Abstract
Green algal species of spherical cell shape are generally considered to belong to the genus Chlorella, which are mostly freshwater or terrestrial organisms. Phylogenetic studies have shown that this genus is polyphyletic and belongs to different classes. However, until now, only freshwater or terrestrial strains have been studied. Here we investigated 11 strains of ‘marine’ Chlorella deposited in public culture collections, which we studied using an integrative approach. These strains were largely isolated from marine rock pools and brackish estuaries. SSU and ITS regions of the nuclear encoded ribosomal DNA were sequenced, ribosomal secondary structures were analysed and cell morphology, salinity tolerance and reproduction were examined. Our results showed that the marine strains are also of polyphyletic origin. Surprisingly, three marine isolates belong to Chlorella vulgaris according to the phylogenetic analyses, but showed a high phenotypic plasticity. Whereas these strains showed the typical morphology of C. vulgaris under freshwater conditions, they increased the cell shape and formed cell packages under marine conditions. In contrast, the other investigated strains showed no changes after changing the media. Two of the investigated strains belong to the genus Chloroidium, and those remaining represent a new genus, Droopiella.
Key words:
Introduction
The genus Chlorella was described by Beijerinck (Citation1890) for small (<10 µm) coccoid unicellular green algae isolated from a freshwater habitat. Since the establishment of the type species, Chlorella vulgaris Beijerinck, more than 100 species of Chlorella have been described, largely of freshwater or terrestrial origin. Fott & Novakova (Citation1969) and Andreyeva (1975) reduced the number of species to eleven and nine, respectively. Many of the others were transferred to a section of doubtful species because of incomplete descriptions. Andreyeva (Citation1975) included all described marine species in this section. Twelve species of Chlorella have been described from brackish and marine habitats (a summary is given in ). Unfortunately, no cultures are available for most of these species in public culture collections. One of the first described species, Chlorella salina described by Kufferath (Citation1919), differs only by its occurrence in the marine environment from Chlorella vulgaris. Morphologically both species are similar (see ). Other marine species are Chlorella spaerckii, described by Ålvik (Citation1934) and C. marina, C. ovalis, C. salina, and C. stigmatophora, all described by Butcher (Citation1952). C. capsulata, originally described by Guillard et al. (Citation1975), was transferred to the genus Schizochlamydella Korshikov by Watanabe (Citation1977). Wolf et al. (Citation2003) demonstrated based on partial SSU rDNA sequences that this species belonged to the Oocystaceae, which was confirmed by Stenclová et al. (Citation2017). The morphology and taxonomic status of all described marine species are summarized in .
Table 1. Morphological features of Chlorella species isolated from marine and brackish habitats.
George (Citation1957) raised the question, whether the strains isolated by Butcher are truly marine species, or freshwater organisms, which entered the marine ecosystem through river estuaries and were then capable of surviving in brackish or fully marine seawater. The results of his culturing experiment supported this hypothesis.
Several strains used for feeding marine fish larvae in hatcheries have also been called marine Chlorella. They were found to be useful due to their small size (2–4 µm diameter) and ease of culture. These strains were informally named ‘Chlorella’ due to their green coccoid appearance. However, several strains assigned as ‘marine Chlorella’ (UTEX 2341, CCAP 211/46, and CCAP 211/78) represented eustigmatophytes based on morphological, physiological and phylogenetic analyses (Fawley et al., Citation2015; Gladu et al., Citation1995; Maruyama et al., Citation1986) and were therefore not included in this study.
The taxonomic revision of the genus Chlorella is still an on-going process. Phylogenetic studies demonstrated that the freshwater species are closely related to genera with a distinct morphology (see summary in Krienitz & Bock (Citation2012) and the references therein). Bock et al. (Citation2011) demonstrated that several strains originally assigned to the genus Dictyosphaerium also belong to the genus Chlorella. As a result of this study, the generic description was emended. Fourteen species are accepted as true Chlorella (Bock et al., Citation2011). Since then, only one additional species has been described (C. thermophila, Ma et al., Citation2015). In the last 10 years the focus has been on the systematics and biodiversity of terrestrial Chlorella and Chlorella-like species. The usage of an integrative approach described in detail in Darienko et al. (Citation2016) has revealed that the diversity of Chlorella-like organisms is much higher than expected and led to the taxonomic revision of these taxa, which are not closely related to the type species of Chlorella. Considering terrestrial strains in public culture collections and new isolates, the genera of the Watanabea clade were taxonomically revised and new genera were described (see Darienko & Pröschold, Citation2019a, Citation2019b and references therein). The genus Auxenochlorella was revised by Darienko & Pröschold (Citation2015), and new genera were described (see Darienko et al., Citation2016 and references therein).
In contrast to the aquatic and terrestrial environments, very little is known about the occurrence and biology of Chlorella-like organisms in brackish and marine habitats. Despite the original descriptions of marine taxa (), only the genus Marinichlorella has been described based on the morphology and phylogeny of a newly isolated strain from seawater of the Korean sea (Aslam et al., Citation2007). The aim of this study was to answer the question whether marine Chlorella strains are ‘true’ Chlorella or whether they belong to other phylogenetic lineages. To address this question, we studied several strains using an integrative approach, investigating those held in public culture collections and which originated from brackish or marine environments.
Materials and methods
Origin and phenotypic plasticity, culture conditions and light microscopy of the investigated strains
The origins of the investigated strains are given in Table S1 (see online supplemental material, which is available from the article’s Taylor & Francis Online page at http://dx.doi.org/10.1080/14772000.2019.1690597). The strains were selected from the Culture Collection of Algae and Protozoa in Oban, Scotland (CCAP; https://www.ccap.ac.uk), the Culture Collection of Algae in Göttingen, Germany (SAG; http://sagdb.uni-goettingen.de), and the Culture Collection of Algae at the University of Texas in Austin, USA (https://utex.org). To study the phenotypic plasticity, the strains were cultivated in three culture media: (1) under freshwater conditions in modified Bold Basal medium (3N-BBM+V; medium 26a in Schlösser, Citation1997); (2) under brackish condition in 3N-BBM+V with 50% sterile filtered seawater; and (3) under marine condition 3N-BBM+V with 100% sterile filtered seawater. All investigated algae were grown under standard culture conditions (20 °C, 12 h light: 12 h dark cycle, light intensity of 50 μmol photons/m2s). For morphological observations of the 2-week old cultures, an Olympus BX-60 microscope was used (Olympus, Tokyo, Japan), and micrographs were taken with a ProgRes C14plus camera using the ProgRes CapturePro imaging system (version 2.9.0.1), both from Jenoptik, Jena, Germany.
DNA extraction, PCR, and sequencing
DNA extraction, PCR, and PCR purification and sequencing were conducted using the detailed protocol described in Darienko et al. (Citation2016). The genomic DNA of the strains was extracted using the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) and following the instructions provided by the manufacturer. The SSU and ITS rDNA were amplified in PCR reactions using the Taq PCR MasterMix Kit (Qiagen, Hilden, Germany) with the primers (Eurofins, Cologne, Germany) listed in Table S2 (see supplemental material online). Several of the investigated strains contained introns in the SSU rDNA. Therefore, additional primers were used to complete the SSU and ITS rDNA sequences of the selected strains (Table S2, see supplemental material online). The sequences are available in the EMBL, GenBank and DDBJ sequence databases under the accession numbers given in Table S1 (see supplemental material online) and .
