Abstract
The roles of Biological Resource Centres (BRCs), such as the Culture Collection of Algae and Protozoa (CCAP), have extended beyond their traditional maintenance and provision of curated microorganisms to the user community. A major driver for change has been the exponential increase in metagenomics and environmental sequencing data over the last few years. This has underlined a critical requirement for molecular information on reference biological materials, which would allow better taxonomic interpretation, greater biological understanding and additional exploitation of these data. This is especially relevant for the relatively poorly studied protists (algal/autotrophic as well as heterotrophic) and prokaryotic cyanobacteria, which despite their huge biodiversity, reflected in the genomic data that has been generated, are under-represented in BRCs worldwide. Here we describe the functionalities of the Culture Collection of Algae and Protozoa (CCAP) KnowledgeBase (http://www.ccap.ac.uk), developed as a one-stop shop for quality-controlled biological material, hyperlinked to manually curated molecular, bibliographic and taxonomic information. This has been built around the CCAP live collection, which constitutes one of the most genotypically diverse collections in the world with representatives of all the major eukaryotic lineages and the cyanobacteria.
Introduction
The term ‘protist’ encompasses a phenotypically and genotypically diverse range of mostly unicellular eukaryotic organisms. Many such protistan taxa are capable of photoautotrophic growth, and are generally included as ‘algae’; however, many can grow mixotrophically and there are numerous heterotrophic protistan lineages that include free-living, commensal and parasitic organisms. Over the last two decades or so, molecular approaches have helped elucidate their phylogenetic relationships at higher taxonomic levels and revealed that protists include a much greater diversity of lineages than was previously appreciated (Keeling et al., Citation2005; Baldauf, 2008; Archibald, Citation2011). From the perspective of systematics it is reasonable to assert that protists are the most diverse eukaryotic group of organisms, but compared with macroscopic plants and animals, many protistan groups remain poorly characterized. This is not only because of their microscopic size and the lack of obvious defining characters in some taxa, but is also due to difficulties in isolation, cultivation and long-term maintenance. Their diversity has been further substantiated by molecular surveys and metagenomics initiatives that have consistently highlighted the extent of cryptic or unknown protistan diversity (e.g. Massana et al., Citation2006; Kim et al., Citation2011). Furthermore, DNA barcoding, whole genome and polyphasic approaches, using both phenotypic and genotypic characters, have shown that many traditional ‘morpho-taxa’ are comprised of a number of phylogenetically distinct clades (Müller et al., Citation2005; Degerlund et al., Citation2012; Rybalka et al., Citation2013).
In addition to their obvious evolutionary biodiversity and phylogenetic importance, autotrophic and heterotrophic protists play key roles in ecosystem function. Algae, including cyanobacteria drive a number of global biogeochemical processes, contributing > 50% of the world's photosynthetic activity and form the base of the food chain for > 70% of worldwide biomass (Falkowski et al., Citation2004; Chavez et al., Citation2011). Similarly, the multifaceted contributions of heterotrophic protists to nutrient cycles is increasingly being recognized including their key roles as consumers (Christaki et al., Citation1999; Hartmann et al., Citation2012), degraders/saprotrophs (Kimura et al., Citation2001) and pathogens (Chambouvet et al., Citation2008; Gachon et al., Citation2010). These tropic interactions are clearly crucial to ensuring ecosystem function and stability (Berninger, 1991; Steiner et al., Citation2006). At the base of pelagic marine food webs, microzooplankton dominate trophic interactions and biogeochemical processes. Thus, their responses to a changing ocean environment have potentially enormous implications for ocean ecosystem functioning (Caron et al., 2013). The term ‘algae’, formerly used in relation to organisms that were not something (fungi, liverworts, mosses, ferns, flowering plants, etc.) is now known to encompass a polyphyletic group of organisms that includes some of the smallest (e.g. the prokaryotic cyanophyte Prochlorococus, about 0.5 μm in diameter) and the largest (e.g. the phaeophyceaen seaweed Macrocystis, reaching 60 m in length) photosynthetic organisms on Earth. Their diverse evolutionary/systematic background is reflected in varied metabolic pathways, e.g. with species of coccolithophores, diatoms and unicellular green algae acquiring iron through various mechanisms (Morrissey & Bowler, Citation2012), biochemical adaptations such as unique and varied cell-wall carbohydrates (Popper et al., Citation2011), and an extraordinary halogen metabolism (Küpper et al., Citation2011). In the future, linking the genetic, molecular and environmental signatures of algae/protists to real specimens that can be manipulated experimentally, or used in mesocosm experimentation, will be crucial to the understanding of the processes involved. Furthermore, the elucidation of functional relationships between different taxa and within species will have implications, not only for blue-skies science/fundamental research, but also for potential applications in manipulating global biogeochemistry (‘geoengineering’) and, less controversially, biotechnology. It is this latter application that is the focus of considerable recent academic and commercial interest (Williams & Laurens, Citation2010; Milledge, Citation2011).
