Abstract
The genus Bicyclus is one of the largest groups of African butterflies, but due to the generally cryptic nature and seasonal variation of adult wing patterns, there has been a lot of systematic confusion. With a large research community working with the model species Bicyclus anynana there has been increasing interest in the evolutionary history of the genus. A previous phylogeny started to unravel interesting patterns, but only included 61% of the then known species. With a range of new species having been described in the last decade there has been a need for an updated phylogeny for the genus. We present the most complete phylogeny of Bicyclus yet, including 93% of the currently 103 recognized species and make a range of taxonomic revisions. We revise the status of four previous subspecies and synonymized taxa that in the light of the new genetic data are raised to species level. We also subsume two subspecies and describe a new species, Bicyclus collinsi sp. nov., based on both genetic and morphological evidence. A further new taxon is identified, but not described at this point due to lack of morphological data. Our phylogeny lays a solid foundation for better understanding the evolution of Bicyclus and highlights key species-groups and complexes with intriguing ecological patterns making them prime candidates for future studies.
http://zoobank.org/urn:lsid:zoobank.org:pub:2F775351-097E-4CD7-8F8F-A90B26D52DE8
Introduction
Bicyclus (Bush Browns) is by far the most species-rich genus of African Satyrinae (Nymphalidae) butterflies, and among the largest of all butterfly genera on the African mainland (Ackery, Smith, & Vane-Wright, Citation1995). However, due to the rather cryptic nature of many of the species, the high degree of seasonal variation, and the fact that most species were described over a prolonged period without any systematic revision, there has been substantial confusion and numerous misunderstandings with regard to accurate species-specific morphological features and the distributional patterns of many species.
The first known mention of any taxon now placed in Bicyclus comes from Pieter Cramer's (Citation1779) description of a new species of butterfly that he named Papilio dorothea. Translated from the original Dutch and French the translated description reads: “One can notice this nymph with eyes as a singularity, it has a small tuft of hair, ash-grey in colour, at the outer edge of the lower wings, near the knuckles, extending along the lower edge of the upper wings. The beautiful mixture of colours and the multitude of small and large eyes on the wings makes this little butterfly look beautiful. They live on the Coast of Guinea, Sierra Leone.” Whilst a majority of observers today might not agree that Bicyclus have a beautiful mixture of colours, Cramer did comment on the row of ventral eyespots (found in all members of the genus) that have been the focus of much contemporary research. The development and seasonal plasticity of these eyespots perhaps account for why B. anynana has become established as a butterfly model organism (e.g., Brakefield, Beldade, & Zwaan, Citation2009). Cramer also noted the small black distal androconial brush, but apparently missed the larger more basal one often hidden under the forewing in a set specimen. Androconia were often overlooked in early taxonomic work on Bicyclus, but as species numbers increased and identification became more challenging they gradually gained importance in species descriptions. More recently the pheromonal functions of Bicyclus androconia (Nieberding et al., Citation2008) and their links to speciation process (Bacquet et al., Citation2015) have been the focus of new research.
Following this first description, a gradual stream of new species was described over the subsequent two centuries. As the colonial presence in inland Africa increased, much material was made available to taxonomists working in Europe. Since seasonally plastic variation was not understood at that time, a plethora of names were given to a range of seasonal morphs of many species. This meant that by 1955, a total of 159 names had been assigned to what we now consider to be 75 species and subspecies of Bicyclus. In Citation1871, the genus Bicyclus was proposed by Kirby as a replacement name for the genus Idiomorphus when this name was found to be preoccupied by a genus of beetles. Initially, Bicyclus only included a small number of morphologically distinct large blue-banded species. The majority of the current members of Bicyclus were instead classified as Mycalesis, a very large genus which at that time was considered to have representatives throughout the whole Old World tropics. Several authors had pointed out the need for a thorough revision of the African members of the genus (e.g. Aurivillius, Citation1925; Moore, Citation1880), but it was not until the mid 1950s that organized work was started with the specific goal of revising all African Mycalesis. This work was mainly carried out by Michele Condamin, who between 1958 and 1971 published a series of papers reorganizing all African species, and describing a range of new species. In Citation1961, Condamin moved most African species of Mycalesis into the genus Bicyclus, the only two species that were not incorporated in this new definition of Bicyclus were placed in the new genus Hallelesis (Condamin, Citation1961), as they showed a unique androconial configuration. In Citation1973, Condamin summarized all of his earlier work in an impressive monographic treatment covering all Bicyclus as he defined them, and this work still remains the main reference for the genus. At that time Bicyclus included 77 recognized species, and a further 26 subspecies. With an apparent tendency to support subspecific separations, Condamin in fact described half of these 26 subspecific taxa himself.
One important part of Condamin's work was to clear up the vast range of invalid names. Condamin listed a total of 101 junior synonyms, the majority of which were officially recognized as such for the first time. To organize the large number of species more effectively he created 29 species-groups, all named after the oldest valid name assigned to the species forming each group. Since Condamin's revision the genus has stayed relatively stable. Even if a sizeable number of new species have been described, all have fitted neatly into the framework of Condamin's original species-groups. His final contribution to the systematic work of Bicyclus came a decade later (Condamin, Citation1983) with the description of two Tanzanian species, B. kiellandi and B. tanzanicus.
Later, with the publication of his book on Tanzania's butterfly fauna, Kielland (Citation1990) described two montane endemics, B. simulacris and B. uzungwensis (each with an additional subspecies). In Citation1996, Libert described new montane taxa from Cameroon in the species B. amieti and B. ewondo, as well as the subspecies B. graueri choveti. In Citation1998, Congdon, Collins and Kielland described the species B. mahale from Tanzania increasing the total number of recognized species to 84. By that time the species B. anynana had emerged as a model organism for studies of topics such as evo-devo (French & Brakefield, Citation1995) and adaptive phenotypic plasticity (Brakefield & Larsen, Citation1984). With a growing interest for comparative studies across the genus with regard to patterns found in B. anynana, the first molecular phylogeny of the genus was published by Monteiro and Pierce in 2001, including 51 species (61% of the then described species). At the time of its publication, Monteiro and Pierce (Citation2001) presented the largest molecular phylogeny constructed so far for an African butterfly genus. This phylogeny was later employed for investigations of the roles of sexual and natural selection within the genus (Oliver, Robertson, & Monteiro, Citation2009), as well as for studies of the evolution of seasonal polyphenism (Brakefield & Frankino, Citation2009), and the origin of sexual dimorphism (Oliver & Monteiro, Citation2011).
In the last decade there has been another burst of systematic work within the genus. In Citation2007 Vande weghe described B. ivindo from Gabon. The year after, Collins and Larsen (Citation2008) described the two montane endemics B. sealeae (from Bioko) and B. pareensis (from Pare Mountains in Tanzania). The following year Vande weghe (Citation2009) described B. larseni and B. wakaensis. In the same paper he also raised B. bergeri to a full species (it had previously been treated as a subspecies of B. ephorus) and described the new subspecies, B. graueri kota. In Citation2012 Brattström described the species B. brakefieldi from Democratic Republic of Congo. Recently Brattström, Aduse-Poku, Collins, and Brakefield (Citation2015) and Brattström, Aduse-Poku, Collins, Di Micco de Santo, and Brakefield (Citation2016) published two revisions of Bicyclus species groups describing six further species: B. ottossoni and B. vandweghei in 2015 and B. elishiae, B. heathi, B. sigiussidorum, and B. subtilisurae in 2016. In the 2015 paper all subspecies of B. ignobilis were also synonymized with the nominate form. As a result, prior to the work presented in this paper, the total number of recognized species of Bicyclus stood at 97, with a further 27 subspecies.