Fig. 1 Molecular phylogeny of the Trebouxiophyceae based on SSU rDNA sequence comparisons. The phylogenetic tree shown was inferred using the Maximum likelihood method based on the data sets (1784 aligned positions of 69 taxa) using PAUP 4.0b10. For the analyses, the best model was calculated by Modeltest 3.7. The setting of the best model was given as follows: TrN + I + G (base frequencies: A 0.2468, C 0.2322, G 0.2778, T 0.2432; rate matrix A-C 1.0000, A-G 2.1652, A-U 1.0000, C-G 1.0000, C-U 5.1240, G-U 1.0000) with the proportion of invariable sites (I = 0.5997) and gamma shape parameter (G = 0.5342). The branches in bold are highly supported in all analyses (Bayesian values > 0.95 calculated with MrBayes; bootstrap values > 70% calculated with PAUP using Maximum likelihood, Neighbour-joining, Maximum parsimony and RAxML using Maximum likelihood). The new sequences presented in this study are highlighted in bold.
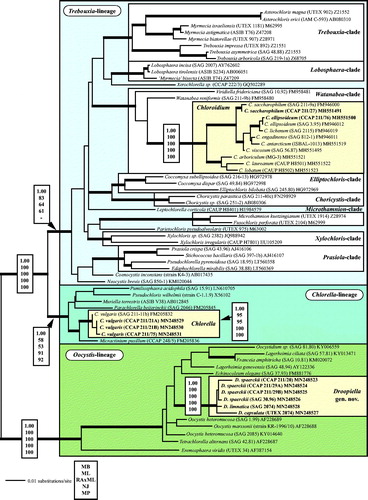
Fig. 2 (2.1) Molecular phylogeny of the genera Chlorella and Micractinium based on SSU and ITS rDNA sequence comparisons. The phylogenetic tree shown was inferred using the Maximum likelihood method based on the data sets (2602 aligned positions of 13 taxa) using PAUP 4.0b10. For the analyses, the best model was calculated by Modeltest 3.7. The setting of the best model was given as follows: GTR + I + G (base frequencies: A 0.2256, C 0.2541, G 0.2751, T 0.2452; rate matrix A-C 0.7854, A-G 1.2408, A-U 1.0687, C-G 0.4617, C-U 3.2928, G-U 1.0000) with the proportion of invariable sites (I = 0.7774) and gamma shape parameter (G = 1.2073). The branches in bold are highly supported in all analyses (Bayesian values > 0.95 calculated with MrBayes; bootstrap values > 70% calculated with PAUP using Maximum likelihood, Neighbour-joining, Maximum parsimony and RAxML using Maximum likelihood). (2.2) TCS haplotype network inferred from ITS-1, 5.8S, and ITS-2 rDNA sequences of Chlorella vulgaris. This network was inferred using the algorithm described by Clement et al. (Citation2002). Sequence nodes corresponding to samples collected from different habitats. The accession numbers of all haplotypes are summarized in Table S3 (see supplemental material online).
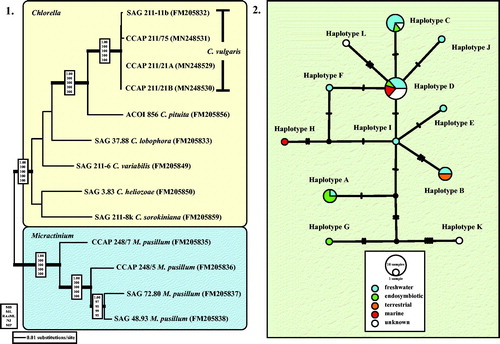
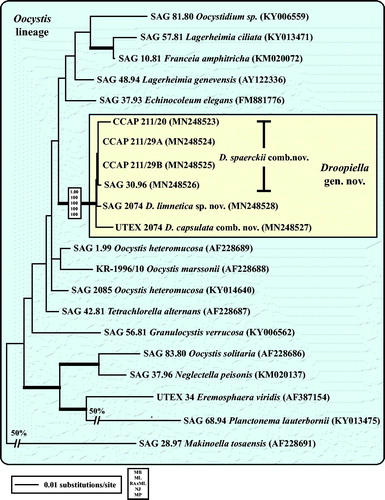
Fig. 3 Molecular phylogeny of the Oocystis lineage (Trebouxiophyceae) based on SSU rDNA sequence comparisons. The phylogenetic tree shown was inferred using the Maximum likelihood method based on the data sets (1778 aligned positions of 23 taxa) using PAUP 4.0b10. For the analyses, the best model was calculated by Modeltest 3.7. The setting of the best model was given as follows: TrN + I + G (base frequencies: A 0.2498, C 0.2263, G 0.2752, T 0.2487; rate matrix A-C 1.0000, A-G 1.5365, A-U 1.0000, C-G 1.0000, C-U 4.1953, G-U 1.0000) with the proportion of invariable sites (I = 0.5925) and gamma shape parameter (G = 0.6827). The branches in bold are highly supported in all analyses (Bayesian values > 0.95 calculated with MrBayes; bootstrap values > 70% calculated with PAUP using Maximum likelihood, Neighbour-joining, Maximum parsimony and RAxML using Maximum likelihood).
Phylogenetic analyses
The SSU rDNA sequences of all strains were aligned according to their secondary structures. The ITS-1 and ITS-2 sequences of all strains were folded according to the protocol described in detail in Darienko et al. (Citation2016). The new SSU sequences were included in a data set of 69 SSU rDNA sequences of all representative lineages belonging to the Trebouxiophyceae (1784 bp). The new SSU and ITS rDNA sequences belonging to the Chlorella clade were analysed in a data set (13 taxa, 2602 bp), which included closely related species of Chlorella vulgaris and Micractinium isolates, the sister of the genus Chlorella, as an outgroup. The sequences of strains belonging to the Oocystis clade were included in a larger data set (23 taxa, 1778 bp) representing all lineages of this clade.
For all data sets the best evolutionary models were calculated with the program Modeltest 3.7 (Posada, Citation2008) using the Akaike Information Criterion (Akaike, Citation1974). The settings of the best models are given in the figure legends. The following methods were used for the phylogenetic analyses: distance, Maximum parsimony, Maximum likelihood, and Bayesian inference. Programs used included PAUP version 4.0a165 (Swofford, Citation2002), RAxML version 7.0.3 (Stamatakis, Citation2006), and MrBayes version 3.2.3 (Ronquist et al., Citation2012).
ITS-2 secondary structures and ITS-2/CBC approach
The secondary structures of ITS-2 sequences were folded using the computer programmfold version 3.6 (Zuker, Citation2003), which used the thermodynamic model (minimal energy). Details on folding constraints and further methodology are given in the protocol of Darienko et al. (Citation2018). The folding results were then used for species delimitation within the newly erected genus Droopiella (see below). For the ITS-2/CBC approach, the conserved region of ITS-2 was extracted following the procedure as described in Darienko et al. (Citation2018). The barcodes for each species were compared to detect compensatory base changes (CBCs), hemi-CBCs (HCBCs), insertions/deletions, and single or unpaired bases.
Distribution and haplotype network analyses
Network analyses were performed to detect and determine the distribution of haplotypes among populations of Chlorella vulgaris. To establish an overview about the distribution of C. vulgaris, the ITS haplotypes of this study and the study of Müller et al. (Citation2005) were used for the BLASTn search (100% coverage, >97% identity; Altschul et al., Citation1990). The metadata covering the habitat of each entry in GenBank for the different haplotypes were collected. To construct the haplotype network, we used the TCS network tool (Clement et al., Citation2000, Clement et al., Citation2002) implemented in PopART version 1.7 (Leigh & Bryant, Citation2015).