Biological Resource Centres (BRCs) such as the Culture Collection for Algae and Protozoa (CCAP) are de facto depositories of ‘biological standards’ holding taxonomic and other live reference strains on which much of the associated published science and industrial standards are built (Day & Stacey, Citation2008). They provide multiple services, ranging from the provision of quality-controlled material for research, commercial and teaching purposes and conservation of patented strains, to the ex situ conservation of biodiversity (Gachon et al., Citation2007). CCAP is the largest European BRC for algae and other protists. The collection comprises more than 2800 publicly available strains: 950 marine algae, 1550 freshwater algae and 300 protozoa. Phylogenetically, the collection is extremely diverse, holding representative taxa from all of the major protistan lineages (). Over 300 strains are derived from designated taxonomic type material, i.e. the actual strain/specimen to which the scientific name of that organism is formally attached when it was first described in the scientific literature. These cultures may have been subsequently maintained by serial transfer for many decades and may be described as ‘authentic strains’ by some workers (Day et al., Citation2010). The genotypic and phenotypic stability of these strains in particular are of crucial importance to the scientific community and recommendation on the application of long-term conservation of new holotype specimens, as cryopreserved non-growing specimens, have been made that will ensure their availability to the scientific community (Day et al., Citation2010).
Table 1 Overview of the phylogenetic diversity of CCAP strain holdings1.
The provision of cultures from BRCs is of little value unless these cultures are accompanied by information on their identity, provenance and characteristics. Information resources grow with time as work on cultures is published and individual collections, and microorganism database organizations such as StrainInfo at http://www.straininfo.net/, will provide more comprehensive data. Here, we focus on the description of the functionalities of the CCAP online repository (http://www.ccap.ac.uk), which we have transformed from a simple online catalogue into a one-stop-shop (CCAP KnowledgeBase) for quality-controlled biological material hyperlinked to manually curated molecular, bibliographical and taxonomic information.
System overview
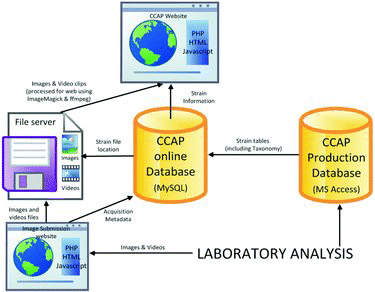
The CCAP online database is used to service the publicly accessible CCAP website and contains a subset of information, principally strain data, synchronized from the restricted CCAP Production database ( when strain data are added or amended. The online database resides on a MySQL database system and also contains the metadata for the strain-related digital images and videos which are stored on a separate internal file server. Images and videos of strains are submitted through a set of internally accessible web pages which are also used to manage the files and maintain the metadata. The forward-facing CCAP website then uses the information in the online database to display strain details and any related images or videos on the relevant webpages, specifically the search result page and the individual strain information pages. Images are watermarked and labelled ‘on the fly’ for web display. Both the internal and external web pages used to display and manage the strain information are written in a mixture of PHP, Javascript and HTML.
