A comprehensive insight into how each part of a protein work can result in a better understanding of its role in cells and biological activities. According to the classical structure-function paradigm, proteins following the translation process and based on their amino acid sequence, acquire a three-dimensional (3D) structure that channels them to a special function; a concept which proposed by Christian Anfinsen and is known as Anfinsen’s postulate or thermodynamic hypothesis [Citation1]. However, not all proteins need an ordered 3D structure to acquire a function. Numerous studies have revealed that a large number of protein segments lack in a defined 3D structure. These segments are called intrinsically disordered regions (IDRs) and their function relies on an unstructured conformation (disorder-function paradigm) [Citation2,Citation3]. The hydrophobic interactions between amino acids with long-distance in a linear polypeptide sequence are responsible for stable 3D structure [Citation4]. Hence, decrease in the content of hydrophobic amino acids combined with the increase in the number of charged and polar residues represent some of the characteristic features of a disordered polypeptide [Citation5] ().
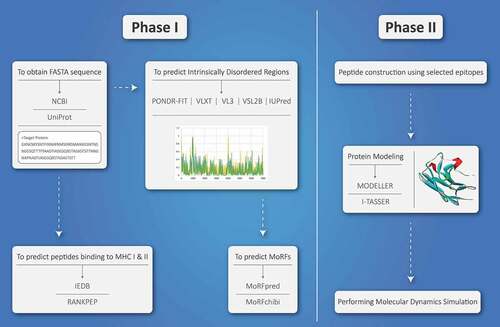
Figure 1. The diagram of vaccine design process using intrinsically disordered regions. Phase I, the desired protein sequence needs to be obtained from NCBI or UniProt servers. This sequence must be analyzed for the presence of intrinsically disordered regions (using PONDR-FIT, VLXT, VL3, VSL2B, and IUPred servers), MoRF regions (using MoRFpred and MorFchibi), and MHC I and II binding epitopes (IEDB and RANKPEP). Phase II, after reaching to the potential epitopes, the peptide sequence should be constructed. Moreover, three-dimensional structure of the final peptide must be predicted (using MODELER or I-TASSER). Furthermore, molecular dynamics simulation is used for evaluating the stability of final vaccine construct.
If the entire protein sequence was disordered, it would be referred as an intrinsically disordered protein (IDP) [Citation6]. Initially, it was assumed that IDRs are passive components in a protein and responsible for linking the structured domains. Interestingly, their role in various protein-mediated functions has been well established today. IDRs participate in signaling, gene regulation, and posttranslational modifications [Citation7,Citation8]. Furthermore, entirely disordered proteins and proteins with disordered regions often serve as important hubs in protein-protein interaction (PPI) networks [Citation9].
Along with all the characteristics of IDRs, their ability to stimulate the immune response makes them an attractive candidate for vaccine development. Generally, a new generation of peptide vaccines contain only the effective immunodominant epitopes for inducing a robust immune response. Therefore, the idea of using IDRs epitopes in developing peptide vaccines has been implemented in recent years [Citation10]. Since disordered epitopes are usually linear, it is ideal to employ them in peptide vaccine design. Furthermore, IDRs are abundant in proteomes of various pathogens and are especially common in viruses. There is a continual competition between viruses and their hosts. It should be noted that virus efforts are always in the path of defeating the host immune system. However, hosts are not given up, and they try to modify their strategy. Since this competition occurs at the molecular levels, IDRs can be a winning card here. Both viruses and hosts use these regions and proteins to conquer this war [Citation11]. Curiously, intrinsic disorder region is utilized by viruses to infect the host [Citation12], and by human antiviral proteins, proteins associated with innate immune response, and defense response to the virus [Citation11].