Results
Molecular phylogeny of marine Chlorella
All investigated strains belong to three independent lineages of the Trebouxiophyceae (). The strains CCAP 211/27 and CCAP 211/76, originally assigned as Chlorella marina and C. ellipsoidea, are members of the Watanabea clade and represent the two species Chloroidium saccharophilum and C. ellipsoideum, respectively (see details in Darienko et al., Citation2018). The strains CCAP 211/21A and CCAP 211/21B both assigned as Chlorella ovalis and CCAP 211/75 C. sp. are 99.7% identical in sequence with the authentic strain of Chlorella vulgaris (Chlorella clade). All remaining strains (CCAP 211/20, CCAP 211/29 A, CCAP 211/29B, SAG 30.96, SAG 2074, and UTEX 2074) form a well-supported clade in the Oocystis clade.
To confirm that the three strains belong to Chlorella vulgaris, we analysed the SSU and ITS rDNA sequences using two different methods. The first method, a phylogenetic analysis of a concatenated data set representing the closest relatives of Chlorella vulgaris clearly showed that those strains belong to C. vulgaris (Fig. 2.1). However, the SSU and ITS rDNA sequences are not 100% identical. Therefore, the second method tested all available ITS rDNA sequences in GenBank and compared them in a TCS network to detect if the marine strains belong to a separate genotype. As shown in Fig. 2.2, the ITS network represented the distribution of 12 haplotypes (A-L) among Chlorella vulgaris. The marine isolates CCAP 211/21A, CCAP 211/21B, and CCAP 211/75 belong to the haplotypes D and H, respectively. Whereas haplotype D, the most common one, contains isolates of different habitats (freshwater, endosymbionts, marine), haplotype H is exclusively represented by a marine strain so far showing that the marine strains were distributed in a different pattern.
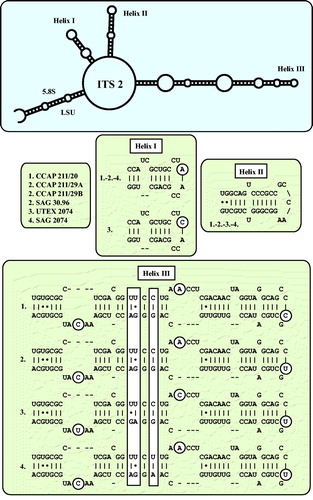
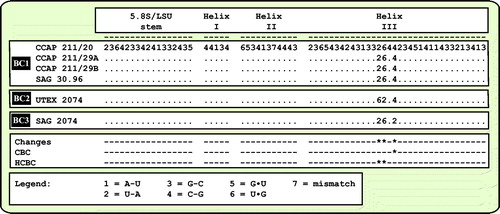
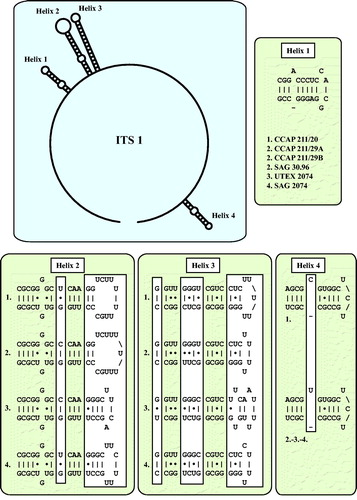
As already demonstrated in , the remaining marine strains representing the new genus Droopiella (see description below) were members of the Oocystis clade, which is confirmed by phylogenetic analyses of a SSU data set containing additional taxa of this clade (). To decide how many species the six strains represent, we applied the ITS-2/CBC approach, which was introduced by Darienko et al. (Citation2016). The secondary structures of ITS-1 and ITS-2 are given in . The comparison of these structures showed few changes among the strains in helices 2-4 of ITS-1 and in helices I-III of ITS-2 (highlighted in white boxes in ). Among the conserved region of ITS-2 (ITS-2 Barcode), the strains differed only by one CBC and two HCBCs. The strains CCAP 211/29 A, CCAP 211/29B, and SAG 30.96 were identical in ITS rDNA sequences and belong therefore to the same species, Droopiella spaerckii (see emended diagnosis below). The authentic strain of Chlorella stigmatophora showed only few differences in the ITS and belong therefore to the same species of Droopiella. D. spaerckii is closely related to the strain UTEX 2074, authentic strain of Chlorella capsulata, which differed by two HCBCs in Helix III of ITS-2. Only SAG 2074 originally assigned as Amphikrikos nanus has one CBC in Helix III compared with the other strains and represents a new species according to the ITS-2/CBC approach. We describe this species below as Droopiella limnetica ().
Fig. 4 ITS-1 secondary structures of the Droopiella strains investigated in this study. The variable regions are marked in white boxes.
Morphology and phenotypical plasticity of investigated strains
George (Citation1957) questioned the existence of true marine Chlorella and highlighted the possibility that Chlorella species isolated from seawater originated from freshwater habitats. To address this question, we cultivated the investigated strains under both freshwater and marine conditions. The three marine isolates, as shown above, were almost identical in sequence with the freshwater strain CCAP 211/11B of Chlorella vulgaris. Surprisingly the three marine strains (CCAP 211/21A, CCAP 211/21B, and CCAP 211/75) cultivated in marine medium looked morphologically different compared with the original description of Chlorella vulgaris (). The differences are summarized as follows:
Figs. 7–18 Morphology and phenotypic plasticity of Chlorella vulgaris strains under freshwater (7, 10, 13, 16), brackish (8, 11, 14, 17) and marine conditions (9, 12, 15, 18). 9. CCAP 211/75; 10–12. CCAP 211/21A; 13–15. CCAP 211/21B; 16–18. CCAP 211/11B; scale bar = 20 µm.
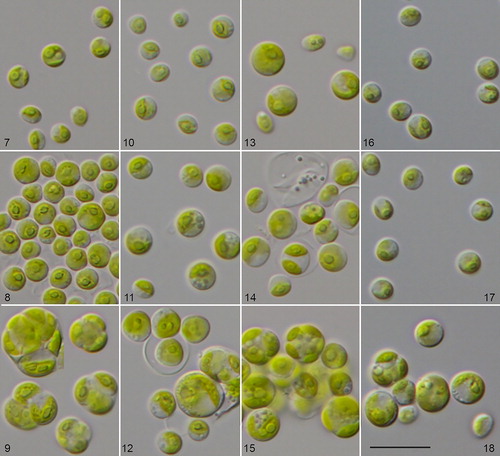
All three strains were almost identical in their morphology. The cells were often gathered in packages of 4–8 or more cells. Solitary cells were rarely present. Cells were 10–12 µm in diameter, cell packages had dimensions of 11.2 × 18.6 − 15.7 × 21.6 µm. Cell walls were relatively thick. Chloroplasts were cup-shaped covering almost the whole cell. Pyrenoids were large and usually clearly visible, surrounded by 2–4 large starch grains. Mucilage production was not observed. Reproduction was by autospores, which were produced in large numbers (8–16). Young cells often remained in the sporangium. The freshwater strain CCAP 211/11B also changed its morphology after 2 weeks in culture in marine conditions. Cells usually increased in size and reached on average 10–11 µm in diameter. Cell walls thickened, and cells started to form 4–8-celled packages. In contrast, cells of the same age cultivated on 3N-BBM+V medium were 6.0–7.2 µm in diameter and usually solitary. The marine strains in 2-week old cultures on 3N-BBM+V agar had similar cell dimensions (4.9–6.7 µm in diameter) and became solitary, as is typical for Chlorella vulgaris in freshwater habitats ().
