;1:134.
- Negri D, Blasi M, LaBranche C, et al. Immunization with an SIV-based IDLV expressing HIV-1 Env 1086 clade C elicits durable humoral and cellular responses in rhesus macaques. Mol Ther J Am Soc Gene Ther. 2016;24:2021–2032.
- Slifka MK, Amanna I. How advances in immunology provide insight into improving vaccine efficacy. Vaccine. 2014;32:2948–2957.
- Kanekiyo M, Joyce MG, Gillespie RA, et al. Mosaic nanoparticle display of diverse influenza virus hemagglutinins elicits broad B cell responses. Nat Immunol. 2019;20:362.
- Greiner V, Bou Puerto R, Liu S, et al. CRISPR-mediated editing of the B cell receptor in primary human B cells. iScience. 2019;12:369–378.
- Hartweger H, McGuire AT, Horning M, et al. HIV-specific humoral immune responses by CRISPR/Cas9-edited B cells. J Exp Med. 2019;216:1301–1310.
- Lin Y-C, Pecetta S, Steichen JM, et al. One-step CRISPR/Cas9 method for the rapid generation of human antibody heavy chain knock-in mice. Embo J. 2018;37:e99243.
- Voss JE, Gonzalez-Martin A, Andrabi R, et al. Reprogramming the antigen specificity of B cells using genome-editing technologies. Kurosaki T, Storz G, Yasuda T, editors. eLife. 2019;8:e42995.
- Cirelli KM, Carnathan DG, Nogal B, et al. Slow delivery immunization enhances HIV neutralizing antibody and germinal center responses via modulation of immunodominance. Cell. 2019;177:1153–1171.e28.