With almost 100 species identified and with interest in the evolutionary history of the genus coinciding with the availability of more cost-effective sequencing methods, we aimed to produce as complete a molecular phylogeny as possible for the whole genus Bicyclus, with the goal of facilitating future ecological work within a phylogenetic framework. In addition to attempting to acquire samples from all recognized species, we also included a set of samples from potential undescribed taxa. This was a result of our previous work on the systematics of the genus which had made us aware of several morphologically distinct populations that almost certainly represent new, or previously suppressed, species in need of verification. Finally, we also aimed to include more than one specimen from as many species as possible to detect potential cryptic species.
Materials and methods
Taxon sampling, DNA extraction, and sequencing
Most samples were either collected by the authors and colleagues during field expeditions, or were obtained from the collections at the African Butterfly Research Institute (ABRI) in Nairobi, Kenya. We aimed to assemble samples from all 97 species described at the time, and where possible, more than one specimen per species. DNA sequences, and where possible, aliquots of extracts used in the previous study on Bicyclus relationships (Aduse-Poku et al., Citation2015; Monteiro & Pierce, Citation2001) were obtained from the authors and used in the present study with further genes sequenced from some of the aliquots. We also reevaluated the species identification for some of these species using photographs provided by the authors.
Condamin (Citation1973) considered B. istaris to be a widely distributed species occurring in all parts of the tropical forest block of Africa. Despite this assertion we could not find any positively verified records across a range of major collections (listed in Brattström et al., Citation2016) from the central forest belt of Cameroon, Republic of Congo, Equatorial Guinea, Gabon, nor western Democratic Republic of Congo. By investigating male genitalia of B. istaris from across these western and eastern areas and by comparing them to samples from the other two members in the sophrosyne-group (B. lamani and B. sophrosyne) it seemed likely that B. istaris contained two separate species. We therefore included samples from both parts of its distribution in our phylogeny. We also added a sample from a morphologically odd population of B. mesogena, with a light yellow brush in cell RS on the hindwing compared with the normally black or dark brown brush found in all previously described species, in the mesogena-group.
Despite our best efforts, samples from seven previously described species failed to produce any useful sequences. These missing species are: B. bergeri, B. condamini, B. kiellandi, B. nachtetis, B. similis, B. suffusa, and B. vansoni. Our total dataset contains 166 successfully amplified individual samples representing 90 (93%) of the 97 currently recognized species of Bicyclus. We also included a further five specimens from the two species in the genus Hallelesis, and eight exemplar taxa from the closely related South-east Asian genus Mydosama as outgroups.
We extracted genomic DNA from one or two legs per individual using QIAgen's DNEasy extraction kit. For older samples we used thoracic tissue. Field-collected samples that had been stored in 99.9% ethanol were air dried overnight before extraction. Up to a total of 10 molecular markers; one mitochondrial (cytochrome c oxidase subunit I, CO1) and nine nuclear (carbamoyl phosphate synthetase domain protein, CAD; Ribosomal Protein S5, RpS5; Ribosomal Protein S2, RpS2; wingless, wgl; cytosolic malate dehydrogenase, MDH; glyceraldehyde-3-phosphate dehydrogenase, GAPDH; Elongation factor 1 alpha, EF-1α; and Arginine Kinase, ArgKin and isocitrate dehydrogenase, IDH) gene regions, were amplified and sequenced for each of the exemplar taxa. We used primer-pairs obtained from Wahlberg and Wheat (Citation2008), and included the universal forward/reverse tail which facilitated sequencing.
All PCRs were performed in a 20 µL reaction volume. The thermal cycling profile for COI, Wingless and the second half of EF-1α (Al-EfrcM4) primer pairs was 95 ºC for 7 min, 40 cycles of 95 ºC for 30 s, 50 ºC for 30 s and 72 ºC for 1 min followed by a final extension period of 72 ºC for 10 min. The thermal cycling profile for CAD, IDH, MDH, GAPDH, RpS5, RpS2, and the first half of EF-1α (Starsky-Monica) differed only in an elevated annealing temperature of 55 ºC, compared with 50 ºC in the previous thermal cycling profile. All successful PCR products were cleaned of single-stranded DNA and unused dNTPs using EXO-SAPIT, and sent to Macrogen Services in Amsterdam for Sanger sequencing. The resultant DNA sequences of targeted gene regions were manually aligned using the software BioEdit 7.2.5 (Hall, Citation1999). We assessed individual sequence properties using MEGA v6 (Tamura, Stecher, Peterson, Filipski, & Kumar, Citation2013).
Phylogenetic analysis
Phylogenetic analyses were performed using both Maximum likelihood (ML) and Bayesian Inference (BI) methods, implemented in RAxML-HPC2 v8.0.24 (Stamatakis, Citation2014) and MrBayes v3.2 (Ronquist & Huelsenbeck, Citation2003), respectively. The phylogenetic analyses were performed on the CIPRES Science Gateway v3.3 (Miller, Pfeiffer, & Schwartz, Citation2010). To improve phylogenetic resolution of our multi-gene dataset, PartitionFinder (Lanfear, Calcott, Ho, & Guindon, Citation2012) was used to select the optimal gene partitioning schemes and the best-fit model of nucleotide substitution for each partitioned dataset at the codon level. For the ML analysis, we used reversible jump- Markov Chain Monte Carlo (MCMC) and GTRCAT model for the rapid bootstrapping phase, and GTRGAMMA for the final best scoring ML tree. We performed 1000 Maximum Likelihood (ML) pseudo-replicate analyses for the boot-strapping estimation of nodes under auto Majority Rule Criterion (autoMRE). For the BI analysis, we ran two parallels of four chains (three heated and one cold) using MCMC randomization for 10 million generations. The trace files generated by the Bayesian MCMC were analysed using TRACER v1.6 (Rambaut, Suchard, Xie, & Drummond, Citation2014) to inspect whether the effective sample sizes of the posterior distribution of the tree likelihoods and model parameters estimates were above the desired threshold of 200.
Measurements of morphological characters
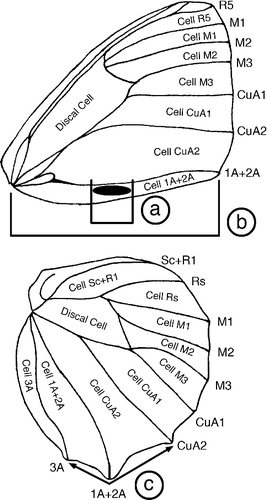
Upon finding an unexpected semi-cryptic species in the angulosa-group (see Results) with only limited morphological divergence compared with its closest relatives we decided to collect quantitative data for a set of morphological characters to investigate if they could be used in the description of this new species. Because colour is hard to quantify in museum specimens with a variable amount of wear and age, we focused on measuring the size of the distal forewing androconia and the prominence of the hindwing tail at Vein 1A+2A to investigate how well these two traits could be used to separate the two groups and a closely related species in the same complex. To analyse a range of photographed specimens from museums and private collections we used the software Fiji (Schindelin et al., Citation2012). Traits measured were: (1) Relative androconial size calculated as the ratio of the total length of the androconial patch along Vein 1A+2A, divided by the length of a straight line drawn from the base of the wing (where all the major veins merge) up to the point where Vein 1A+2A terminates at the wing margin (). (2) Prominence of hindwing tail, quantified by taking an angular measurement with the end points placed at the marginal end of Vein CuA2 and Vein 3A, and the vertex positioned at the end of Vein 1A+2A. We placed the landmarks at the edge of the wing surface where the cilia originate as many specimens had damaged cilia or they were completely missing (). The morphological data were analysed with the R Statistical Package v 3.1.2 (R Core Team, Citation2015). One-way ANOVAs were used to analyse the difference in the two measured traits between the investigated populations and species, whilst Tukey's HSD test was used to examine which groups were different from each other.