The other investigated strains showed no morphological changes under marine and freshwater conditions (). The cells of the strain CCAP 211/27 () originally described as Chlorella marina showed similar morphology to the authentic strain SAG 211-9a of Chloroidium saccharophilum. Strain CCAP 211/76 () is similar in morphology to the authentic strain SAG 3.95 Chloroidium ellipsoideum. Detailed morphological descriptions of both species are given in Darienko et al. (Citation2010, Citation2018).
Figs. 19–22 Morphology and phenotypic plasticity of Chloroidium saccharophilum (19–20) and C. ellipsoideum (21–22) under freshwater (19, 21), and marine conditions (20, 22). 19–20. CCAP 211/27; 21–22. CCAP 211/76; scale bar = 10 µm.

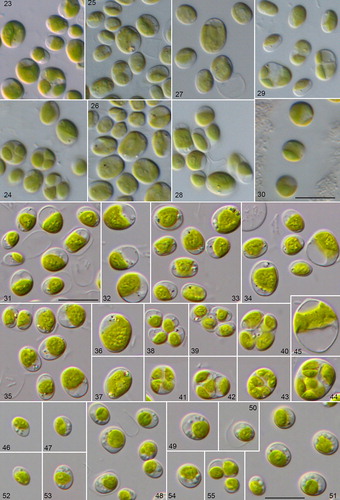
Figs. 23–55 Morphology and phenotypic plasticity of the different Droopiella species. (23-28) D. spaerckii and (29-30) D. capsulata under freshwater (23,25,27,29), and marine conditions (24,26,28,30). Strains: (23-24) CCAP 211/20; (25-26) CCAP 211/29A; (27-28) CCAP 211/29B; (29-30) UTEX 2074; (31-45) D. limnetica sp. nov. (SAG 2074) under freshwater conditions; (46-55) D. spaerckii (SAG 30.96) under brackish conditions; scale bar = 10 μm.
The morphology and phenotypic plasticity of the other investigated strains are described in detail in the species diagnoses of the newly erected genus Droopiella (see below). These strains also showed no morphological differences when cultivated under both conditions (marine and freshwater; ).
Discussion
Are there any true marine Chlorella species? Polyphyly of brackish and marine Chlorella-like organisms
The answer to this question is demonstrated in . Chlorella strains isolated from brackish and marine habitats were distributed in three independent lineages of the Trebouxiophyceae. Only three investigated strains are ‘true’ Chlorella and belong to Chlorella vulgaris, the type species of the genus. Interestingly, these strains showed a high phenotypic plasticity so that they could not be identified as Chlorella vulgaris or even assigned to this genus if cultivated under marine conditions. González et al. (Citation2013) also found a strain of C. vulgaris in an estuarine coastal environment in Chile (called Baker strain), which showed similar morphological differences as shown for our strains. They also isolated another strain (called Coliumo strain), which clearly belong together with two investigated strains (CCAP 211/27 and CCAP 211/76) to the genus Chloroidium, a genus which occurs in almost all kinds of habitats including marine (Darienko et al., Citation2010, Citation2018). The six remaining strains investigated in this study are members of the Oocystis clade and represent their own genus, Droopiella. Originally, these isolates were assigned either to the genus Amphikrikos Korshikov or to Schizochlamydella Korshikov (Korshikov, Citation1953). Neither assignment fit any of the investigated strains, which is discussed in detail in the section ‘Taxonomic consequences’ below. Summarizing, our study showed that Chlorella-like organisms are not often reported despite their being distributed worldwide. For example, Aslam et al. (Citation2007) described a new genus Marini-chlorella from the South Korean Sea, which belongs to the Parachlorella clade. Marinichlorella kaistiae cultivated in marine media showed almost no morphological differences to Chlorella vulgaris, which was grown under freshwater conditions. It seems that Chlorella-like organisms of brackish and marine habitats are often overlooked.
Historical background of marine Chlorella taxa
Our results raised the question of the affiliation of the described Chlorella species. As presented in , eleven taxa could be found in the literature. Unfortunately, only two authentic strains of marine Chlorella species were available in public culture collections (CCAP 211/20 Chlorella stigmatophora and UTEX 2074 C. capsulata), and both belong to the new genus Droopiella. In the following section, we try to evaluate the described taxa given in the literature and to correct some taxonomic errors. This evaluation was often difficult because the descriptions were often poor. In addition, morphological techniques have dramatically improved over the last 100 years.
Chlorella salina versus Chlorella gloriosa
Kufferath (Citation1919) described Chlorella salina from a seawater sample collected in Westende, Belgium. According to the description, this alga has a cell size of 3.0–6.0 µm, which sometimes can reach 8.0 µm, and possesses a cup-shaped chloroplast with a pyrenoid surrounded by starch grains. It reproduces by 2–4 autospores. Kufferath mentioned that C. salina is similar to C. vulgaris; however, he described it as a new species because of its occurrence in the marine habitat. Similar, Butcher (Citation1952) isolated a strain of Chlorella-like organism from a seawater sample from a shell fish tank at Conway, North Wales. He probably was not aware of Kufferath’s description and named it C. salina. Butcher (Citation1952) also highlighted the morphological similarity to C. vulgaris. He observed cells of 4.0–7.0 µm in diameter, with a saucer-shaped chloroplast containing the pyrenoid surrounded by starch grains, which reproduces by eight autospores. The only small difference he observed was a slightly different shape of the chloroplast and the occurrence in a marine habitat.
According to our opinion, in both cases we are dealing with the same organism. Calvo-Pérez Rodó & Molinari-Novoa (Citation2015) highlighted that the name C. salina was already introduced by Kufferath (Citation1919) and proposed the new name Chlorella gloriosa Molinari & Calvo-Perez for Chlorella salina sensu Butcher. Unfortunately, the authors did not check the type descriptions of both organisms and made no morphological comparisons of both descriptions. As a consequence, Chlorella salina sensu Kufferrath is identical to C. salina sensu Butcher. The erection of the new name Chlorella gloriosa Calvo-Pérez Rodó & Molinari-Novoa is therefore superfluous. Despite the morphological similarity of C. salina with C. vulgaris, we cannot conclude here whether C. salina is a synonym of C. vulgaris because of the different morphology of C. vulgaris under a marine environment. The strain CCAP 211/25 assigned as Chlorella salina Butcher has a net-like chloroplast containing a large central pyrenoid, a cell size of 15–25 µm, and reproduces by formation of zoospores. This morphology does not correspond to the type description of Butcher (Citation1952). This was already observed by the ultrastructural investigations of this strain by Rascio et al. (Citation1980d).
Chlorella ovalis Butcher (Citation1952)
This species was found in a brackish habitat (water sample, River Crouch, Essex). The cells have an oval to ellipsoidal cell shape, and are 3.0 × 5.0 – 5.0 × 10.0 µm in size. The chloroplasts are parietal, band-like and slightly lobed, without a pyrenoid. Reproduction occurs through forming eight autospores. The iconotype of this species showed autosporangia containing autospores of unequal sizes. All these features are typical for the genus Chloroidium, in particular for C. saccharophilum. The only exception is the lack of pyrenoid for Chlorella ovalis. However, as shown for Chloroidium saccharophilum, this species contains a chloroplast with a single pyrenoid without visible starch grains. Therefore, the presence of a pyrenoid could be easily overseen. In contrast, the strains CCAP 211/21A and CCAP 211/21B, assigned as Chlorella ovalis, have a clearly visible pyrenoid in the chloroplast and reproduce by equal-sized autospores. This does not correspond to the original description of Chlorella ovalis by Butcher. As shown above, both strains belong to C. vulgaris.