Knowledgebase content, organization and features
The KnowledgeBase has grown from the collection's traditional hard-copy catalogues (e.g. Tompkins et al., Citation1995) and the later online catalogue, and therefore has a strain-centric structure. A stable, sequential accession number designates each strain available from the collection. This is associated with a scientific name, which may be updated or amended, for example as the nomenclature evolves, or new studies allow better resolution of the taxonomy (e.g. Pröschold et al., Citation2001; Smirnov et al., Citation2011). Information on strain deposition, culturing methods and strain maintenance is easily accessible via dedicated tabs. The KnowledgeBase is updated at least once a month, with key changes or major strain acquisitions highlighted in the ‘News’ section. Users can contact the CCAP by phone, letter, e-mail, or via an online enquiry form and requests are responded to by the CCAP curatorial staff.
Search functions
The KnowledgeBase is fully searchable, using either a simple or an advanced search function. A simple search allows retrieval of strains belonging to a specific taxon, or information on strain numbers when the user knows them. The advanced search functionality is especially useful for the identification of related strains belonging to a given taxonomic group, or to design more complex queries based on biogeographical or habitat information. In all queries, the search output is a list of matching entries that the user can further explore individually.
Core strain information
Each strain available from the collection is associated with a standard page record (). It contains core information about the person who isolated the strain, the geographical origin and habitat of the organism, maintenance information, storage method within the CCAP (cryopreservation, serial sub-culture, or both), and possible co-occurring organisms (such as bacteria) in the culture. One field is dedicated to other important information, such as older names and alternative denominations, or whether the strain might be available from other BRCs. Extensive explanations about the scope of each section and the meaning of abbreviations are available in the ‘key to strain data’ page.
Figs 2–6. Screenshots of the KnowledgeBase entry for the authentic type culture Chlorella vulgaris CCAP 211/11B. 2, All core information about this strain is directly available on the entry page. Hyperlinks to: 3, the corresponding microscopy pictures; 4, AlgaeBase entry; 5, European Nucleotide Archive; 6, exemplar peer-reviewed paper from the literature references listed on Fig. 2 are circled in blue, red, black and green, respectively.
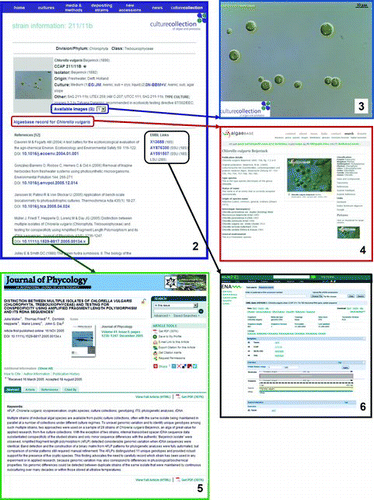
More recently, microscopy pictures, and, when appropriate, videos, have been added to illustrate the typical morphological features of some strains (). Finally, each strain page record is hyperlinked to a list of references that have been generated using it and hypertext links can take you to individual relevant publications (). This task is on-going, but we are aware of 80–100 peer-reviewed publications generated with CCAP material annually.
Links with other databases
Molecular information: hyperlinking with the European Nucleotide Archive
Until recently, very few molecular marker sequences were available for the organisms held in CCAP; also, the available molecular information was scattered in the literature and was not systematically linked with the source biological material (see in Gachon et al., Citation2007). We have therefore undertaken a systematic programme of generating molecular signatures, from which DNA barcodes can be deduced, of our strain holdings, mostly focusing on obtaining sequences for the small ribosomal subunit and internal transcribed spacers of the ribosomal RNA operon (rbcL), with these data made publicly available on the National Center for Biotechnology Information (NCBI)/GenBank, European Nucleotide Archive (ENA) and DNA Data Bank of Japan (DDBJ). Where available, we are also adding sequences derived from CCAP strains that have been made publicly available by other laboratories deposited in the ENA. Over 1350 CCAP strain page records are now hyperlinked with at least one ENA accession number (). Our primary objective is to obtain an accurate and reliable DNA identification of strain holdings, typically using ribosomal operon sequences, in order to guide our users in their selection of biological material. This undertaking also provides the foundations for a state-of-the-art molecular-enabled taxonomic revision of the CCAP's holdings. It is envisaged that by employing both molecular data, as well as conventional phenotypic information, definitive names can be confirmed or revised for individual strains in the collection; this in turn will result in enhancement of quality assurance within the collection.