Traditionally, the higher antigenicity is dependent on the higher complexity of the antigen. In addition, some ideas challenge the ability of disordered epitopes to induce a powerful antibody response. This quarrel is based on the fact that IDRs only acquire a specific conformation when they interact with their target protein, such as an antibody. Consequently, in this situation despite the specificity of the binding, the affinity of interaction is frequently low. Nevertheless, in the signaling pathways the signal needs to be turned on and off quickly; therefore, the ability to change conformations makes IDPs to act as important mediators in many cases. Another feature of antigens is flexibility, which causes immune induction; in this context, various B-cell epitopes have been determined to be disordered with the ability to induce tailored immune responses [Citation13,Citation14]. A study demonstrated that disordered epitopes are smaller compare to ordered epitopes, which makes them suitable to interact efficiently with antibodies. Additionally, applying a large dataset of protein antigens revealed that disordered epitopes are authentic targets for being recognized by antibodies [Citation15].
Molecular recognition features (MoRFs) are particular types of functional IDRs that are disorder-based binding sites, which undergo disorder-to-order transition at interaction with their cognate proteins [Citation16]. These segments exist in IDRs and upon binding to the target molecule, they play an important role in effective interaction. These conformational changes are one of the main causes of argument about the immunogenicity of IDRs, accordingly, makes them inappropriate for vaccine development.
Several online servers and algorithms have been developed for prediction of IDRs and MoRF regions, including PONDR-FIT, VLXT, VL3, VSL2B, IUPred, and MoRFPred. Some studies have applied the disordered regions for developing efficient epitope vaccine ().
Table 1. The list of vaccines that employed disordered epitopes
It should be noted that IDRs are widely found in Plasmodium species that cause malaria. Two popular vaccines against Plasmodium falciparum were developed using IDRs. An effective one is RTS,S that is based on circumsporozoite protein (CSP). It is a major surface protein of Plasmodium falciparum sporozoite. To develop RTS,S vaccine the C-terminal of CSP is conjugated to the hepatitis B surface antigen. CSP contains various disordered epitopes, which are the targets of the antibodies stimulation after injecting RTS,S [Citation21]. Another vaccine against Plasmodium falciparum with the presence of disordered epitopes in its structure is MSP2-C1. This vaccine uses merozoite surface protein 2 (MSP2) that contains numerous intrinsically disordered regions [Citation22].
Expert opinion
The presence of MoRFs can affect the affinity of antigen-antibody interactions. There is a high possibility that the interaction between disordered epitopes and antibodies is done with reduced affinity. However, using a mixture of ordered and disordered epitopes in a peptide vaccine construct may modulate the recognition of disordered epitopes by antibodies. The presence of low-complexity regions and the ability to change the conformation should not remove disordered epitopes from the list of candidates for peptide vaccine development. Given that several peptide vaccines contain disordered epitopes, we should implement more studies to understand the nature of IDRs and their potential for designing vaccines. Since IDRs are abundant in numerous pathogens and viruses, their potential as vaccine candidates should not be neglected. If a fundamental solution to the use of IDRs in vaccine design is reached, it is possible that the efficiency of the final vaccine will improve. Notably, the intrinsic disorder in these proteins allows for frequent mutations without significant consequences, and they are a target for positive selection at least in mammals; this is an inherent weakness of IDR-based vaccine.
Anyhow, to achieve the best use of disordered epitopes in vaccine development, there is a need for further studies and trials.
Author contributions
All authors contributed to the conceptualization, design, writing, and editing of the article. All authors have read, reviewed, and approved the final paper.
Declaration of interest
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Reviewer disclosures
Peer reviewers on this manuscript have no relevant financial or other relationships to disclose.
Additional information
Funding
References
- Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973 Jul 20;181(4096):223–230.
- Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol. 1999 Oct 22;293(2):321–331.
- Kriwacki RW, Hengst L, Tennant L, et al. Structural studies of p21Waf1/Cip1/Sdi1 in the free and Cdk2-bound state: conformational disorder mediates binding diversity. Proc Natl Acad Sci U S A. 1996 Oct 15;93(21):11504–11509.