Fig. 1. Bicyclus wing with venation nomenclature. Names of cells are given inside the cells whilst names of veins are given immediately to the right or below the wing margin at the terminal point of each vein. Also shown is a schematic for three morphological measurements taken for specimens from the auricruda-group. Relative androconial size was calculated by dividing the length of the outer forewing androconial patch (a) with the total distance from the wing base to the distal end of vein 1A+2A (inner androconial patch not shown). Hindwing tail angle (c) was calculated as the angle formed when placing landmarks at the distal ends of veins CuA2, 1A+2A & 3A.
Results
Dataset
The nucleotide alignment of the 10-gene concatenated data matrix from our 95 species of Bicyclus (including newly discovered species, see below) with the outgroups (Hallelesis and Mydosama) (supplemental data S1, see online supplemental material, which is available from the article's Taylor & Francis Online page at http://dx.doi.org/10.1080/14772000.2016.1226979) included, contained 7735 base pairs of which 36.9% and 28.8% of sites were variable and parsimony informative, respectively. The best partitioning schemes and the optimal evolutionary models for each of the partitioned datasets are listed in supplemental data S2 (see supplemental material online). The effective sample size (ESS) for all the parameters of the independent MCMC runs was higher than 200. All sequences are available on GenBank (accession numbers are listed in supplemental data S1, see supplemental material online).
Phylogenetic inference
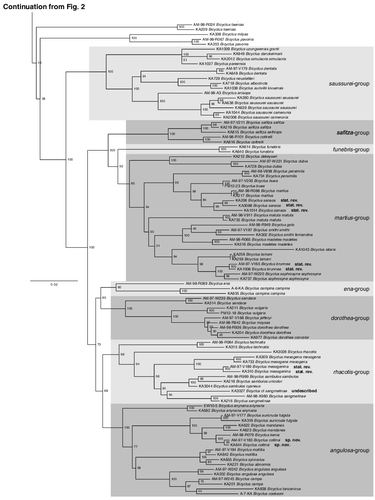
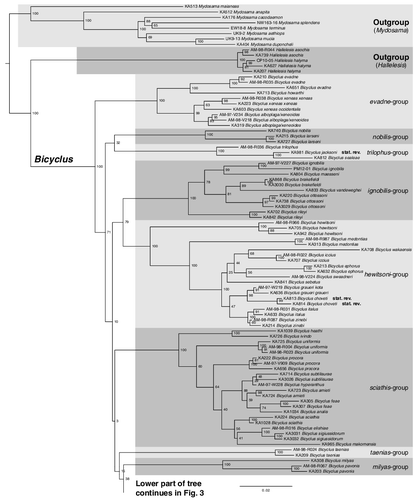
The trees produced by both the Maximum likelihood (, ) and the Bayesian Inference (supplemental data S3, see supplemental material online) methods were largely congruent. In both analyses, we recovered Bicyclus as a well-supported monophyletic group (BS = 100, PP = 1) with Hallelesis as its sister group. The evadne-species group (including B. alboplaga, B. evadne, B. howarthi, B. xeneas, and B. xeneoides) was retrieved as the sister clade to all other Bicyclus (BS = 100, PP = 1). Following the branching off of the evadne-group the next set of branches were largely unsupported in both the ML and the BI trees. However, despite the general basal uncertainty, the traditional nobilis-, trilophus-, ignobilis-, hewitsoni-, medontias-, italus-, sciathis-, taenias-, and milyas-groups within this part of the phylogeny were all supported as monophyletic groupings (). Following this section of the tree, we recovered, with high support, and in both analyses, a large clade of 53 Bicyclus species (). The taxa in this clade represent 20 of the 29 traditional species-groups as delimitated by Condamin (Citation1973) and the phylogenetic relationships among the different classic species-groups in the lower clade were generally all well supported, but some of them were found to be either paraphyletic or polyphyletic. A revision of the Bicyclus species-groups is presented further below in a separate section of the discussion. Based on our presented phylogenetic hypothesis (, & supplemental data S3, see supplemental material online) we merge two subspecies into one, while raising three others to full species level (see next section for details). We also reinstate one species previously treated as a junior synonym, change the name of one previously described species, describe a new semi-cryptic species, and find a new species in need of formal description. Our phylogeny also highlights groups of closely related species in need of more investigation in the dorothea- and alboplaga-complex. A complete checklist of all recognized species of Bicyclus following our presented revision is available in supplemental data S4, see supplemental material online.
Fig. 2. Phylogenetic relationships of the genus Bicyclus (with samples from Mydosama and Hallelesis used as outgroups) as inferred by Maximum likelihood. The numbers at the nodes are the nodal support values of 1000 bootstrap runs. The phylogenetic tree is split in two parts due to size with the outgroups and the first species-groups of Bicyclus shown in and the remaining species-groups shown in . The grey blocks show our updated species groupings (originally defined by Condamin, Citation1973).
Revised subspecies
A few subspecies showed either a large genetic distance to other representatives of the same species, or were grouped with samples of entirely different species. We hereby raise the following three taxa (previously treated as subspecies) to full species: Bicyclus choveti Libert, Citation1996 stat. rev. (, formerly B. graueri choveti), Bicyclus jacksoni Condamin, Citation1961 stat. rev. (, formerly B. trilophus jacksoni), and Bicyclus sanaos (Hewitson, 1866) stat. rev. (, formerly B. martius sanaos). We also sink the subspecies Bicyclus saussurei angustus Condamin, 1971 syn. nov. as our samples (KA638 & KA639) failed to separate with regards to the nominate species (KA260) (). We also subsume Bicyclus evadne elionas (Hewitson, 1866) syn. nov. based on morphological evidence. It was described as a separate subspecies with a distribution from Ivory Coast and further eastwards across the whole forest zone with a pale apical band setting it apart from the nominate species with a fully dark female being found in Sierra Leone and Liberia and the westernmost parts of Ivory Coast. Examining 276 female specimens kept at the African Butterfly Research Institute, Nairobi, Kenya (ABRI) collected across the complete distribution of the species, and combining this with field observations in Liberia revealed no clear border between the two colour patterns. Almost all female Bicyclus evadne found in Ghana and eastwards have the pale patch, but specimens being almost totally dark do occur. In western part of the distribution specimens are on average darker, but both types are found in sympatry with variable frequencies with no clear separation as previously reported by Condamin (Citation1973).
New species among the candidate samples
The sequenced samples from what was traditionally (after Condamin, Citation1973) treated as B. istaris showed a deep genetic divergence between a specimen from Liberia and a pair of specimens from north-eastern DRC (Kivu) and Uganda, verifying our suspicion that these should be separated into two distinct species. After investigating all older names assigned to this group and studying available type material we are confident that the types of Monotrichitis sophrosyne brunnea Jackson, 1951, are the same species as our eastern samples of ‘B. istaris’. Condamin (Citation1973) treated brunnea as a junior synonym of B. istaris, but based on our phylogeny and morphological investigations we hereby raise this taxon to full species status as Bicyclus brunnea (Jackson, 1951) stat. rev. ().
During our collection of samples we realized that the species B. mahale was more widespread than previously thought. It was described as a Tanzania endemic restricted to a small forested area in the extreme west of the country. However, among our samples of B. mesogena from Uganda we found multiple specimens better matching the morphology of B. mahale than typical B. mesogena. While investigating a range of historic types of Bicyclus in Berlin we found that the species described as Mycalesis mesogenina Grünberg, 1912, and later treated as junior synonym of B. mesogena by Condamin (Citation1973) perfectly matched the morphology of B. mahale. Compared with B. mesogena the samples belonging to B. mahale/mesogenina have a different forewing shape with a rounder outer margin, a much more developed androconial brush at the end of the hind wing Discal Cell, and a reduced androconial brush at the basal part of Cell RS. The genitalia appear to be somewhat variable, but with two main morphological types, matching the androconial types. The main difference is the shape of the rear part of the paddle-like structure formed at the tip of the valves. By investigating these morphological differences across a large series of specimens we found that the two morphological groups frequently occurred in sympatry over a wide region of the central forest belt, with no evidence that B. mahale is different to M. mesogenina. We therefore reinstate Bicyclus mesogenina stat. rev. () as a valid name, and in the same process subsume Bicyclus mahale syn. nov.
