Abstract
The culture broth of endophytic Streptomyces sp. AB100, isolated from the shoots of medicinal plant Atropa belladonna (L.) was investigated for the presence of antibacterial compounds. After initial testing followed by bioactivity-guided fractionation, six new piperazic acid (PA)-containing congeners of two known peptides, JBIR-39 and JBIR-40, were identified by HR-MS/MS and NMR analyses. Only the dehydroxylated hexapeptidic derivatives with unusual incorporation of four PA moieties exhibited weak antibacterial activity against Gram-positive test organism Bacillus subtilis. A 16S rDNA-based phylogenetic tree of known Streptomyces spp. producing PA-containing hexapeptides isolated from different habitats and endophyte Streptomyces AB100 showed considerable diversity, suggesting that these metabolites may play an important environmental role beyond their antibacterial activity.
1. Introduction
Actinomycetes produce more than 40% of all known bioactive natural products of microbial origin (Berdy Citation2012), and impressive 76% of all original natural products used as anti-infective agents are derived from actinomycetes (Gomez-Escribano and Bibb Citation2014). For decades representatives of the actinomycete genus Streptomyces, have been a rich source of antibacterial natural products. Streptomycetes have been isolated from diverse ecological habitats, deep sea (Kamjam et al. Citation2017), desert soils (Mohammadipanah and Wink Citation2015), marine sediments and sponges (Indraningrat et al. Citation2016), insects, as, for example, ants (Liu et al. Citation2016), to name just a few. A promising source of new Streptomyces spp. with the potential to produce novel antibiotic substances are medicinal plants, where these bacteria dwell as endophytes (Martinez-Klimova et al. Citation2017; Golinska et al., Citation2015). Endophytes have an intimate symbiotic relationship with their host plants and are known to provide growth promoting metabolites, insect and pest repellents, antimicrobials against plant pathogens, as well as metabolites protecting their hosts against abiotic stresses (Ryan et al. Citation2008).
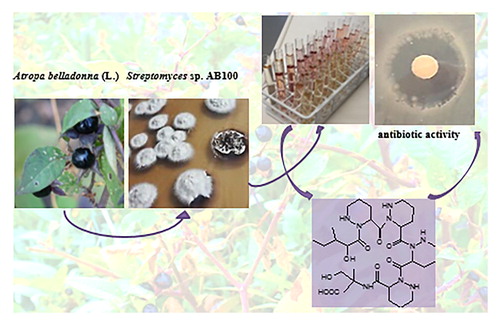
In the course of our search for new sources of antibacterial compounds we isolated endophytic bacteria from different organs of the medicinal atropine- and scopolamine-producing plant Atropa belladonna (L.). Our interest was drawn to a Streptomyces isolate capable of producing hexahydropyridazine-3-carboxylic acid (piperazic acid, PA) containing antimicrobial peptides. PA-containing hexapeptides have been found to be synthesised by Streptomyces spp. derived from soil, marine sediments or deep-sea sediment samples (Isshiki et al. Citation1990; Hosaka et al. Citation2009; Miller et al. Citation2007; Liu et al. Citation2015; Wei et al. Citation2012), all of them exhibiting antibacterial activity against Gram-positive bacteria, but no activity against Gram-negative bacteria (Oelke et al. Citation2011). No endophytic Streptomyces sp. has so far been reported to produce PA containing hexapeptides.
Here, we describe isolation and identification of the bioactive Streptomyces strain, production, purification and structure elucidation of the PA containing peptides and initial tests on their antimicrobial activity.