Taxonomic information: AlgaeBase
AlgaeBase (http://www.algaebase.org) has grown over the last 18 years into the most comprehensive and up-to-date online resource on algal taxonomy available. It currently includes > 133 000 species and infraspecific names, of which 36 000 are the names of taxonomically accepted species, > 17 000 images, > 50 000 bibliographic items and > 220 000 distributional records (Guiry & Guiry, Citation2013). Its content is very relevant to CCAP, as it includes freshwater, terrestrial and marine microalgae and macroalgae, and prokaryotic cyanobacteria, together with achlorophyllous protists including chromists, dinoflagellates and euglenoid flagellates.
AlgaeBase entries for algae are linked to a number of biological resource centres and repositories of cultures, including CCAP, which has > 2400 strains linked to their taxonomic data on AlgaeBase. We have introduced reciprocal systematic links from CCAP strain records to AlgaeBase (), in order to provide users of the KnowledgeBase with additional expert taxonomic information. It should be noted, however, that this information is given for indicative purposes only. Indeed, AlgaeBase records are linked to the scientific name assigned to a given CCAP strain, and therefore are subject to the accuracy of its current taxonomic assignation. As explained above, the delineation and taxonomic placement of many protistan taxa remains imperfect; hence, with the exception of ex-type material (commonly described as authentic strains in algal collections), the identification accompanying CCAP strain accessions is a useful guide, but does not constitute a definitive taxonomic reference.
Future directions
Our objective is to add further value to the information available on the CCAP biological material by progressing its integration into a manually curated platform. We hope that our efforts to develop a user-friendly interface will support the breadth of activities of our users, including systematic research, biotechnological exploitation and teaching. The CCAP KnowledgeBase is continuously updated, and we therefore especially welcome feedback from our users, including taxonomic updates, pictures or publications generated with material obtained from the CCAP. Arguably, comprehensive ‘Knowledge-bases’ are of increasing value to the scientific community and curated links between live material and bioinformatics data are of vital importance, particularly in the light of concerns over errors (taxonomic and others) in data published in the major depositories of molecular data (Bridge et al., Citation2003; Tindall, Citation2007). Usage statistics indicate a steady increase in visitors interrogating the CCAP KnowledgeBase having increased from ∼26 400 in 2008 to ∼35 400 in 2012 (data from Google Analytics; http://www.google.com/analytics). Further developments currently under consideration include additional links to other data types, such as metabolomics or chemical fingerprinting. We also aim to further underpin the exponential development of metagenomics and genomics of unconventional and novel model organisms; in particular with the cross-referencing of environmental molecular data to specified live culturable material (del Campo & Massana, Citation2011). Our objective is to ensure that the CCAP KnowledgeBase is ‘fit for purpose’ for 21st century science with improved searchability, better connectivity to other relevant resources and all data are underpinned by live cultures available to the scientific community worldwide.
Acknowledgements
We are grateful to all former and current SAMS staff and visiting scientists who contributed to this project and those who gave us feedback on the KnowledgeBase. Author contributions: writing (CMMG, JGD, FCK, MDG, KG); data generation and knowledgebase development (CMMG, SH, UEMA-D, DB, CNC, AC, RGD, JF, SG, CR-M, AV, MDG, KG, JGD), CCAP KnowledgeBase concept (FCK, CNC, SG, MB, JGD). This work was supported by the UK Natural Environment Research Council (Oceans 2025 programme NF3 – CCAP), NERC NC funding for the CCAP; and the European Commission ASSEMBLE Integrated Infrastructures Initiative (grant agreement no. 227799). A large fraction of the sequence data in CCAP were generated by the GenePool Natural Environment Research Council Biomolecular Analysis Facility at the University of Edinburgh (allocation no. MGF154).
Associate Editor: Elliot Shubert
References
- Adl, S.M., Simpson, A.G.B., Lane, C.E., Lukes, J., Bass, D., Bowser, S.S., Brown, M.W., Burki, F., Dunthorn, M., Hampl, V., Heiss, A., Hoppenrath, M., Lara, E., Le Gall, L., Lynn, D.H., McManus, H., Mitchell, E.A.D., Mozley-Stanridge, S.E., Parfrey, L.W., Pawlowski, J., Rueckert, S., Shadwick, L., Schoch, C.L., Smirnov, A. & Spiegel, F.W. 2012. The revised classification of eukaryotes. Journal of Eukaryotic Microbiology 59, 429–514.