Not unexpectedly, we recovered B. mesogena and B. mesogenina as sister species, but with a rather deep divergence. In the same broad clade around B. mesogena and B. mesogenina we also found evidence for a further new species, represented by a single sample taken from a group of morphologically slightly different ‘B. mesogena’. Compared with the typical morphology of the species they had a distinctly yellow androconial brush in the basal part of Cell RS on the hindwing (this brush is normally much darker in the other species in the group). We have seen a handful of specimens from across the central forest belt of Africa with this morphology, but the majority have been several decades old and failed to produce any useful DNA sequences. Based on this single specimen's location in our phylogeny we are now certain this taxon represents a new, yet undescribed, species. However, due to a lack of comprehensive material we will hold back the formal description of this species, temporarily calling it B. cf. sangmelinae (), while a full revision of the combined mesogena- and sambulos-group is being prepared (Brattström et al. unpublished data).
New semi-cryptic species in the classic auricruda-group
The most surprising finding in our phylogeny was a deep divergence within B. mandanes with the morphologically clearly differentiated species B. kenia retrieved as sister to samples initially identified as B. mandanes from Kenya and Uganda (labelled B. collinsi in , ). This cluster is in turn sister to another cluster also identified as B. mandanes represented by samples from Ghana and Togo. A further specimen from Cameroon also clustered with these latter samples, but due to low gene coverage this individual was removed from the final tree. Based on this result suggesting that B. mandanes as classified by Condamin (Citation1973) represents at least two species, we investigated a large number of specimens from across the entire distribution to try to identify morphological differences separating the samples into two or more distinct groups. We found the following four traits that appeared to be consistent with the phylogenetic results. (1) On the ventral surface of the forewing there are two patches of androconial scales in Cell 1A+2A. Both of these forewing patches can be observed on the dorsal surface of the wing as a bulge, occasionally with a slightly different colour than the dark brown base colour. The more distal of these two patches shows a substantial variation in size between specimens, whilst the inner appears more constant in size. (2) The andoroconial brush at the base of the discal cell of the dorsal hindwing has either a yellow or white/light grey tint to the tip. (3) The discal band is either white and quite well delineated or more yellow with a diffuse outline. (4) The hindwing has a small tail-like projection at the end of Vein 1A+2A that appeared to vary in prominence with the sample location. No noticeable male genital difference was found across the whole range, but this was investigated with a limited number of samples as the valve tips of all species in the classic auricruda-group are extremely fragile and many were damaged prior to dissection.
Fig. 4. Panel to the left show a simplified phylogenetic tree for the three species closest related to Bicyclus mandanes. The plots to the right show the result of two morphological measurements taken from the same three species, as well as samples from Cameroonian specimens (bottom row) thought to belong either to B. mandanes or B. collinsi. The filled symbols represent measurements from the types of the following taxon names: mandanes (diamond), graphidhabra (triangle), kenia (square), and collinsi (circle).
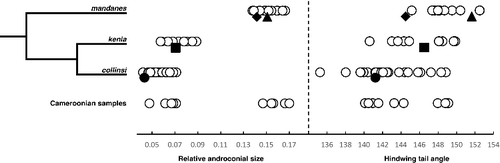
All investigated males from West Africa (excluding Cameroon), Republic of Congo and Angola (including a specimen from the original type series of Mycalesis mandanes) had a large forewing androconia, a more yellow hindwing brush tip, a better developed and more white apical band, and also appeared to have a less prominent hindwing tail. In the following text we refer to this group as the ‘Large Androconial Type’. In contrast, all samples from eastern Democratic Republic of Congo, South Sudan, Uganda, Kenya, Tanzania, and Rwanda had small forewing androconia, a white tipped hindwing brush, generally a more diffuse and yellowish apical band, and a more pronounced hindwing tail. We therefore refer to this group as the ‘Small Androconial Type’. Samples from Cameroon appeared to match the morphology of either of these groups, but importantly we never found both types at the same exact location and no specimens appeared to be intermediate between the two types. We only found samples from the ‘Small Androconial Type’ in the Bamenda Highlands (Santa and Acha-Tugi) of western Cameroon. However, the ‘Large Androconial Type’ is found in the same general area (but not at the same location) both at high and intermediate altitudes as well as in the southern lowland forest. It appears that the two types are at least parapatric, but our data are too limited to rule out the possibility of sympatric areas. There appears to be a genuine gap in the distribution throughout the whole of Gabon, as previously reported by Vande weghe (Citation2010). Nor could we find any records from the western parts of Democratic Republic of Congo despite investigating the world's largest holding of material from the region, kept in Tervuren, Belgium. Condamin and Soltani (Citation1980) recorded B. mandanes from the Central African Republic, but we have not seen any vouchers and therefore we do not know what morphological group these records belong to. For some locations we only have female specimens, but as the androconial characters appear to be the main morphological difference between the species we are currently not able to accurately identify the systematic position of these vouchers.
Whilst our phylogeny and morphological surveys indicate that B. mandanes in reality represents two separate species, the correct application of the original name is somewhat less straightforward. A full report on the available types and the history of the potential available names for this new species is given in supplemental data S5 (see supplemental material online). From our survey of the auricruda-group we concluded that the large androconial type matches the description and type of B. mandanes and that the only potentially available name that could be used for new species in the group, Dichothyris graphidhabra Karsch, 1893, belongs to the same morphological group (). Therefore, we name this new species Bicyclus collinsi Aduse-Poku sp. nov.
Bicyclus collinsi sp. nov.
HOLOTYPE: Voucher ID: SZS-KW-007, DNA extract code: KA644 (Sáfián, S., Collins, S.C., Horváth, Á & Collins, M.; 21-23.02.2009). Deposited at ABRI.
TYPE LOCALITY: Kenya, Kakamega Forest, Ron do Retreat-Lirhanda Hill, Yala River Reserve.
ADDITIONAL MATERIAL: Sequenced, but morphologically not investigated, sample from Uganda, Kibale NP. Voucher ID: AM-97-V183 (Antonia Monteiro).
HABITAT AND DISTRIBUTION: Not fully resolved as description is based on genetic data, but the morphological survey presented above suggest that the species should occur in all but the wettest of forested areas in Western Kenya, Uganda, NW Tanzania, the northern and eastern parts of the Democratic Republic of Congo, South Sudan, possibly also Central African Republic and the Republic of Congo. There are also presumably isolated populations in the Bamenda Highlands in W Cameroon.
ETYMOLOGY: Named after Steve C. Collins, founder of the African Butterfly Research Institute. Without his assistance we would never have been able to access such a complete set of samples giving a geographic overview of the whole genus Bicyclus.
DIAGNOSIS: Comparing the two genetic samples of this species to those of B. mandanes shows a deep divergence between these morphologically otherwise rather similar species. There are also strong suggestions that androconial differences found in classically defined B. mandanes when comparing different areas of Africa (see Remarks) is going to be linked with the genetic separation.