- Chothia C. Hydrophobic bonding and accessible surface area in proteins. Nature. 1974 Mar 01;248(5446):338–339.
- Schalch DS, Gonzalez-Barcena D, Kastin AJ, et al. Plasma gonadotropins after administration of LH-releasing hormone in patients with renal or hepatic failure. J Clin Endocrinol Metab. 1975 Nov;41(5):921–925.
- van der Lee R, Buljan M, Lang B, et al. Classification of intrinsically disordered regions and proteins. Chem Rev. 2014 Jul 9;114(13):6589–6631.
- Darling AL, Uversky VN. Intrinsic disorder and posttranslational modifications: the darker side of the biological dark matter. Front Genet. 2018;9:158.
- Wright PE, Dyson HJ. Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol. 2015 Jan;16(1):18–29.
- Dunker AK, Cortese MS, Romero P, et al. Flexible nets. The roles of intrinsic disorder in protein interaction networks. Febs J. 2005 Oct;272(20):5129–5148.
- Ramamurthy M, Sankar S, Abraham AM, et al. B cell epitopes in the intrinsically disordered regions of neuraminidase and hemagglutinin proteins of H5N1 and H9N2 avian influenza viruses for peptide‐based vaccine development. J Cell Biochem. 2019;120(10):17534–17544.
- Xue B, Uversky VN. Intrinsic disorder in proteins involved in the innate antiviral immunity: another flexible side of a molecular arms race. J Mol Biol. 2014 Mar 20;426(6):1322–1350.
- Xue B, Blocquel D, Habchi J, et al. Structural disorder in viral proteins. Chem Rev. 2014 Jul 9;114(13):6880–6911.
- Olugbile S, Kulangara C, Bang G, et al. Vaccine potentials of an intrinsically unstructured fragment derived from the blood stage-associated Plasmodium falciparum protein PFF0165c. Infect Immun. 2009;77(12):5701–5709.
- Adda CG, MacRaild CA, Reiling L, et al. Antigenic characterization of an intrinsically unstructured protein, plasmodium falciparum merozoite surface protein 2. Infect Immun. 2012;80(12):4177–4185.
- MacRaild Christopher A, Richards Jack S, Anders Robin F, et al. Antibody recognition of disordered antigens. Structure. 2016 Jan 05;24(1):148–157.
- Oldfield CJ, Cheng Y, Cortese MS, et al. Coupled folding and binding with alpha-helix-forming molecular recognition elements. Biochemistry. 2005 Sep 20;44(37):12454–12470.
- Foquet L, Hermsen CC, van Gemert GJ, et al. Vaccine-induced monoclonal antibodies targeting circumsporozoite protein prevent Plasmodium falciparum infection. J Clin Invest. 2014 Jan;124(1):140–144.
- Flück C, Smith T, Beck HP, et al. Strain-specific humoral response to a polymorphic malaria vaccine. Infect Immun. 2004 Nov;72(11):6300–6305.
- Cai X, Zheng W, Pan S, et al. A virus-like particle of the hepatitis B virus preS antigen elicits robust neutralizing antibodies and T cell responses in mice. Antiviral Res. 2018 Jan;149:48–57.
- Esposito X, Zheng V, Musi V, de Chiara C, et al. Structure of the C-terminal domain of Neisseria heparin binding antigen (NHBA), one of the main antigens of a novel vaccine against Neisseria meningitidis. Journal of Biological Chemistry. 2011 Jan;286(48): 41767–41775.
- Kaslow DC, Biernaux S. RTS,S: toward a first landmark on the Malaria Vaccine Technology Roadmap. Vaccine. 2015 Dec 22;33(52):7425–7432.
- Genton B, Betuela I, Felger I, et al. A recombinant blood-stage malaria vaccine reduces Plasmodium falciparum density and exerts selective pressure on parasite populations in a phase 1-2b trial in papua new guinea. J Infect Dis. 2002 Mar 15;185(6):820–827.