2. Results and discussion
2.1 Isolation and identification of streptomyces sp. AB100 from Atropa belladonna (L.) shoots
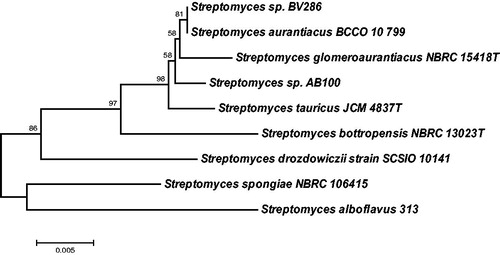
Endophytic Streptomyces sp. AB100 was isolated from Atropa belladonna (L.) shoots as described in detail in the experimental section. It was classified as Streptomyces by RDP (Ribosomal Database Project) classifier based on its 16S rDNA sequence. A BLAST search of this sequence revealed Streptomyces sp. strain BV286 with GenBank accession no. MF511794 and Streptomyces aurantiacus strain BCCO 10_799 with GenBank accession no. KP718496 as having the highest sequence identities to the query Streptomyces sp. AB100 (99% identities). Phylogenetic analysis using neighbor-joining method with bootstrap support based on 1000 replicates, which included 16S rDNA gene sequences from the closest related type strains, suggested that Streptomyces sp. AB100 differs from previously isolated streptomycetes (). For the reasons described below we also included three 16S rDNA sequences of Streptomyces spp. known to produce PA-containing hexapeptides into the phylogenetic tree. Those included Streptomyces drozdowiczii SCSIO 10141 (GenBank accession no. JX101439) isolated from the deep sea sediment and producing cyclic peptides marformycins (Liu et al. Citation2015), Streptomyces spongiae NBRC 106415 (GenBank accession no. NR108117) isolated from a marine sponge Haliclona sp. producing linear peptides JBIR-39 and JBIR-40 (Kozone et al. Citation2011) and Streptomyces alboflavus 313 (GenBank accession no. FJ032029.1) isolated from Chinese soil and producing cyclic peptides NW-G10 and NW-G11 (Wei et al. Citation2012). Though all four strains, including Streptomyces sp. AB100, were isolated from diverse ecological habitats and seem not to belong to the same species (). However, they share the ability to produce PA, to modify it and to include it into the hexapeptides, probably using non-ribosomal peptide synthases (NRPS) as biosynthetic machinery.
Figure 1. Phylogenetic tree of the16S rDNA sequences from three isolates of Steptomyces sp. known to produce PA-containing hexapeptides and isolate AB100 from Atropa belladonna shoot. The tree was built with the neighbour-joining algorithm usingMega7.0.18 (Kumar et al. Citation2016).
2.2 Piperazic acid containing peptides
Isolate Streptomyces sp. AB100 was cultivated in a range of media providing different carbon and nitrogen sources at neutral pH7 and at pH10.Basic pH was shown to activate secondary metabolite production in some Streptomyces in a previous study by Zhu et al. (Citation2014) (see ‘Experimental’ section for details). Crude extracts were tested for antibacterial activity against three indicator strains: Gram-positive Bacillus subtilis and Gram-negative Pseudomonas fluorescens and Escherichia coli. All crude extracts exhibited antibacterial activity against B. subtilis (Supplementary information Figure S1), while none of them was active against the Gram-negative indicator strains. Crude extract from medium 5254/pH7 showed the largest inhibition zone (8 mm) in agar diffusion assays against B. subtilis. Therefore, the medium 5254/pH7 was used for an up-scaled cultivation, preparation of the crude extract, its fractionation and purification of secondary metabolites.
HR-MS/MS analysis of the active extract revealed the presence of peptidic compounds, mainly hexa- and pentapeptides. The main product was the known PA-containing linear peptide JBIR-39 (Supplementary information Figure S3) with a calculated sum formula of C30H51N9O10, measured m/z 698.3850 [M + H]+, calculated for [C30H52N9O10]+ m/z 698.3832, Δm/z = −2.6 ppm. This compound was purified and its structure was confirmed by NMR and MS/MS measurements (Supplementary material). The MS/MS fragmentation pattern was congruent with the described fragmentation pattern for JBIR-39, which was originally isolated together with JBIR-40 from the culture broth of the marine sponge-derived S. spongiae NBRC 106415 (Kozone et al., Citation2011). JBIR-39 contains a methylserine moiety, a γ-hydroxyl PA moiety, three PA moieties and an isoleucic acid moiety. The isoleucic acid moiety is replaced by a valinic acid moiety in JBIR-40 (Kozone et al. Citation2011).
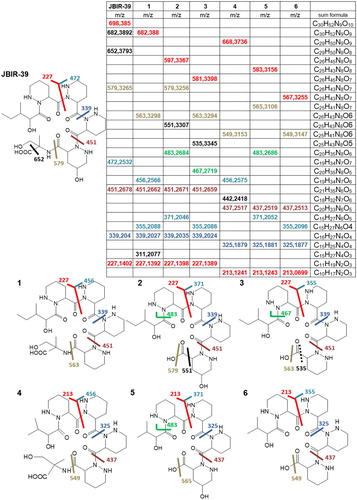
Based on the calculated sum formula and HR-MS/MS fragmentation patterns () compared to JBIR-39, 6 potentially new compounds (compounds 1–6, ) and the known JBIR-40 (Supplementary information Figure S3) were identified as congeners of JBIR-39. Compounds 2, 3, 5 and 6 are most likely linear pentapeptidic derivatives or degradation products of JBIR-39 or JBIR-40. They lack the C-terminal methylserine moiety, which is indicated by the reduced m/z values, the calculated sum formula and the MS/MS fragments m/z 483 for compounds 2 and 5, m/z 467 for compound 3 and m/z 549 for compound 6 (see for explanation). The very similar compounds 2 and 3 or 5 and 6, respectively, most likely differ by their hydroxylation status. Compounds 3 and 6 seem to be dehydroxylated 2 or 5, respectively. The dehydroxylation leads to the lack of fragments m/z 579 or m/z 565 and to the occurrence of fragments m/z 563 for compound 3 and m/z 549 for compound 6 ().