Description
Holotype (): Forewing length 24 mm. Wing margin scalloped and hindwing showing a small tail at the end of vein 1A+2A; dorsal wing surface dark brown; yellowish white apical band on forewing with diffuse outline; two minute blind eyespots in cells CuA1 and M1; ventral wing surface with a heavily dentate discal band across both wings; ventral wing pattern is formed of dark and warm hues of brown and some more purple areas; the pattern is broken up by a multitude of fine dark striae; there is a well-developed eyespot with minute white pupil and thin orange outer ring on the forewing in cell CuA1 and a smaller spot of similar colour, but with better developed pupil in call M1; the ventral hindwing have a marginal row of small eyespots with less prominent outer rings; individual spots are found in cell Rs, M1, and CuA and two spots present in cell CuA2; the ventral forewing have two androconial patches placed immediately below vein 1A+2A in an ivory coloured area extending across all but the most marginal parts of Cell 1A+2A; the inner of these patches well developed and the outer much reduced; both of these patches are also visible on the dorsal surface as small bulges in the wing surface; the dorsal hindwing have four androconia brushes; the discocellular brush is light brown at the base turning gradually towards white at the tip, it covers an androconial patch along the base of vein Rs; the brush at the base of cell CuA2 is dark brown with a slightly lighter tip and covers a patch of shiny graphite grey scales at the base of vein CuA2; two further brushes not linked to any androconial patches are found at the distal end of the discal cell and the base of cell Rs, the former being of the same dark brown colour as the wing and the latter having a light brown tip.
REMARKS: To investigate in more detail patterns of the morphological features noted in our museum survey, we measured a set of traits using individual photographs of 62 male specimens. As accurate measurements of colour could not be obtained in collected specimens of various ages (and our photographs were not always colour calibrated), we focused our quantitative comparisons on the FW androconia and hindwing tail. We compared all samples of the two morphological groups, in the following text treated as belonging to the two species B. mandanes (Large group, N = 15) and B. collinsi (Small group, N = 25). However, we kept all samples from Cameroon as two separate groups: ‘Large-Cam’ (N = 6) and ‘Small-Cam’ (N = 5). We also included samples from the related species B. kenia (N = 12) as a comparison. Due to damage on some wings we could not measure the androconia in one specimen, or the tail-angle in two other specimens. The ratio of the forewing androconia (F4,57 = 322.5, P < 0.0001) as well as the hindwing tail angle (F4,56 = 9.653, P < 0.0001) was significantly different between the groups. The tail angle measurements showed a substantial amount of overlap between groups, but B. collinsi (excluding Cameroon) remains significantly different from non-Cameroonian B. mandanes (Tukey's HSD: P < 0.0001) and B. kenia (Tukey's HSD: P < 0.01). The ranges of the relative androconial size measurements showed no overlap between either species (with Cameroon excluded). The post hoc test identified all groups as significantly different (Tukey's HSD: P > 0.001), except for the Cameroonian samples. The ‘Small-Cam’ group could not be separated from B. collinsi (Tukey's HSD: P = 0.734) or B. kenia (Tukey's HSD: P = 0.330), while the ‘Large-Cam’ group could not be separated from B. mandanes (Tukey's HSD: P = 0.861). The results are summarized in
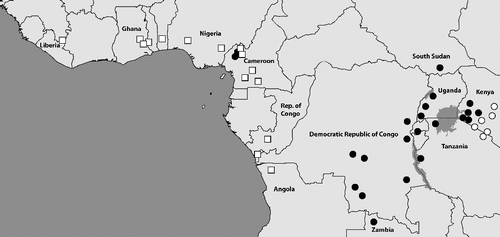
Whilst more genetic evidence is required to fully link these differences in morphology to the two species, we are confident that the androconial differences will remain valid when more samples are sequenced across the whole distribution. It is also possible that the small androconial types in Cameroon, that we currently identify as B. collinsi, represent small remaining relict populations, possibly of subspecific status, from a time when this morphological type was far more widespread. A similar pattern of occurrence where predominantly East African species have isolated subspecies in the Cameroon and Nigeria highlands is quite common, for example Neptis ochracea milbraedi Gaede, 1915 and Metisella midas malda (Evans, 1937). shows the locations of all investigated specimens (and some literature sources).
Discussion
The dorothea-complex
The three species, B. dorothea, B. jefferyi, and B. moyses, show a limited genetic difference () and morphologically they are rather similar. Based on the lack of genetic differentiation alone one could argue that these should all be merged as a single variable species, or alternatively treated as subspecies. There are, however, several reasons why these almost certainly represent real species.
Males of all three species can generally be separated by small, but consistent, differences in wing patterns. Condamin and Fox (Citation1964) and Condamin (Citation1973) describe the morphology in detail and the following description summarizes their main observation. The male of B. dorothea has a distinct pale, almost fully white, basal dorsal area on all four wings, with broad blackish borders overlaid by a bluish sheen in fresh specimens. The underside is white and the usual banding pattern found in all closely related species is broken up into small individual striate markings. Bicyclus moyses has a uniform brown tone across the entire dorsal wing surface, but with a clear violet sheen. The dorsal colouration also tends to be rather weak so that some of the pattern elements of the ventral side are visible through the wings. The underside has a clear banding pattern and is generally more brown. Bicyclus jefferyi is also uniformly brown on the dorsal surface, but with a stronger often darker colouration and with a faint bronze coloured sheen. The underside is similar to B. moyses, but the contrast between the discal band and the general ventral colour is less marked and the banding pattern is less obvious. The difference is less marked in females, but generally they can also be separated into three distinct groups matching the general morphology of the males. The males of the three species have androconial brushes on the dorsal hindwing, one in the basal discal cell, one in Cell Rs and one in CuA2. The two former brushes are linked to androconial patches which lie directly under the brushes. However, in B. dorothea the brush and patch in Cell Rs is generally weakly developed, and the brush itself often drops off during the life time of individual specimens. The discocellular brush is light brown to yellow in B. dorothea, and darker in B. moyses, whilst in B. jefferyi it is almost black.
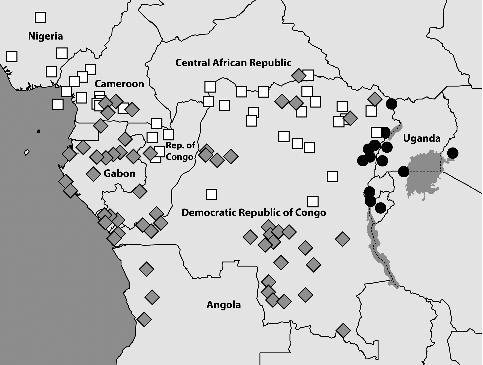
To better understand the distributional patterns of the three species we screened the collections of the African Butterfly Research Institute, Nairobi, Kenya (ABRI) and Musée Royal de l'Afrique Central, Tervuren, Belgium (MRAC). We found numerous samples of B. dorothea from across the whole forest zone of West Africa including Cameroon extending further east along the northern parts of the Republic of Congo, southern Central African Republic, and Northern Democratic Republic of Congo. There are also a few records from further south, but in general the distribution appears to be more to the north of the central African forest zone. Bicyclus dorothea concolor is endemic to the island of Bioko and shows a quite large genetic distance compared with the nominate subspecies found on the mainland. It is morphologically distinct in that the female has the same wing pattern as the male. The distribution of B. moyses overlaps with B. dorothea in southern Cameroon and areas further east, but it is also well documented much further south from Gabon, Northern Angola, and the more southern parts of the Congolese forest zone. Both species are linked to wet forests, but appear to prefer more open areas in these habitats and can be found in heavily fragmented forests. Bicyclus jefferyi has a more eastern distribution and is found in eastern Democratic Republic of Congo, most of Uganda, Western Kenya, North-western Tanzania, Rwanda, and Burundi. It is generally found in savannah habitats and occasionally found in open areas in fragmented forest habitats, but unlike its two close relatives it is not dependent on the proximity to forest. This means the all three species have substantial areas where they are fully allopatric, but B. dorothea and B. moyses also occur sympatrically over large areas (). Bicyclus jefferyi is probably never found in exactly the same location as the other two species due to differences in habitat use, but we do not know if B. dorothea and B. moyses utilize slightly different habitats in areas where they occur in apparent sympatry.
