ABSTRACT
Introduction
The importance of biomarkers for pharmaceutical drug development and clinical diagnostics is more significant than ever in the current shift toward personalized medicine. Biomarkers have taken a central position either as companion markers to support drug development and patient selection, or as indicators aiming to detect the earliest perturbations indicative of disease, minimizing therapeutic intervention or even enabling disease reversal. Protein biomarkers are of particular interest given their central role in biochemical pathways. Hence, capabilities to analyze multiple protein biomarkers in one assay are highly interesting for biomedical research.
Areas covered
We here review multiple methods that are suitable for robust, high throughput, standardized, and affordable analysis of protein biomarkers in a multiplex format. We describe innovative developments in immunoassays, the vanguard of methods in clinical laboratories, and mass spectrometry, increasingly implemented for protein biomarker analysis. Moreover, emerging techniques are discussed with potentially improved protein capture, separation, and detection that will further boost multiplex analyses.
Expert commentary
The development of clinically applied multiplex protein biomarker assays is essential as multi-protein signatures provide more comprehensive information about biological systems than single biomarkers, leading to improved insights in mechanisms of disease, diagnostics, and the effect of personalized medicine.
KEYWORDS:
1. Introduction
Proteins are important mediators and actors in biology. Through their multiple functional roles as enzymes, cellular signaling components, neurotransmitters, cofactors or structural components, proteins affect the metabolic state and activity of cells, tissues, and organisms. Over the last century, proteins have been extensively studied, particularly their level of expression, modification, and interaction, as well as the dynamics involved. This understanding has been used to select proteins as potential drug targets and biomarkers to drive development of innovative therapeutic drugs. More recently, differences in protein biology associated with different pathologies, in some cases in combination with genetic variations, have been used to drive personalized medicine, whereby individually optimized therapies are based on these molecular changes [Citation1].
Personalized medicine is from its conception driven by biomarkers that provide a comprehensive view of a specific human biological system. These biomarkers include different types of molecules (proteins, DNA, RNA, metabolites), cellular and tissue morphologies, and functional read-outs [Citation2]. One of the first clinical demonstrators of biomarker-driven patient stratification toward higher clinical success was the combination of trastuzumab with the HER-2/neu immunohistochemistry assay [Citation3], followed by other successes in oncology and beyond. As a consequence of increasing biological understanding, researchers have a rapidly growing need to measure multiple functional biomarkers, preferably in a simultaneous way. This review focuses on analytical methods that allow multiplex quantification of proteins and can potentially be used to quantify protein biomarkers in the clinic.
The overall workflow for multiplex protein analysis is in several aspects similar to singleplex protein analysis: 1) define the context of use, 2) select the proteins to be studied, 3) develop sample preparation methods that fit the measurement of the selected proteins, 4) develop analytical methods to measure the selected protein parameters, 5) develop data analysis methods to interpret the data in a biological context. However, in multiplex protein analysis inter-protein effects can complicate assay development and application. In addition, the need for stringent quality control of assay reagents to avoid research irreproducibility [Citation4] is increased when multiple proteins are targeted in one assay. For that reason, researchers need to carefully select the analytical platform for multiplex protein tests.
Recently, researchers including us urged for the need to define best biomarker practice guidelines to bridge translational innovation gaps and improve the field of biomarker research and development [Citation5,Citation6]. We subsequently formed the pan-European COST CliniMARK consortium with academic and industrial biomarker scientists to define best biomarker practices. Among the items addressed, there was a strong focus on translational protein biomarker assays. In fact, despite the widespread knowledge of singleplex protein analyses, there is still limited information on the various methods and platforms available for clinical multiplex protein testing. Furthermore, regulatory guidelines for multiplex protein diagnostic tests are emerging but still have to find their way to research and health-care communities.
Here, we compare several techniques for multiplex analysis of proteins and include a pragmatic head-to-head comparison of multiplex analytical approaches. We focus on soluble (or solubilized) proteins using assay platforms with clinical potential. Immunoassay and mass spectrometry-MS methods are reviewed as the predominant platforms, and emerging methods with potential for further development into clinical application are also presented. In addition to analytical qualities, pre-analytical aspects are also essential in biomarker research. In this review, pre-analytical aspects are only considered in relation to a few specific analytical approaches. More general and comprehensive information about the importance of pre-analytical techniques is described elsewhere [Citation7].