- Archibald, J.M. 2011. Newly identified and diverse plastid-bearing branch on the eukaryotic tree of life. Proceedings of the National Academy of Sciences USA 108, 1496–1500.
- Baldauf, S.L. 2008. An overview of the phylogeny and diversity of eukaryotes. Journal of Systematics and Evolution 46, 263–273.
- Berninger, U.G., Finlay, B.J. & Kuuppoleinikki, P. 1991. Protozoan control of bacterial abundances in fresh-water. Limnology and Oceanography 36, 139–147.
- Bridge, P.D., Roberts, P.J., Spooner, B.M. & Panchal, G. 2003. On the reliability of published DNA sequences. New Phytologist 160, 43–48.
- Caron, D.A. & Hutchins, D.A. 2013. The effects of changing climate on microzooplankton grazing and community structure: drivers, predictions and knowledge gaps. Journal of Plankton Research 35, 235–252.
- Chambouvet, A., Morin, P., Marie, D. & Guillou, L. 2008. Control of toxic marine dinoflagellate blooms by serial parasitic killers. Science 322:1254–1257.
- Chavez, F.P., Messie, M. & Pennington, J.T. 2011. Marine primary production in relation to climate variability and change. Annual Review of Marine Science 3, 227–260.
- Christaki, U., Jacquet, S., Dolan, J.R., Vaulot, D. & Rassoulzadegan, F. 1999. Growth and grazing on Prochlorococcus and Synechococcus by two marine ciliates. Limnology and Oceanography 44: 52–61.
- Day, J.G. & Stacey, G.N. 2008. Biobanking. Molecular Biotechnology 40, 202–213.
- Day, J.G., Pröschold, T., Friedl, T., Lorenz, M. & Silva, P.C. 2010. Conservation of microalgal type material: approaches needed for 21st century science. Taxon 59, 3–6.
- Degerlund, M., Huseby, S., Zingone, A., Sarno, D. & Landfald, B. 2012. Functional diversity in cryptic species of Chaetoceros socialis Lauder (Bacillariophyceae). Journal of Plankton Research 34, 416–431.
- Del Campo, J. & Massana, R. 2011. Emerging diversity within Chrysophytes, Choanoflagellates and Bicosoecids based on molecular surveys. Protist, 162, 435–448.
- Falkowski, P.G., Katz, M.E., Knoll, A.H., Quigg, A., Raven, J.A., Schofield, O. & Taylor, F.J.R. 2004. The evolution of modern eukaryotic phytoplankton. Science 305, 354–360.
- Gachon, C.M.M., Day, J.G., Campbell, C.N., Pröschold, T., Saxon, R.J. & Küpper, F.C. 2007. The Culture Collection of Algae and Protozoa (CCAP): a biological resource for protistan genomics. Gene 406, 51–57.
- Gachon, C.M.M., Sime-Ngando, T., Strittmatter, M., Chambouvet, A. & Kim, G.H. 2010. Algal diseases: spotlight on a black box. Trends in Plant Science 15, 633–640.
- Glücksman, E., Snell, E.A., Berney, C., Chao, E.E., Bass, D. & Cavalier-Smith, T. 2011. The novel marine gliding zooflagellate genus Mantamonas (Mantamonadida ord. n.: Apusozoa). Protist 162, 207–221.
- Guiry, M.D. & Guiry, G.M. 2013. AlgaeBase. World-wide electronic publication, National University of Ireland, Galway. http://www.algaebase.org; accessed 7 May 2013.
- Hartmann, M., Grob, C., Tarran, G.A., Martin, A.P., Burkill, P.H., Scanlan, D.J. & Zubkov, M.V. 2012. Mixotrophic basis of Atlantic oligotrophic ecosystems. Proceedings of the National Academy of Sciences USA 109, 5756–5760.