Fig. 7. Central African samples locations (generally verified through voucher photos) for the three species in the dorothea-complex: B. dorothea (white squares), B. moyses (grey diamonds), and B. jefferyi (black circles). Most locations in Gabon are sourced from Vande weghe (Citation2010) while remaining data generally acquired from photos of voucher specimens in the collections of ABRI and MRAC. Some additional data are taken from fieldwork notes and photos by the authors and collaborators. The distribution of B. dorothea extends further to the west throughout all forested areas of West Africa (not shown), but in this region the two other species are fully absent.
The male sex pheromones differ between B. dorothea and B. jefferyi (Bacquet et al., Citation2015), but we unfortunately have no such data for B. moyses. Preliminary studies of male genitalia fail to find any discernible differences. Breeding B. dorothea in the lab in a range of temperature conditions generates the normal wet/dry season plasticity of eyespots commonly found in Bicyclus, but the underside banding pattern remains faint and the upperside always shows the typical white basal area (Brattström, unpublished data). This suggest that the morphological difference between B. dorothea and B. moyses is not due to a plastic response in the same species mediated by microclimate from potential different habitat use. The three species in the dorothea-complex could possibly represent ecotypes from the same widespread ancestral species currently in the early stages of full separation, and it is likely that there is still considerable gene flow between them. Potential hybrid specimens which fall morphologically in between B. dorothea and B. moyses are occasionally found across the sympatric part of their range, but they always make up a very small proportion of any large collection.
The alboplaga-complex
As with the three species in the dorothea-complex (see previous section), the two species B. alboplaga and B. xeneoides show a limited genetic difference (). This is surprising as they are described as two morphologically distinct species. Both of these taxa are morphologically (as well as genetically) well separated from their sister species B. xeneas due to the lack of an androconial brush on the dorsal forewing at the base of CuA1 and as a result of their far more irregular discal band on the ventral side of both wings. Bicyclus alboplaga was described from a series of three males based on an unusually prominent light dorsal apical band on the forewing and the colour of the outer of the two costal androconial brushes found in all three species. This brush is rust red in B. alboplaga, while in the two other species it is black (and also heavily reduced in B. xeneas). The ventral discal band was also noted as being more irregular than in B. xeneoides. Our presented tree shows almost no genetic difference between specimens, including one male matching the morphology of B. xeneoides and two females matching the morphology of B. alboplaga. A further three males (one B. xeneoides and two B. alboplaga) that were removed from the final tree (due to a lower average gene coverage) also showed limited genetic differences from the other specimens, and the clustering did not follow either morphology or geography.
This unexpected result led us to investigate the basic external morphology of a large set of samples deposited at ABRI, collected along the entire distributional range of both B. alboplaga and B. xeneoides. In total we screened 303 males and 185 females. Our survey found a large number of specimens which fell outside the classical morphological designations. It also identified no clinal patterns. Specimens with intermediate brush colours were frequently found, as well as males with a strong apical patch as expected from B. alboplaga, combined with a black outer brush as expected from B. xeneoides. The underside band pattern also showed a wide variation that did not follow the predictions based on the original descriptions. Only in the most extreme western (Nigeria) and eastern (Uganda and Tanzania) was morphology consistent and all males were similar to B. xeneoides as traditionally defined. We have not been able to conduct a full investigation of genitalia characters, but examinations of a smaller set of 13 samples across the range it appears the valves are gradually wider the further east a sample originates, regardless of whether the wing morphology would place the specimens as B. alboplaga or B. xeneoides. This pattern combined with an apparent lack of clear genetic separation of the morphological types further supports the conclusion that we are dealing with an unusually morphologically variable species.
We do not advocate here the formal synonymy of the name B. xeneoides as we believe a more detailed investigation is necessary. Our survey of wing pattern covering 488 samples across the whole range shows that the original description of the two species, B. alboplaga and B. xeneoides, do not cover all the natural variation we find within this complex, although the genetic difference is still negligible with the markers used in our current study. Given that androconial colour is usually a very stable trait in the identification of Bicyclus species (as evidenced by the example of the undescribed taxa B. cf. sangmelinae), this complex is most intriguing and a future detailed study would be most welcome.
Revision of Condamin's classic species-groups
We consider Condamin's (Citation1973) concept of species-grouping to remain largely useful and advocate its continued usage. However, our phylogeny shows that some of his species groupings are redundant or do not represent natural groups. Based mainly on the molecular evidence presented here, but also on examinations of androconia and male genitalia we hereby propose a few revisions of Condamin's species group arrangement. We aimed to find ways to lower the number of species-groups, and also removed monospecific groups by merging them with their sister groups when the relationships were strongly supported. Any species-group found not to be monophyletic was always revised. The following discussion roughly follows the tree in , working through the groups from top down. A complete listing of the revised species-group members is found in supplemental data S4 (see supplemental material online).
The evadne-group remain as defined by Condmain (Citation1973), but as noted above we believe future work will show that B. xeneoides represents the same species as B. alboplaga. The nobilis- and trilophus-groups were both originally described as monospecific, but with the addition of two newly described species (B. sealeae and B. larseni) and our raising of B. jacksoni to full species status they now contain two and three species, respectively. They are both fairly distinct from other groups based on androconia and male genitalia. Taking this into account, as well as their lack of good support with any other group, we feel they should remain as independent groups at present.
The ignobilis-group has had a recent full revision with the addition of three new species (Brattström et al., Citation2015). This group is recovered as sister to a well-supported clade containing the original three, hewitsoni-, italus-, and medontias-groups that all share a similar morphology. Although there are deep basal divergences within this clade in our tree, they nevertheless together constitute a well-supported clade (BS = 100, PP = 1). These species are morphologically distinct compared with other Bicyclus. In addition to their bluish dorsal transverse bands, they also share a distinct wing pattern and male genital morphology. The two species in which larval morphology is documented (B. medontias and B. italus) show distinct differences to all other known Bicyclus larvae and the host plants are Zingiberaceae rather than grasses. To our knowledge there are no ecological characters that clearly separate either B. hewitsoni or B. medonitas from any of the other species previously assigned to the italus-group, and therefore we hereby merge all of these taxa in the new hewitsoni-group. This arrangement helps reduce the number of groups, and also removes two monospecific groups. We did not include samples from B. bergeri in our phylogeny, but its placement within this group is not in doubt.
The sciathis-group remains as defined by Condamin (Citation1973), but has been expanded recently with a full systematic revision that included description of four new species (Brattström et al., Citation2016).
The monospecific taenias-group and the two species forming the milyas-group have a weak support with regards to their location within the tree and we have therefore kept these groups as they were originally designated.
Three of Condamin's species groups: the monospecific anisops-group, the saussurei-group (B. saussurei, B. suffusa, and B. dentata) and the danckelmani-group show a grouping that requires some modification. The danckelmani-group was originally described with six members (B. danckelmani, B. neustetteri, B. matuta, B. persimilis, B. albocinctus, and B. aurivillii) and with later descriptions a further three species were included (B. pareensis, B. simulacris, and B. uzungwensis). Seven of these cluster together, but with the members of the anisops- and saussueri-group. However, the original groups are not recovered as monophyletic groups and we therefore merge all of the members of these three groups (excluding B. matuta and B. persimilis that are recovered in a totally different cluster redefined as the martius-group; see below) as our redefined saussurei-group. We have no genetic data from B. suffusa, but morphologically it is extremely close to B. saussurei and we feel confident of its placement in this group.