2. Sample selection and preparation
Validation of protein biomarkers requires the use of a robust assay to analyze hundreds, if not thousands of samples, standards, and calibrators. Since the final goal is the implementation of the test in the clinic, it has to be highly reproducible, capable of standardization, scalable for automation and high throughput, and implementable for users of diverse skills. Hence, moving into the validation phase is not only about confirming a good or excellent biomarker response in relation to disease, but also about demonstrating the potential to translate the procedures to open-access platforms and multi-user interfaces. Sample preparation should be simple, preferably comprising a few steps to minimize experimental variations and analytically validated. Appropriate internal and external controls should be included to enable quality assessment when comparing batch-to-batch analyses over time. With this in mind, we provide a guide to the preferable sample sources and preparation methods for biomarker validation studies (, ).
Figure 1. (a) – represents different ELISA formats and configurations. The resulting immune-complex is detected through an enzymatic reaction to produce a color, chemiluminescent, or fluorescent signal. (b) – represents ‘in cell ELISA’ for detection of intracellular analytes. (c) – represents ELISpot for detection of analytes secreted by cells. (d) – represents different strategies for immunoassay multiplexing. The bead-based approach relies on analyte-specific color-coding of fluorescent capture beads, while the label-based approach relies on the use of analyte-specific labels attached to the detection antibodies. The spot-based approach relies on physically separated analyte-specific spots.
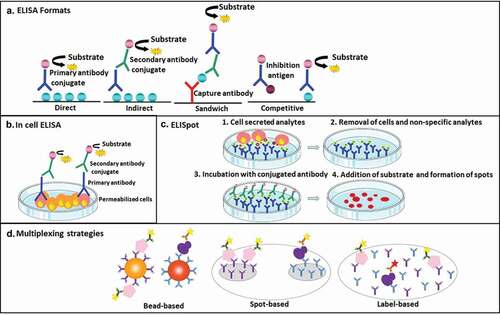
Figure 2. Representation of common MS-based proteomics approaches, i.e. targeted proteomics or data-dependent proteomics. In both bottom-up strategies, peptide detection is applied for protein identification or quantification. Generally, proteins are enriched, mainly by precipitation, and further denatured, reduced, alkylated, and digested chemically or enzymatically into peptides. After separating the peptides by liquid chromatography (LC) or capillary electrophoresis (CE), the ionization process converts the peptides into gas-phase ions, which enables the mass spectrometer to separate the ionized peptides based on their m/z ratios. Targeted proteomics (SRM/MRM/PRM) allows absolute quantification of selected proteins if stable isotope standards are included in the analysis. In classical label? free data-dependent acquisition (DDA) proteomics or ‘shotgun’ proteomics, peptide ions are specifically selected for fragmentation based on their detection intensity in precursor ion scans collecting MS/MS spectra for as many peptides as possible, which are identified against a proteomics database.
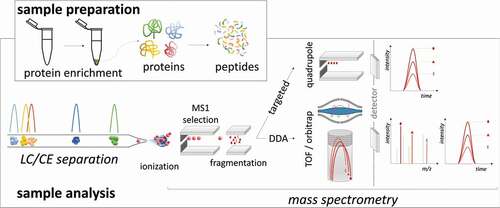
Figure 3. Decision matrix for the biomarker validation workflow using MS-based proteomics (font size defined by the number of papers found using keyword searches [biomarker + validation or verification + protein ± clinical + specific term, e.g. tissue or top-down] in Pubmed). The red-circled keyword is ‘organoid,’ a newly emerging 3D primary cell-based sample proving valuable in personalized biomarker validation, but representing only 0.036% of the ‘sample type’ papers at the current time.
![Figure 3. Decision matrix for the biomarker validation workflow using MS-based proteomics (font size defined by the number of papers found using keyword searches [biomarker + validation or verification + protein ± clinical + specific term, e.g. tissue or top-down] in Pubmed). The red-circled keyword is ‘organoid,’ a newly emerging 3D primary cell-based sample proving valuable in personalized biomarker validation, but representing only 0.036% of the ‘sample type’ papers at the current time.](/cms/asset/2c3fc3f2-5993-45d1-81bf-e62aa85d81ca/ieru_a_1763174_f0003_oc.jpg)
Selection of the sample type is critical. Whereas solid tissues are ideal for biomarker discovery since they are the source of biochemical alterations, they fall short in the validation stage. Solid tissues are inherently complex comprising multiple cell types, presence of blood and can vary substantially from patient to patient. Physical (homogenization, cryo-pulverization, extrusion, and sonication), chemical (detergents, chaotropic salts), and/or enzymatic (matrix proteases) treatment steps are required to extract proteins from solid tissues creating a source of experimental variation. Hence, quantitative interpretation of protein expression in solid tissues is challenging and impractical for longitudinal studies. Newly emerging strategies to isolate and clonally grow specific primary cells as organoids to simulate tissue microenvironments is gaining traction in understanding the molecular processes of disease for individual patients and lends itself ideally for proteomics analysis [Citation8]. Due to the relatively small size of these primary cells, similar extraction methods to those used for cell line cultures can be employed. As yet, organoids are not commonly used for biomarker validation but rather to select therapeutic agents.