Chlorella marina Butcher (Citation1952)
This species was described from a culture of marine flagellate from Port Erin (Irish Sea). This alga is characterized by ovoid cell shape, and is 4.0 × 7.0 – 6.0 × 10.0 µm in size. The chloroplast is parietal, and finely granulated. A pyrenoid was not observed. Reproduction occurs through forming eight autospores. According to Butcher, this organism is similar to Chlorella ovalis Butcher, but differs in the presence of many lipid drops and a ‘granulated’ chloroplast, which was the reason for the separation into the new species. The strain CCAP 211/27 corresponds to the original description. Our phylogenetic analyses revealed that this strain belongs to Chloroidium saccharophilum (Darienko et al., Citation2018). As highlighted above, this species contains a naked pyrenoid, which can be inconspicuous without a special staining or using DIC. Rascio & Casadoro (Citation1981) confirmed the presence of a naked pyrenoid by studying the ultrastructure of the same strain. Therefore, it is possible that Butcher had overlooked the pyrenoid. The features he used for the separation of Chlorella marina are controversial and not sufficient at the present time. In our opinion Chlorella ovalis and Chlorella marina both belong to Chloroidium saccharophilum and represent later synonyms.
Chlorella nordstedtii Printz (Citation1938)
This species was described from a biofilm on a floating piece of a wooden trunk in the sea. Printz (Citation1938) himself was not sure if this alga was marine or brackish, and would survive in a marine habitat. The cells had ovoid to spherical cell shape and sometimes reached a size of 10.0–14.0 µm. Reproduction is by autospores, 8–16 per sporangium. Printz wrote that C. nordstedtii is similar to C. ellipsoidea (now Chloroidium ellipsoideum). The special feature for the separation from C. ellipsoidea was a deeply lobed chloroplast containing a prominent pyrenoid resulting in an asteroid appearance in his opinion. The other peculiarity of this alga was the thin and delicate cell wall, which also partially dissolves after the autospores are released. In all investigations, Printz used fresh water with the addition of ‘a little bit’ of seawater for cultivation. As demonstrated by Darienko et al. (Citation2010, Citation2018), the chloroplast morphology of Chloroidium ellipsoideum is quite plastic. Some of the isolates had deeply lobed chloroplasts, which were partially removed from the cell wall and looked similar to the asteroid chloroplast (Darienko et al., Citation2010). This variability in the chloroplast morphology does not correspond to the results of the phylogenetic analyses. In our opinion, Chlorella nordstedtii could be a later synonym of Chloroidium ellipsoideum.
Chlorella spaerckii Ålvik (Citation1934) versus Chlorella stigmatophora Butcher (Citation1952)
Chlorella spaerckii was described by Ålvik (Citation1934) from oyster breeding tanks in Norway, but he was sure that the organism was widely used for feeding of oysters in the UK and Germany. According to the type description, the algae are ellipsoidal to spherical in shape, and 2.5 × 2.8 – 5.0 × 7.0 µm in size. Chloroplasts are parietal, slightly lobed to perforated. The pyrenoid is absent. Unfortunately, no type cultures are available. Later, Butcher (Citation1952) found in a similar habitat two Chlorella species, which he designated as Chlorella spaerckii and a new species Chlorella stigmatophora. According to his observations, the latter strain has a strong similarity to the C. spaerckii, but possessed a brownish spot, which he called ‘stigma’. For the same reasons, Butcher did not discuss the presence or absence of a pyrenoid in the comparison of both species. The presented figures of both organisms showed many similarities, but were not identical. The first discrepancy was a presence of a perforated chloroplast by C. spaerckii sensu Ålvik. It is possible that Ålvik’s culture was not uni-algal and contained additional algae. Another explanation could be that he observed some old cell stages where the chloroplasts were penetrated by vacuoles, which led him to a wrong interpretation. The second discrepancy is the absence of a pyrenoid by C. spaerckii. On the other hand, there are some morphological features such as cell shape, presence of brownish spots, orientation of autospores and their release from the sporangium, which show high similarity. Rascio et al. (Citation1980a, Citation1980b) investigated the ultrastructure of both species using TEM. They confirmed that brownish spots in the chloroplast of Chlorella stigmatophora are not the stigmata. They also found that Chlorella spaerckii (either CCAP 211/29A or CCAP 211/29B, it remains unresolved which strain was investigated) has the pyrenoid. Both strains also have similar cell wall structure and do not contain sporopollenin. Their investigations correspond with our recent morphological observations. Summarizing, we can partially confirm that Chlorella spaerckii Ålvik (sensu Ålvik and sensu Butcher), and Chlorella stigmatophora Butcher are morphologically identical. The absence of a pyrenoid in C. spaerckii is probably because of low resolution of light microscopy in that time.
Marine Chlorella species, which should be excluded from the genus Chlorella
Chlorella nana Andreoli, Rascio & Casadoro (Citation1978)
According to the type description, this alga has a smooth cell wall containing the sporopollenin. Cells are solitary and small (3.0–4.0 µm in size), containing a bilobed chloroplast without a pyrenoid. Reproduction is by two or four autospores. Interestingly, the four-celled autosporangia are often in the form of a package. Unfortunately, the type culture is unavailable in public culture collections. In our opinion, this species is not a member of the genus Chlorella because of the lack of a pyrenoid. The presence of a pyrenoid is one of the characteristic morphological features, and is also reflected in molecular phylogeny (Krienitz & Bock, Citation2012). The only genus in the Chlorella-clade containing chloroplast without a pyrenoid is the genus Meyerella (Fawley et al., Citation2005). This organism was found in freshwater lakes in Minnesota (USA) and as an endosymbiont in Paramecium species (Kreutz et al., Citation2012; Lanzoni et al., Citation2016).
Chlorella peruviana Chacón (Citation1980). According to the type description, this species is also lacking a pyrenoid and should therefore be excluded from the genus Chlorella.
Taxonomic consequences
Chloroidium saccharophilum (W. Krüger) Darienko, Gustavs, Mudimu, Rad-Menéndez, Schumann, Karsten, Friedl & Pröschold, Citation2010: European Journal of Phycology 45(1): 92, .
Synonyms: Chlorella marina Butcher, Citation1952: Journal of the Marine Biological Association of the United Kingdom 31: 181, pl. I: ; Chlorella ovalis Butcher, Citation1952: Journal of the Marine Biological Association of the United Kingdom 31: 181, pl. I: .
Chloroidium ellipsoideum (Gerneck) Darienko, Gustavs, Mudimu, Rad-Menéndez, Schumann, Karsten, Friedl & Pröschold, Citation2010: European Journal of Phycology 45(1): 91–92, .
Synonym: Chlorella nordstedtii Printz, Citation1938: Botaniska Notiser 1938: 82, .
Droopiella gen. nov.
()
Description: Alga is unicellular, short-cylindrical to almost spherical in cell shape. Cell wall smooth without any warts, spines, and bristles. Chloroplast parietal, cup-shaped to saucer-shaped, filling around half to 2/3 of cell volume. Pyrenoid is located in the middle of the chloroplast, surrounded by several little starch grains. Vegetative cells are surrounded by thin mucilaginous envelope. Reproduction is by autospore formation. Two, four, or eight autospores are usually produced per cell and released by rupturing of the mother cell wall on one side. The remains of the autosporangia cell wall could be observed in culture in form of sacs.