As previously shown by Monteiro and Pierce (Citation2001) we also found that Condamin's (Citation1973) placement of B. anynana together with B. safitza and B. cottrelli was incorrect. Bicyclus anynana is instead retrieved with high support as sister to a large clade containing the auricruda-, mollitia-, and angulosa-group. The redefined safitza-group, therefore, only includes B. safitza and B. cottrelli. The monospecific funebris-group is retained as it has a long independent evolutionary history and it shows distinctly different genitalia from related species.
The remaining two species of Condamin's (Citation1973) danckelmani-group (B. matuta and B. persimilis) that failed to cluster with the other original members of this group (see above) instead clustered with three other species-groups in a well-supported clade (BS = 92, PP = 1). This clade includes all members of the dubia-, martius-, madetes, and sophrosyne-groups. The martius-group used to be called the sanaos-group until Larsen (Citation2003) changed the name of its oldest member. To keep the number of groups to a reasonable level we are merging this clade as our revised martius-group. We also treat the species B. similis as a member of our larger group based on genitalia morphology and the peculiar lack of a discal cell brush androconia, a trait shared by B. buea. Many other members of our redefined martius-group show a strong reduction in size of this androconia further supporting the placement of B. similis.
We retrieved B. ena as the sister taxon to the campina-group rather than the dorothea-group where it was placed by Condamin (Citation1973). We therefore expand the original campina species-group to include B. ena, but as B. ena is an older name we rename the group into the ena-group. This group also includes two species for which we have no sequences (B. condamini and B. vansoni) as their male genitalia are extremely similar to B. campina. The dorothea-group is recovered with strong support, but with the monospecific vulgaris-group nested within it. We therefore move B. vulgaris to our redefined dorothea-group.
To further reduce the number of species-groups, we combine the rhacotis-, mesogena-, and sambulos-group as the revised rhacotis-group. We have not been able to include any sequence data from one member, B. nachtetis, but its morphology is very close to B. technatis and we feel confident about its placement.
As indicated above we retrieved B. anynana with high support (BS = 100, PP = 1) as a sister taxon to three species groups: auricruda-, mollitia-, and angulosa-groups and we therefore redefine the angulosa-group to include all of these taxa. No sequence data were available for the very rare and local species B. kiellandi, but Condamin's (Citation1983) morphological data place it firmly in his classic angulosa-group so we consider it a member of our larger redefined group.
Conclusions
To our knowledge, the current study represents the most comprehensive phylogenetic analysis for a large African butterfly group in terms of taxon sampling and the number of markers used. We have sampled both species of Hallelesis and 93% of all 103 currently recognized Bicyclus species and used up to 10 molecular markers totalling ∼7.5kb of DNA nucleotide sequence. Only seven described Bicyclus species (B. bergeri, B. condamini, B. kiellandi, B. nachtetis, B. similis, B. suffusa, and B. vansoni) are not represented in our tree, but with the exception of B. similis, their traditional species groups are all represented in our data matrix.
Our phylogeny greatly improves upon the hypothesis presented by Monteiro and Pierce (Citation2001) which was based on three molecular markers and 51 Bicyclus species. We now have a more robust understanding of the evolution of the group, establishing with confidence (except for some early nodes) the relationships within and among most of the different species-groups of Bicyclus. The key focus for future phylogenetic work in the genus is likely to be the recent radiations within the individual species-groups. With this phylogeny we now have a solid framework for future ecological studies, but it also shows the potential of the Bicyclus system for a range of more detailed studies. Further, it highlights the importance of including multiple samples of each recognized species as well as careful investigation of geographic distributions when performing the selection of samples for sequencing. The identification of the new species, B. collinsi, or the ‘rediscovery’ of B. brunnea would have been unlikely without such precautions.
A few groups remain in need of thorough revisions at the species-group level, with the evadne, rhacotis, and martius-groups currently being investigated (Brattström et al. unpublished). The dorothea-complex would be an ideal candidate group for genomic studies of patterns of selection in presumably early stages of speciation. Given the relative abundance of these species it would be possible to collect samples from populations across the whole range of all three species with multiple sympatric, parapatric, and allopatric comparisons. It is possible that a similar pattern exists within the alboplaga-complex, but given that the variation in this complex is less structured combined with a relative scarcity of the involved taxa that would be a much harder task. As B. dorothea can be raised in a laboratory environment it would also be possible to conduct behavioural studies to investigate potential hybrid avoidance and patterns in mate choice to better understand the evolutionary history in this group.
Supplemental data
Supplemental data for this article can be accessed here: http://dx.doi.org/10.1080/14772000.2016.1226979.
Supplemental data S1: List of all sampled specimens with location data and gene coverage included
Download MS Excel (30.5 KB)Supplemental data S2: The best partitioning schemes and optimal evolutionary models for each dataset.
Download PDF (70.1 KB)Supplemental data S3: Bayesian Inference tree for the genus Bicyclus (with samples from Mydosama and Hallelesis used as outgroups) constructed by Mr. Bayes 3.2.
Download PDF (554.9 KB)Supplemental data S5: Systematic checklist (with complete synonymic listing) of currently known Bicyclus species (October 2016)
Download PDF (135 KB)Supplemental data S5: Notes on the taxonomic history and available types in the aucricruda-group
Download PDF (2.6 MB)Acknowledgements
Blanca Huertas (BMNH), Ugo Dall'Asta, Alice-Marie Busset and Jurate de Prins (MRAC), and Steve C. Collins (ABRI) kindly gave us access to the museum collections. Teresa Di Micco de Santo assisted in the investigation of the material kept at ABRI. Gunvi Lindberg provided photographs of type material deposited at the Swedish Museum of Natural History in Stockholm. Antonia Monteiro kindly shared aliquots of previously sequenced specimens, as well as photos of many important voucher specimens. Freerk Molleman and Szabolcs Sáfián provided specimens for the molecular analysis. David C. Lees provided location data for material in the dorothea-complex. We would also like to thank all the people who collected the material now deposited in the investigated museum collections. Eli Harji assisted with proofreading and language editing. Finally, we like to thank Michel Condamin, without his impressive monographic treatment of Bicyclus it is doubtful the systematics of the genus would be as well resolved as it is now. Logistical support in Nigeria was provided by A.P. Leventis Ornithological Research Institute (APLORI). This is APLORI contribution number 113.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Ackery, P. R., Smith, C. R., & Vane-Wright, R. I. (1995). Carcasson's African butterflies - an annotated catalogue of the Papilionides and Hesperoidea of the Afrotropical region. London: The Natural History Museum.
- Aduse-Poku, K., Brattström, O., Kodandaramaiah, U., Lees, D. C., Brakefield, P. M., & Wahlberg, N. (2015). Systematics and historical biogeography of the Old World butterfly subtribe Mycalesina (Lepidoptera: Nymphalidae: Satyrinae). BioMed Central Evolutionary Biology, 15, 167.
- Aurivillius, P. O. C. (1925). Familie: Satyridae, Gattung: Mycalesis [Family: Satyridae, Genus: Mycalesis]. In A. Seitz (Ed.), Die Gross-Schmetterlinge Der Erde [Macrolepidoptera of the world] (Vol. 13, pp. 84–97). Stuttgart: Alfred Kernen.
- Bacquet, P. M. B., Brattström, O., Wang, H-L., Allen, C. E., Löfstedt, C., Brakefield, P. M., & Nieberding, C. M. (2015). Selection on male sex pheromone composition contributes to butterfly reproductive isolation. Proceedings of the Royal Society of London B, 282, 20142734. doi:10.1098/rspb.2014.2734
- Brakefield, P. M., Beldade, P., & Zwaan, B. J. (2009). The African Butterfly Bicyclus anynana: A model for evolutionary genetics and evolutionary developmental biology. Cold Spring Harbor Protocols, 2009, 1–9. doi:10.1101/pdb.emo122
- Brakefield, P. M., & Frankino, W. A. (2009). Polyphenisms in Lepidoptera: Multidisciplinary approaches to studies of evolution. In N. Ananthakrishnan & D. W. Whitman (Eds.), Phenotypic plasticity in insects. Mechanisms and consequences (pp. 121–152). Plymouth: Science Publishers.