Consequently, within the current regimen of acquiring clinical samples, which is aligned to immunoassay-based and mass spectrometry strategies, validation translates to more accessible sources such as biofluids. There are more than 30 biofluids, the most common being blood, cerebral spinal fluid (CSF), urine, saliva, tears, sweat, ear wax, interstitial fluid, and amniotic fluid. Biofluids or liquid biopsies require fewer preparation steps and are suitable for longitudinal collection from patients as part of continuous surveillance. They can also be a source of extracellular vesicles (e.g. exosomes) which contain molecular (protein, lipids, nucleic acids) information regarding specific diseases. Hence, liquid biopsies although surrogate for solid tissues, are more advantageous in the majority of validation studies.
Standardized procedures for collecting liquid biopsies are critical for immunoassay and proteomics analyses and data interpretation. Implementation of a collection protocol requires clear understanding between the clinician (e.g. consenting, patient reassurance), patient/volunteer (e.g. willingness to participate, fasting) and analyst (e.g. sample traceability, standard operating procedures). Where possible this should be within a controlled environment (e.g. storage for sample stability), even for readily collectible fluids such as saliva or urine. In addition, accurate, anonymized clinicopathological and patient personal data are essential to determine biomarker response related to the studied disease. A further consideration is the implementation of these procedures across different collection locations, whether these are hospitals, community centers, or at home. Even within the same type of settings, such as phlebotomy suites, understanding the variables (for example, different practitioners, different makes of storage vessel, transfer time from patient to storage location) are important variables, the tolerances of which need to be measurable and acceptable for a biomarker assay to viable. Ultimately, anonymized patient records, not only clinical data but also relevant personal data, are essential to enable interpretation of biomarker levels proportionate to disease specificity and not to unrelated confounding factors.
3. Immunoassays
3.1. ELISA
Enzyme-linked Immunosorbent Assay (ELISA) is a quantitative analytical method that measures antigen–antibody reaction via a color/light/fluorescent signal obtained by using an enzyme-linked conjugate and an enzyme substrate. This allows the measurement of the concentration of molecules in biological fluids or biological systems [Citation9]. The technology was originally developed in 1971 as a follow-up on radioactivity-based assays [Citation10]. It is a widely used method for research and clinical diagnostic purposes in different disciplines such as immunology, cancer, infectious diseases, and inflammation. It is a sensitive and specific test that produces rapid high-throughput results, with the advantage that it does not require complex equipment or radioactive labels.
ELISA assays can be found in different formats ()). The most commonly used ‘sandwich’ ELISA format requires two antibodies specific for different epitopes of the antigen; a capture antibody and a detection antibody. The antigen-antibody complex can be visualized via color change, chemiluminescence, or fluorescent signals using an enzyme-linked conjugate (e.g. HRP – Horse Radish Peroxidase) and an appropriate substrate (e.g. TMB – 3,3ʹ,5,5ʹ-tetramethylbenzidine). In some cases, the visualization is based on a labeled secondary antibody being specifically bound to a primary detecting antibody [Citation11]. The sandwich format offers higher sensitivity (pg/mL up to μg/mL) and specificity than direct and indirect ELISAs (using just one antibody) and enables the widespread use in preclinical and clinical laboratories. In addition, competitive ELISA [Citation12] and in-cell ELISA (quantification of a target protein in cultured cells with a primary antibody; )) [Citation13] are techniques based on the same principle but with specific applications and throughput. The ELISPOT (Enzyme-linked Immunospot Assay) is a highly sensitive and quantitative sandwich immunoassay for measurement of proteins secreted by plated cells attached to supportive matrixes [Citation14] ()).
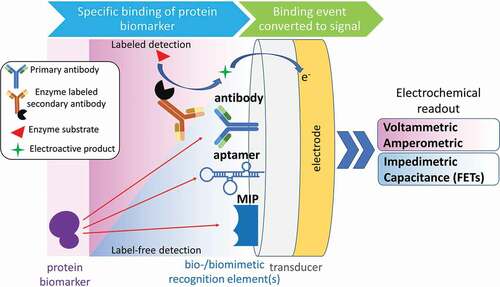
Figure 4. Labeled- and label-free strategies for electrochemical sensing of protein biomarkers. Various bio- or biomimetic recognition elements ensure the selective binding of the target protein, followed by the transduction of the binding event into a measurable electrochemical signal (MIP – molecularly imprinted polymer; FET – field-effect transistor).