Diagnosis: Differs from other genera of Trebouxiophyceae by the SSU-ITS sequences and smooth cell wall.
Etymology: The genus was named in honour of Dr Michael Droop (1918–2011) for his contributions to phycology. Michael Droop learned the techniques of micro-algal culture from his mentor, EG Pringsheim, in Cambridge. After obtaining his PhD he worked for the Scottish Marine Biological Association (later SAMS) at Millport in the Firth of Clyde in Scotland and then near Oban in Argyll. He published papers on algal nutrition (the Cell Quota method) and built up a collection of several hundred strains of pure algae until his retirement in 1982. For more details, see Leadbeater (Citation2006).
Type species (designated here): Droopiella spaerckii (Ålvik) Darienko, Rad-Menéndez, Campbell & Pröschold comb. nov.
Comment: The investigated strains belonging to Droopiella were originally assigned to the genera Amphikrikos and Schizochlamydella. The type species of both genera are not available for further investigations and are freshwater species. The diagnostic feature of Amphikrikos is the presence of two rows of warts on the cell surface. The ultrastructural investigations of our strains by Rascio et al. (Citation1980a, Citation1980b, Citation1980c) showed that all strains have smooth cell walls without any ornamentation. The type species of Schizochlamydella, S. delicatula (West) Korshikov, is currently accepted as basionym of Phaeoschizochlamys mucosa, a chrysophycean alga (Bourrelly, Citation1957). The morphological features presented by Korshikov (Citation1953) for Schizochlamydella (release of autospores at lateral site of the autosporangia, cell division always in one plane) differ from those strains here investigated. As a consequence, our strains cannot be assigned to either of these genera.
Droopiella spaerckii (Ålvik) Darienko, Rad-Menéndez, Campbell & Pröschold, comb. nov.
Basionym: Chlorella spaerckii Ålvik, Citation1934: Bergens Museum Årbok 1934 (6): 31, pl. I: (lectotype designated here).
Synonym: Chlorella stigmatophora Butcher, Citation1952: Journal of the Marine Biological Association of the United Kingdom 31: 181, pl. I: .
Emended description (, ): Cells are slightly elongated to spherical, surrounded by a smooth cell wall. Chloroplast parietal cup-shaped containing clearly visible pyrenoid surrounded by starch grains. Some vacuoles and inclusions in the chloroplast could be present. Cells are surrounded by thin delicate mucilage sheet, which is usually visible only for a brief moment by staining with methylene blue or Indian ink and then collapses. Reproduction is by autosporogenesis. Usually 2 to 4 autospores are produced. Liberation of autospores occurs by rupturing of the mother cell wall. SSU and ITS rDNA sequences (GenBank: MN248523) and ITS-2 DNA Barcode: Barcode BC1 in .
Diagnosis: Differs from other members of Trebouxiophyceae by SSU-ITS sequences.
Epitype (designated here): The strain CCAP 211/20 cryopreserved in a metabolic inactive state at the Culture Collection of Algae and Protozoa (CCAP), Oban, Scotland.
Droopiella capsulata (Guillard, Bold & MacEntee) Darienko, Rad-Menéndez, Campbell & Pröschold, comb. nov. ()
Basionym: Chlorella capsulata Guillard, Bold & MacEntree Citation1975: Phycologia 14: 22, .
Synonym: Schizochlamydella capsulata (Guillard, Bold & MacEntee) Watanabe, Citation1977: Journal of Japanese Botany 52(11): 342.
Diagnosis: Differs from the type variety through spherical cell shape, thicker mucilage layer and differences in SSU and ITS sequences (GenBank: MN248527) and ITS-2 DNA Barcode: Barcode BC2 in .
Epitype (designated here): The strain UTEX 2074 cryopreserved in a metabolic inactive state at the Culture Collection of Algae and Protozoa (CCAP), Oban, Scotland.
Comment: As demonstrated in . capsulata and D. spaerckii have the same ITS-2 barcode (BC1); however, the comparison of the SSU rDNA sequences showed several differences such as one CBC in Helix E23_1 and three HCBCs in the helices 11, 15, and E23_2 (Fig. S1, see supplemental material online).
Droopiella limnetica sp. nov.
()
Description: Cells are ovoid, slightly elongated to almost spherical, surrounded by smooth cell wall, 4.2 × 3.3 – 7.6 × 6.4 µm in size. Chloroplast is parietal cup-shaped containing a pyrenoid surrounded by several starch grains. Old cells are mostly broadly ellipsoidal to spherical, sometimes irregular, 7.6 µm in diameter, or 7.9 × 6.7 – 11.2 × 7.8 µm. Such cells often contain one or two large vacuoles and some inclusions in the chloroplast. Reproduction is by autoporogenesis. Autosporangia are 5.1 × 4.1 – 10.8 × 9.0 µm in size. Two to four, sometimes eight autospores are formed per cell. Liberation of autospores is by rupturing of the mother cell wall. SSU and ITS rDNA sequences (GenBank: MN248528) and ITS-2 DNA Barcode: Barcode BC3 in .
Diagnosis: Differs from other species of Droopiella by SSU-ITS sequences.
Holotype: (designated here): The strain SAG 2074 cryopreserved in a metabolic inactive state at the Culture Collection of Algae at the University of Göttingen (SAG), Germany.
Type locality: Lake Balaton, Hungary.
Comment: Stenclová et al. (Citation2017) investigated three of the strains, which were studied here. They assigned the strains SAG 30.96, SAG 2074, and CCMP 245 (= UTEX 2074) to a clade, which they called ‘Granulated Clade 2‘. However, as demonstrated above, all strains do not form any granulations on the cell walls. The authors mentioned that the photographs of CCMP 245 showed granules, which were visible. This is incorrect, no granules are visible on the pictures and we also could not discover any ornamentations of the surface of the cell walls. The SSU rDNA sequences of UTEX 2074 (this study) and CCMP 245 (KY013468 and AY044651-2) are almost identical despite several sequencing errors in the deposited sequences in GenBank. Therefore, as discussed above, these strains cannot be members of the genus Amphikrikos. In addition, the authors assumed that the strains SAG 30.96 and SAG 2074 represent the same species and combined therefore the SSU rDNA sequence of SAG 2074 with the rbcL gene of SAG 30.96 in their concatenated data set for the phylogenetic analyses. As shown here, both strains belong to different species.
Chlorella salina Kufferath, Citation1919: Annales de Biologie Lacustre 9: 7,
Synonyms: Chlorella salina Butcher, Citation1952: Journal of the Marine Biological Association of the United Kingdom 31: 179, pl. 1 [1]: , nom. illeg.; Chlorella gloriosa Molinari & Calvo-Pérez in Calvo-Pérez Rodó & Molinari-Novoa, Citation2015: The Biologist (Lima) 13: 73.
Comment: In our opinion, Kufferath (Citation1919) and Butcher (1952) observed the same alga, which is probably widely distributed in marine habitats around Europe. This organism has similar morphology to Chlorella vulgaris, but probably represents another species, due to the changing morphology of C. vulgaris under marine conditions, which has not been observed for Chlorella salina sensu Kufferath.