- Brakefield, P. M., & Larsen, T. B. (1984). The evolutionary significance of dry and wet season forms in polyphenic tropical Satyrinae. Biological Journal of the Linnaean Society, 22, l–12.
- Brattström, O. (2012). Bicyclus brakefieldi (Nymphalidae, Satyrinae), a new species of Bicyclus from the Congo River basin. Tropical Lepidoptera Research, 22, 62–65.
- Brattström, O., Aduse-Poku, K., Collins, S. C., & Brakefield, P. M. (2015). Revision of the Bicyclus ignobilis species-group (Lepidoptera: Nymphalidae: Satyrinae) with descriptions of two new species. Zootaxa, 4018, 57–79.
- Brattström, O., Aduse-Poku, K., Collins, S. C., Di Micco de Santo, T., & Brakefield, P. M. (2016). Revision of the Bicyclus sciathis species group (Lepidoptera: Nymphalidae) with descriptions of four new species and corrected distributional records. Systematic Entomology, 41, 207–228.
- Collins, S. C., & Larsen, T. B. (2008). Eighteen new species, five new subspecies, and interesting data on other African butterflies - Fourth ABRI research paper. Metamorphosis, 19, 42–113.
- Condamin, M. (1961). Mises au point de synonymie et descriptions de nouveaux Bicyclus (Lepidoptera Satyridae) [Updates on synonymy and descriptions of new Bicyclus (Lepidoptera Satyridae)]. Bulletin de l'I.F.A.N ser.A, 23, 782–799.
- Condamin, M. (1973). Monographie du genre Bicyclus (Lepidoptera Satyridae) [Monograph of the genus Bicyclus (Lepidoptera Satyridae)]. Dakar: IFAN-Dakar.
- Condamin, M. (1983). Deux nouvelles espèces de Bicyclus (Lepdioptera Satyridae) de Tanzanie [Two new species of Bicyclus (Lepidoptera Satyridae) from Tanzania]. Bulletin de l'Institut fondamental d'Afrique noire Série A, 45, 173–187.
- Condamin, M., & Fox, R. M. (1964). Le complexe de Bicyclus dorothea (Lepidoptera Satyridae) [The Bicyclus-dorothea complex (Lepidoptera Satyridae)]. Bulletin de l'Institut fondamental d'Afrique noire Série A, 26, 624–631.
- Condamin, M., & Soltani, B. (1980). Contribution à l'étude des Lépidoptères Satyridae de Centrafrique [Contributions to the study of Central African Satyridae]. Bulletin de l'Institut fondamental d'Afrique noire Série A, 42, 166–180.
- Congdon, C., Collins, S. C., & Kielland, J. (1998). Kielland's Butterflies of Tanzania. Supplement. Nairobi: African Butterfly Research Institute.
- Cramer, P. (1779). De uitlandsche kapellen voorkomende in de drie waereld deelen, Asia, Africa en America [The exotic butterflies occurring in three continents of Asia, Africa and America] (Vol. 3). Amsterdam: S. J. Baalde.
- French, V., & Brakefield, P. M. (1995). Eyespot development on butterfly wings: The Focal signal. Developmental Biology, 168, 112–123.
- Hall, T. A. (1999). BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series, 41, 95–98.
- Lanfear, R., Calcott, B., Ho, S. Y. W., & Guindon, S. (2012). PartitionFinder: Combined selection of partitioning schemes and substitution models for phylogenetic analyses. Molecular Biology and Evolution, 29, 1695–1701.
- Kielland, J. (1990). The butterflies of Tanzania. Melbourne: Hill House.
- Kirby, W. F. (1871). A synonymic catalogue of Diurnal Lepidoptera. London: John van Voorst.
- Larsen, T. B. (2003). The validity and synonymy of the names Bicyclus martius Fabricius, 1793 and B. sanaos Hewitson, 1866 (Nymphalidae; Satyrinae). Entomologists’ Record and Journal of Variation, 115, 95–96.
- Libert, M. (1996). Nouveaux Bicyclus du Cameroun (Lepidoptera, Satyridae) [New Bicyclus from Cameroon (Lepidoptera, Satyridae]. Bulletin de la Société entomologique de France, 101, 201–208.
- Miller, M., Pfeiffer, W., & Schwartz, T. (2010). Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE) (pp. 1–8). New Orleans, LA: IEEE Computer Society.
- Monteiro, A., & Pierce, N. E. (2001). Phylogeny of Bicyclus (Lepidoptera: Nymphalidae) Inferred from COI, COII, and EF-1a Gene Sequences. Molecular Phylogenetics and Evolution, 18, 264–281.
- Moore, F. (1880). On the Asiatic Lepidoptera referred to the genus Mycalesis; with descriptions of new genera and species. Transactions of the Royal Entomological Society of London, 28, 155–178.
- Nieberding, C. M., De Vos, H., Schneider, M. V., Lassance, J-M., Estramil, N., Andersson, J., … Brakefield, P. M. (2008). The male sex pheromone of the butterfly Bicyclus anynana: Towards an evolutionary analysis. Public Library of Science ONE, 3, e2751.
- Oliver, J. C., Robertson, K. A., & Monteiro, A. (2009). Accommodating natural and sexual selection in butterfly wing pattern evolution. Proceedings of the Royal Society of London B, 276, 2369–2375.
- Oliver, J. C., & Monteiro, A. (2011). On the origins of sexual dimorphism in butterflies. Proceedings of the Royal Society of London B, 278, 1981–1988.
- R Core Team. (2015). R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing. Retrieved from http://www.R-project.org (accessed 17 August 2016).
- Rambaut, A., Suchard, M. A., Xie, D., & Drummond, A. J. (2014). Tracer (Version 1.6.). Retrieved from http://beast.bio.ed.ac.uk/Tracer (accessed 17 August 2016).
- Ronquist, F., & Huelsenbeck, J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics, 19, 1572–1574.
- Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., KLongair, M., Pietzsch, T., … Cardona, A. (2012). Fiji: An open-source platform for biological-image analysis. Nature methods, 9, 676–682.
- Stamatakis, A. (2014). RAxML Version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 30, 1312–1313.
- Tamura, K., Stecher, G., Peterson, D., Filipski, A., & Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 30, 2725–2729.
- Vande weghe, G. (2007). Nouvelle espèce de Bicyclus du Gabon (Lepidoptera, Nymphalidae, Satyridae) [New species of Bicyclus from Gabon (Lepidoptera, Nymphalidae, Satyridae)]. Lambillionea, 42, 455–458.
- Vande weghe, G. (2009). Description de nouveaux taxons et contribution à l’étude des Lépidoptères afrotropicaux (Lepidoptera: Nymphalidae, Limenitidinae, Satyrinae; Hesperiidae, Hesperiinae, Lycaenidae, Theclinae) [Description of new taxa and contribution to the study of Afrotropical butterflies]. Entomologia Africana, 14(Suppl.), 1–24.
- Vande weghe, G. (2010). Les papillons du Gabon. [Butterflies of Gabon]. Libreville: Wildlife Conservation Society.
- Wahlberg, N., & Wheat, C. W. (2008). Genomic outposts serve the phylogenomic pioneers: Designing novel nuclear markers for genomic DNA extractions of Lepidoptera. Systematic Biology, 57, 231–242.