Figure 2. Calculated sum formula, measured HR-ESI-MS/MS values and comparison of fragmentation patterns of JBIR-39 and compounds 1–6.
In addition to these pentapeptidic structures, two hexapeptides, compounds 1 and 4, were identified. Compound 1 was purified by Flash chromatography and semi-preparative HPLC and 0.46 mg pure compound were subjected to NMR measurements and HR-MS/MS analysis (, Supplementary information Table S1 and S2, Figures S4 and S5).
Compound 1 has a measured m/z of 682.3880 [M + H]+ and the calculated sum formula C30H51N9O9 indicating it being a reduced derivative of JBIR-39. Extensive NMR analysis and close comparison of 1H and 13C data (see Supplementary information Tables S1 and S2) has revealed that 1 is built up of four identical substituted PA moieties, while JBIR-39 contains only three of them and one hydroxy-substituted PA moiety. This compound was subjected to antibacterial activity tests using the disc diffusion method and showed weak activity against the Gram-positive indicator strain B. subtilis (Supplementary information Figure S1). However, activity was not strong enough to explain the antibacterial activity of the crude extract, which is why further not yet investigated antibacterial substances or activity promoting substances are expected to be present in the crude extract of Streptomyces sp. AB100 grown in 5254 medium at pH7.
Compound 4 with a measured m/z of 668.3736 [M + H]+ and a calculated sum formula of C29H49N9O9 probably is the respective reduced derivative of JBIR-40.
Other PA-containing cyclic hexapeptides contain only three PA moieties, as, for example, the svetamycins (Dardic et al. Citation2017) or NW-G10 and NW-G11 (Wei et al. Citation2012), two PA moieties, as, for example, depsidomycin (Isshiki et al. Citation1990), piperastatin A + B (Murakami, Harada, Takahashi, et al. Citation1996; Murakami, Harada, Yamazaki et al. Citation1996) or pipalamcin (Uchihata et al. Citation2002), or only one PA moiety as in the case for marformycins (Liu et al. Citation2015).
3. Conclusion
Endophytic Streptomyces sp. AB100 was isolated from the medicinal plant Atropa belladonna (L.) shoots. Six new PA containing peptides and two known compounds, JBIR-39 and JBIR-40, were identified by extensive HR-MS/MS and NMR analyses in the culture broth of Streptomyces AB100 grown in a range of media. Two dehydroxylated hexapeptidic PA-containing compounds exhibited antibiotic activity against Gram-positive indicator organism Bacillus subtilis, while hydroxylated hexapeptidic derivatives JBIR-39 and JBIR-40, and all pentapeptidic compounds did not. JBIR-39, JBIR-40, the piperidamycins (Hosaka et al. Citation2009) and the here described new compounds are characterised by the unusual incorporation of four PA modules into the hexa- or pentapeptidic core structure, with even four identical PA moieties in the case of the here described new compounds 1, 3, 4 and 6. The presented data confirms that endophytic Streptomyces spp. are a potential source of new antibiotic substances, and that PA-containing hexapeptides as, for example, JBIR-39 and congeners can be biosynthesized by different streptomycetes from a wide range of ecological habitats.
Bekiesch_AB100_npr_suppl_material_revised2__2_Dateien_zusammengef_gt_.pdf
Download PDF (1.2 MB)Acknowledgements
This study was supported by the University of Vienna. The authors thank Florian Gössnitzer for help with media preparation.
Reference
- Berdy J. 2012. Thoughts and facts about antibiotics: Where we are now and where we are heading. J Antibiot (Tokyo). 65(8):441.
- Dardic D, Lauro G, Bifulco G, Laboudie P, Sakhaii P, Bauer A, Vilcinskas A, Hammann PE, Plaza A. 2017. Svetamycins A-G, unusual piperazic acid-containing peptides from Streptomyces sp. J Org Chem. 82(12):6032–6043.
- Golinska P, Wypij M, Agarkar G, Rathod D, Dahm H, Rai M. 2015. Endophytic actinobacteria of medicinal plants: diversity and bioactivity. Antonie Van Leeuwenhoek. 108(2):267–289.