Conclusion
The phylogenetic analyses of SSU and ITS rDNA sequences revealed that investigated strains belonged to three independent lineages (Watanabea-, Chlorella-, and Oocystis-clades) within the Trebouxiophyceae. The results of our study led us to consider that we should assign two marine strains to the genus Chloroidium, three to the genus Chlorella, and to establish the genus Droopiella for the remaining isolates.
Supplemental data
Supplemental data for this article can be accessed here: https://doi.org/10.1080/14772000.2019.1690597.
Associate Editor: Elliot Shubert
Supplemenrtal Material - Table S3
Download PDF (25.7 KB)Supplemenrtal Material - Table S2
Download PDF (27.4 KB)Supplemenrtal Material - Table S1
Download PDF (25.6 KB)Supplemenrtal Material - Figure S1
Download PDF (290.5 KB)Additional information
Funding
References
- Akaike, H. (1974). A new look at the statistical model identification. IEEE Transactions on Automatic Control, 19, 716–723.
- Altschul, S. F., Gish, W., Miller, W., Myers, E. W., & Lipman, D. J. (1990). Basic local alignment search tool. Journal of Molecular Biology, 215, 403–410.
- Ålvik, G. (1934). Plankton-Algen norwegischer Austernpollen. I. Systematik und Vorkommen der Arten. Bergens Museum Arbok, 6, 1–47.
- Andreyeva, V. M. (1975). Rod Chlorella. Morphologiya, sistematika, princypy klassifikatsii [The genus Chlorella. Morphology, systematics, principal classification]. Leningrad: Nauka, 86 p. (in Russian).
- Andreoli, C., Rascio, N., & Casadoro, G. (1978). Chlorella nana sp. nov. (Chlorophyceae): A new marine Chlorella. Botanica Marina, 21, 253–256.
- Aslam, Z., Shin, W., Kim, M. K., Im, W.-T., & Lee, S.-T. (2007). Marinichlorella kaistiae gen. et sp. nov. (Trebouxiophyceae, Chlorophyta) based on polyphasic taxonomy. Journal of Phycology, 43, 576–584.
- Beijerinck, M. W. (1890). Culturversuche mit Zoochlorellen, Lichenengonidien und anderen niederen Algen. Botanische Zeitung, 48, 725–785.
- Bock, C., Krienitz, L., & Pröschold, T. (2011). Taxonomic reassessment of the genus Chlorella (Trebouxiophyceae) using molecular signatures (barcodes), including description of seven new species. Fottea, 11, 293–312.
- Bourrelly, P. (1957). Recherches sur les Chrysophycees. Morphologie, phylogenie, systematique. Revue Algologique. Mémoire Hors-Série, 1, 1–412.
- Butcher, R. W. (1952). Contributions to our knowledge of the smaller marine algae. Journal of the Marine Biological Association of the United Kingdom, 31, 175–191. doi:10.1017/S0025315400003751
- Calvo-Pérez Rodó, J. D., & Molinari-Novoa, E. A. (2015). A nomenclatural and cultural note on Chlorella peruviana G. Chacón and other species of the genus Chlorella Beij. (Chlorellales, Chlorellaceae). The Biologist (Lima), 13, 71–74.
- Clement, M., Posada, D., & Crandall, K. A. (2000). TCS: A computer program to estimate gene genealogies. Molecular Ecology, 9, 1657–1659.
- Clement, M., Snell, Q., Walker, P., Posada, D., & Crandall, K. (2002). TCS: Estimating gene genealogies. Parallel and Distributed Processing Symposium, International Proceedings, 2, 184.
- Chacón, G. R. (1980). Chlorella peruviana sp. nov. y su ambiente altamente salino. Boletín de la Sociedad Peruana de Botánica, 8, 83–96.
- Coleman, A. W., Suarez, A., & Goff, L. J. (1994). Molecular delineation of species and syngens in volvocacean green algae (Chlorophyta). Journal of Phycology, 30, 80–90.
- Darienko, T., & Pröschold, T. (2015). Genetic variability and taxonomic revision of the genus Auxenochlorella (Shihira et Krauss) Kalina et Puncocharova (Trebouxiophyceae, Chlorophyta. ). Journal of Phycology, 51, 394–400.
- Darienko, T., & Pröschold, T. (2019a). The genus Jaagichlorella Reisigl (Trebouxiophyceae, Chlorophyta) and its close relatives: an evolutionary puzzle. Phytotaxa, 388, 47–68. doi:10.11646/phytotaxa.388.1.2
- Darienko, T., & Pröschold, T. (2019b). Reevaluation and discovery of new species of the rare genus Watanabea and establishment of Massjukichlorella gen. nov. (Trebouxiophyceae, Chlorophyta) using an integrative approach. Journal of Phycology, 55, 493–499. doi:10.1111/jpy.12830
- Darienko, T., Kang, W., Orzechowski, A. K., & Pröschold, T. (2019). Pleurastrosarcina terriformae, a new species of a rare desert trebouxiophycean alga discovered by an integrative approach. Extremophiles, 23, 573–586. doi:10.1007/s00792-019-01108-5
- Darienko, T., Gustavs, L., Mudimu, O., Menendez, C. R., Schumann, R., Karsten, U., … Pröschold, T. (2010). Chloroidium, a common terrestrial coccoid green alga previously assigned to Chlorella (Trebouxiophyceae, Chlorophyta). European Journal of Phycology, 45, 79–95. doi:10.1080/09670260903362820
- Darienko, T., Gustavs, L., & Pröschold, T. (2016). Species concept and nomenclatural changes within the genera Elliptochloris and Pseudochlorella (Trebouxiophyceae) based on an integrative approach. Journal of Phycology, 52, 1125–1145.
- Darienko, T., Lukešová, A., & Pröschold, T. (2018). The polyphasic approach revealed new species of Chloroidium (Trebouxiophyceae, Chlorophyta). Phytotaxa, 372, 51–66.
- Fawley, M. W., Fawley, K. P., & Owen, H. A. (2005). Diversity and ecology of small coccoid green algae from Lake Itasca, Minnesota, USA, including Meyerella planktonica, gen. et sp. nov. Phycologia, 44, 35–48.
- Fawley, M. W., Jameson, I., & Fawley, K. P. (2015). The phylogeny of the genus Nannochloropsis (Monodopsidaceae, Eustigmatophyceae), with descriptions of N. australis sp. nov. and Microchloropsis gen. nov. Phycologia, 54, 545–552.
- Fott, B., & Novakova, M. (1969). A monograph of the genus Chlorella. The fresh water species. In B. Fott (Ed.), Studies in Phycology (pp. 10–74), Prague: Academia.
- George, E. A. (1957). A note on Stichococcus bacillaris Naeg. and some species of Chlorella as marine algae. Journal of the Marine Biological Association of the UK, 36, 111–114.
- Gladu, P. K., Patterson, G. W., Wikfors, G. H., & Smith, B. C. (1995). Sterol, fatty acid, and pigment characteristics of UTEX 2341, a marine eustigmatophyte identified previously as Chlorella minutissima (Chlorophyceae). Journal of Phycology, 31, 774–777. doi:10.1111/j.0022-3646.1995.00774.x
- González, M. A., Pröschold, T., Palacios, Y., Aguayo, P., Inostroza, I., & Gómez, P. I. (2013). Taxonomic identification and lipid production of two Chilean Chlorella-like strains isolated from marine and an estuarine coastal environment. Annals of Botany Plants, 5, 1–12.
- Guillard, R. R. L., Bold, H. C., & MacEntee, F. J. (1975). Four new unicellular chlorophycean algae from mixohaline habitats. Phycologia, 14, 13–24.