- Keeling, P.J., Burger, G., Durnford, D.G., Lang, B.F., Lee, R.W., Pearlman, R.E., Roger, A.J. & Gray, M.W. 2005. The tree of eukaryotes. Trends in Ecology and Evolution 20, 670–676.
- Kim, E., Harrison, J.W., Sudek, S., Jones, M.D.M., Wilcox, H.M., Richards, T.A., Worden, A.Z. & Archibald, J.M. 2011. Newly identified and diverse plastid-bearing branch on the eukaryotic tree of life. Proceedings of the National Academy of Sciences USA 108, 1496–1500.
- Kimura, H., Sato, M., Sugiyama, C. & Naganuma, T. 2001. Coupling of thraustochytrids and POM, and of bacterio- and phytoplankton in a semi-enclosed coastal area: implication for different substrate preference by the planktonic decomposers. Aquatic Microbial Ecology 25, 293–300.
- Küpper, F.C., Feiters, M.C., Olofsson, B., Kaiho, T., Yanagida, S., Zimmermann, M.B., Carpenter, L.J., Luther III, G.W., Lu, Z., Jonsson, M. & Kloo, L. 2011. Commemorating two centuries of iodine research: an interdisciplinary overview of current research. Angewandte Chemie – International Edition 50, 11598–11620.
- Massana, R., Terrado, R., Forn, I., Lovejoy, C. & Pedros-Alio, C. 2006. Distribution and abundance of uncultured heterotrophic flagellates in the world oceans. Environmental Microbiology 8, 1515–1522.
- Milledge, J.J. 2011. Commercial application of microalgae other than as biofuels: a brief review. Reviews in Environmental Science and Biotechnology 10, 31–41.
- Morrissey, J. & Bowler, C. 2012. Iron utilization in marine cyanobacteria and eukaryotic algae. Frontiers in Microbiology 3, 43.
- Müller J., Friedl T., Hepperle, D., Lorenz, M. & Day, J.G. 2005. Distinction of isolates among multiple strains of Chlorella vulgaris (Chlorophyta, Trebouxiophyceae) and testing conspecificity with Amplified Fragment Length Polymorphism and ITS rDNA sequences. Journal of Phycology 41, 1236–1247.
- Popper, Z.A., Michel, G., Hervé, C., Domozych, D.S., Willats, W.G.T., Tuohy, M.G., Kloareg, B. & Stengel, D.B. 2011. Evolution and diversity of plant cell walls: from algae to flowering plants. Annual Review in Plant Biology 62, 567–590.
- Pröschold, T., Marin, B., Schlösser, U.G. & Melkonian, M. 2001. Molecular phylogeny and taxonomic revision of Chlamydomonas (Chlorophyta). I. Emendation of Chlamydomonas Ehrenberg and Chloromonas Gobi, and description of Oogamochlamys gen. nov and Lobochlamys gen. nov. Protist 152, 265–300.
- Rybalka, N., Wolf, M., Andersen, R.A. & Friedl, T. 2013. Congruence of chloroplast- and nuclear-encoded DNA sequence variations used to assess species boundaries in the soil microalga Heterococcus (Stramenopiles, Xanthophyceae). BioMed Central Evolutionary Biology 13, Art. No. 39.
- Smirnov, A.V., Chao, E., Nassonova, E.S. & Cavalier-Smith, T. 2011. A revised classification of naked lobose amoebae (Amoebozoa: Lobosa). Protist 162, 545–570.
- Steiner, C.F., Long, Z.T., Krumins, J.A. & Morin, P.J. 2006. Population and community resilience in multitrophic communities. Ecology 87, 996–1007.
- Tindall, B.J. 2007. Vacuum drying and cryopreservation of prokaryotes. Methods in Molecular Biology 368, 73–98.
- Tompkins, J., Deville, M.M., Day, J.G. & Turner, M.F. 1995. Culture Collection of Algae and Protozoa Catalogue of strains, 6th Edition. Culture Collection of Algae and Protozoa, Ambleside.
- Williams, P.J.L. & Laurens, L.M.L. 2010. Microalgae as biodiesel and biomass feedstocks: review and analysis of the biochemistry, energetics and economics. Energy and Environmental Science 3, 554–590.