- Gomez-Escribano JP, Bibb MJ. 2014. Heterologous expression of natural product biosynthetic gene clusters in Streptomyces coelicolor: from genome mining to manipulation of biosynthetic pathways. J Ind Microbiol Biotechnol. 41(2):425–431.
- Hosaka T, Ohnishi-Kameyama M, Muramatsu H, Murakami K, Tsurumi Y, Kodani S, Yoshida M, Fujie A, Ochi K. 2009. Antibacterial discovery in actinomycetes strains with mutations in RNA polymerase or ribosomal protein S12. Nat Biotechnol. 27(5):462–464.
- Indraningrat AA, Smidt H, Sipkema D. 2016. Bioprospecting sponge-associated microbes for antimicrobial compounds. Mar Drugs. 14(5)
- Isshiki K, Sawa T, Naganawa H, Koizumi Y, Matsuda N, Hamada M, Takeuchi T, Iijima M, Osono M, Masuda T. 1990. Depsidomycin, a new immunomodulating antibiotic. J. Antibiot. 43(9):1195–1198.
- Kamjam M, Sivalingam P, Deng Z, Hong K. 2017. Deep sea actinomycetes and their secondary metabolites. Front Microbiol. 8:760
- Kozone I, Izumikawa M, Motohashi K, Nagai A, Yoshida M. 2011. Isolation of new hexapeptides—JBIR-39 and JBIR-40—from a marine sponge-derived Streptomyces sp. Sp080513SC-24. J Mar Sci Res Dev. 01:101.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Liu J, Wang B, Li H, Xie Y, Li Q, Qin X, Zhang X, Ju J. 2015. Biosynthesis of the anti-infective marformycins featuring pre-NRPS assembly line N-formylation and O-methylation and post-assembly line C-hydroxylation chemistries. Org Lett. 17(6):1509–1512.
- Liu SH, Xu MD, Zhang H, Qi H, Zhang J, Liu CX, Wang JD, Xiang WS, Wang XJ. 2016. New cytotoxic spectinabilin derivative from ant-associated Streptomyces sp. 1H-GS5. J Antibiot. 69(2):128–131.
- Martinez-Klimova E, Rodriguez PK, Sanchez S. 2017. Endophytes as sources of antibiotics. Biochem Pharmacol. 134:1–17.
- Miller ED, Kauffman CA, Jensen PR, Fenical W. 2007. Piperazimycins: cytotoxic hexadepsipeptides from a marine-derived bacterium of the genus Streptomyces. J Org Chem. 72(2):323–330.
- Mohammadipanah F, Wink J. 2015. Actinobacteria from arid and desert habitats: diversity and biological activity. Front Microbiol. 6:1541
- Murakami S, Harada S, Takahashi Y, Naganawa H, Takeuchi T, Aoyagi T. 1996. Piperastatin B: a new selective serine carboxypeptidase inhibitor from Streptomyces lavendofoliae MJ908-WF13. J Enzym Inhib. 11(1):51–66.
- Murakami S, Harada S, Yamazaki T, Takahashi Y, Hamada M, Takeuchi T, Aoyagi T. 1996. Piperastatin A, a new selective serine carboxypeptidase inhibitor produced by actinomycete. I. Taxonomy, production, isolation and biological activities. J Enzym Inhib. 10(2):93–103.
- Oelke AJ, France DJ, Hofmann T, Wuitschik G, Ley SV. 2011. Piperazic acid-containing natural products: isolation, biological relevance and total synthesis. Nat Prod Rep. 28(8):1445–1471.
- Ryan RP, Germaine K, Franks A, Ryan DJ, Dowling DN. 2008. Bacterial endophytes: recent developments and applications. FEMS Microbiol Lett. 278(1):1–9.
- Uchihata Y, Ando N, Ikeda Y, Kondo S, Hamada M, Umezawa K. 2002. Isolation of a novel cyclic hexadepsipeptide pipalamycin from Streptomyces as an apoptosis-inducing agent. J. Antibiot. 55(1):1–5.
- Wei S, Fan L, Wu W, Ji Z. 2012. Two piperazic acid-containing cyclic hexapeptides from Streptomyces alboflavus 313. Amino Acids. 43(5):2191–2198.
- Zhu H, Swierstra J, Wu C, Girard G, Choi YH, van Wamel W, Sandiford SK, van Wezel GP. 2014. Eliciting antibiotics active against the ESKAPE pathogens in a collection of actinomycetes isolated from mountain soils. Microbiology. 160(Pt_8):1714–1725.