- Hoef-Emden, K., Marin, B., & Melkonian, M. (2002). Nuclear and nucleomorph SSU rDNA phylogeny in the Cryptophyta and evolution of cryptophyte diversity. Journal of Molecular Evolution, 55, 161–179.
- Hong, J. W., Kim, O. H., Jo, S.-W., Kim, H., Jeong, M. R., Park, K. M., … Yoon, H.-S. (2016). Biochemical composition of a Korean domestic microalga Chlorella vulgaris KNUA027. Microbiology and Biotechnology Letters, 44, 400–407. doi:10.4014/mbl.1512.12008
- Korshikov, A. A. (1953). Viznachnik prisnovodnihk vodorostey Ukrainsykoi RSR [Vyp] V. Pidklas Protokokovi (Protococcineae). Bakuol’ni (Vacuolales) ta Protokokovi (Protococcales) [The Freshwater Algae of the Ukrainian SSR. V. Subclass Protococcineae. Vacuolales and Protococcales]. (pp. 1–439), Kyjv [Kiev]: Akad. NAUK URSR.
- Kreutz, M., Stoeck, T., & Foissner, W. (2012). Morphological and molecular characterization of Paramecium (Viridoparamecium nov. subgen.) chlorelligerum Kahl 1935 (Ciliophora). Journal of Eukaryotic Microbiology, 59, 548–563.
- Krienitz, L., & Bock, C. (2012). Present state of the systematics of planktonic coccoid green algae of inland waters. Hydrobiologia, 698, 295–326.
- Kufferath, H. (1919). Essais de culture des algues monocellulaires des eaux saumâtres. Annales de Biologie Lacustre, 9, 1–11.
- Ma, S., Han, B., Huss, V. A. R., Hu, X., Sun, X., & Zhang, J. (2015). Chlorella thermophila (Trebouxiophyceae, Chlorophyta), a novel thermo-tolerant Chlorella species isolated from an occupied rooftop incubator. Hydrobiologia, 760, 81–89.
- Lanzoni, O., Fokin, S. I., Lebedeva, N., Migunova, A., Petroni, G., & Potekhin, A. (2016). Rare freshwater ciliate Paramecium chlorelligerum Kahl, 1935 and its macronuclear symbiotic bacterium “Candidatus Holospora parva”. Public Library of Science ONE, 11, e0167928. doi:10.1371/journal.pone.0167928
- Leadbeater, B. S. C. (2006). The ‘Droop Equation’ – Michael Droop and the legacy of the ‘Cell-Quota Model’ of phytoplankton growth. Protist, 157, 345–358. doi:10.1016/j.protis.2006.05.009
- Leigh, J. W., & Bryant, D. (2015). POPART: Full-feature software for haplotype network construction. Methods in Ecology and Evolution, 6, 1110–1116.
- Luo, W., Pröschold, T., Bock, C., & Krienitz, L. (2010). Generic concept in Chlorella-related coccoid green algae (Chlorophyta, Trebouxiophyceae). Plant Biology, 12, 545–553.
- Marin, B., Palm, A., Klingberg, M., & Melkonian, M. (2003). Phylogeny and taxonomic revision of plastid-containing euglenophytes based on SSU rDNA sequence comparisons and synapomorphic signatures in the SSU rRNA secondary structure. Protist, 154, 99–145. doi:10.1078/143446103764928521
- Maruyama, I., Nakamura, T., Matsubayashi, T., Ando, Y., & Maeda, T. (1986). Identification of the alga known as “marine Chlorella” as a member of the Eustigmatophyceae. Japanese Journal of Phycology, 34, 319–325.
- Müller, J., Friedl, T., Hepperle, D., Lorenz, M., & Day, J. G. (2005). Distinction between multiple isolates of Chlorella vulgaris (Chlorophyta, Trebouxiophyceae) and testing for conspecificity using amplified fragment length polymorphism and ITS rDNA sequences. Journal of Phycology, 41, 1236–1247.
- Posada, D. (2008). ModelTest: phylogenetic model averaging. Molecular Biology and Evolution, 25, 1253–1256.
- Printz, H. (1938). Chlorella nordstedtii n. sp., a new submarine alga. Botaniska Notiser, 1938, 77–82.
- Pröschold, T., Darienko, T., Silva, P. C., Reisser, W., & Krienitz, L. (2011). The systematics of "Zoochlorella" revisited employing an integrative approach. Environmental Microbiology, 13, 350–364.
- Rascio, N., Casadoro, G., & Andreoli, C. (1980a). Ultrastructural characterization of marine chlorellae. I. Chlorella ovalis. Botanica Marina, 23, 25–29. doi:10.1515/botm.1980.23.1.25
- Rascio, N., Casadoro, G., & Andreoli, C. (1980b). Ultrastructural characterization of marine chlorellae. II. Chlorella stigmatophora. Botanica Marina, 23, 31–34. doi:10.1515/botm.1980.23.1.31
- Rascio, N., Casadoro, G., & Andreoli, C. (1980c). Ultrastructural characterization of marine chlorellae. III. Chlorella spaerckii. Botanica Marina, 23, 463–465. doi:10.1515/botm.1980.23.1.31
- Rascio, N., Casadoro, G., & Andreoli, C. (1980d). Ultrastructural characterization of marine chlorellae. IV. Chlorella salina. Botanica Marina, 23, 467–469. doi:10.1515/botm.1980.23.1.31
- Rascio, N., & Casadoro, G. (1981). Ultrastructural characterization of marine chlorellae. V. Chlorella marina. Botanica Marina, 24, 291–296.
- Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., … Huelsenbeck, J. P. (2012). MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology, 61, 539–542. doi:10.1093/sysbio/sys029
- Rosenberg, J. N., Kobayashi, N., Barnes, A., Noel, E. A., Betenbaugh, M. J., & Oyler, G. A. (2014). Comparative analyses of three Chlorella species in response to light and sugar reveal distinctive lipid accumulation patterns in the microalga C. sorokiniana. Public Library of Science One, 9, e92460.
- Schlösser, U. G. (1997). Additions to the culture collection of algae since 1994. Botanica Acta, 110, 424–429. doi:10.1111/j.1438-8677.1997.tb00659.x
- Stamatakis, A. (2006). RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics, 22, 2688–2690.
- Stenclová, L., Fučíková, K., Kastovsky, J., & Pazoutová, M. (2017). Molecular and morphological delimitation and generic classification of the family Oocystaceae (Trebouxiophyceae, Chlorophyta). Journal of Phycology, 53, 1263–1282.
- Swofford, D. L. (2002). PAUP* phylogenetic analysis using parsimony (*and other methods), Version 4.0b10. Sunderland, MA, USA: Sinauer Associates.
- Ueda, S. (1927). Neue grüne Alge als Nahrung der Schalenlarven der Auster. Journal of the Imperial Fisheries Institute Tokyo, 23, 78.
- Watanabe, S. (1977). Schizochlamydella sphaerica sp. nov. (Chlorococcales) from Japanese soil. Journal of Japanese Botany, 52, 338–443.
- Wolf, M., Hepperle, D., & Krienitz, L. (2003). On the phylogeny of Radiococcus, Planktosphaeria and Schizochlamydella (Radiococcaceae, Chlorophyta.). Biologia, Bratislava, 58, 759–765.
- Zuker, M. (2003). Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acid Research, 31, 3406–3615. doi:10.1093/nar/gkg595