Although the ELISA is regarded as a singleplex assay format, the method can be made suitable for multiplex protein analysis. Monoclonal, polyclonal, or recombinant antibodies with a defined specificity, sensitivity, and stability can be combined in one test to allow binding of multiple proteins. In addition, the use of different fluorochromes to detect different antibodies enables multiple parallel read-outs, visualizing multiple proteins simultaneously [Citation15]. These multiple readouts are combined in novel immunoassay formats in which different capture antibodies are immobilized on separate particles such as magnetic beads. Protein-specific fluorescence or streptavidin-labeled detection antibodies bind to the specific capture antibody-antigen complexes on a bead, after which each bead-antibody-antigen complex is analyzed by flow cytometry, yielding a multiplex analysis of up to 500 analytes per microwell ()).
The ELISA principle can also be applied for single-cell protein secretome analyses using single-cell barcode chips capturing a panel of 32 pre-specified proteins from a single cell in each microchamber. The cells of interest may be stimulated and sorted before loading the cells on a chip. Chips are analyzed using Isoplex machinery, which contains automated cellular imaging and includes a complete ELISA workflow. The software provides information on the secreted proteins across different categories: homeostatic, proliferative, inflammatory, chemotactic, regulatory, and immune effector functions and provides a polyfunctionality strength index [Citation16] (). Other commercially available methods for multiplex protein detection are reviewed below and summarized in .
Table 1. Multiplex protein detection immunoassay techniques.
3.2. Chemiluminescence
Chemiluminescence detection is more sensitive than chromogenic technology to quantify multiple proteins. Spot-specific chemiluminescence detection by a charge-coupled device (CCD) camera is a plate-based multiplexing mini-array detection approach for high-throughput analysis of proteins. In this multiplex system, antibodies are spotted in a 96-well plate, and well-plate surfaces can be chemically modified to bind proteins. The samples are added to the wells and the target proteins captured by the antibody array. In the next step, biotinylated antibodies, specifically binding to the captured proteins, are added. Streptavidin-HRP conjugate and a chemiluminescent substrate are finally incorporated. A catalyzed oxidation of the substrate generates chemiluminescence at each spot with the captured targeted proteins, which is measured by imaging the entire plate using a compatible CCD camera technology. For the determination of each analyte concentration corresponding protein standards are applied [Citation26,Citation27]. Commercially available planar-based multiplex ELISA kits, requiring chemiluminescent detection for quantification of inflammatory markers in serum, plasma, urine, cell, and tissue lysate from different sources have also been produced [Citation28]. Additionally, chemiluminescence-based single molecular analysis arrays are produced enabling performance of up to a 12-plex assay in a well [Citation26]. ( and ).
Sensitivity of detection is further increased by electrochemiluminescence (ECL) in which the substances are brought to a higher excitation level via an electron transfer reaction. The electron transitions from a high excitation state to a much lower energy state are accompanied by light emission, and the emitted light can be detected, which enables ultra-sensitive analysis [Citation29]. Despite that for many years this phenomenon was considered to be a part of fundamental research without an apparent practical application, ECL detection is now used to develop commercial devices to determine several clinically relevant analytes, including protein biomarkers and antibodies (). An exhaustive overview of ECL bioapplications can be found in [Citation30–Citation32]. Commercial platforms, such as those of Mesoscale Discovery, exploit ECL as their method of detection [Citation33]. Also, the possibilities of using ECL for point-of-care testing [Citation34], microfluidics and paper-based technologies [Citation35] or the combination of ECL with bipolar electrodes [Citation36] have been evaluated at experimental level.
3.3. Lateral flow immunoassays
Lateral flow immunoassays (LFA) are successful systems in Point of care (POC) testing and used for both qualitative and quantitative monitoring in resource-limited or non-laboratory settings, for example in emergency settings [Citation37]. LFA uses similar principles as ELISA, in which the capture antibody is immobilized to form a test line on a solid phase strip, e.g. nitrocellulose membrane, instead of a plastic well. The detection antibody is conjugated to a colorful particle or fluorescent label and this conjugate is allowed to freely react with the target analyte on the strip, when the sample is applied. As capillary action transfers the sample components through the test strip, the complex of the target analyte and detection antibody label-conjugate is halted at the test line by the capture antibody. The presence and/or concentration of the analyte in the sample can be determined by the intensity of the label at the test line [Citation38,Citation39]. Limited multiplexing of LFA can be achieved by incorporating multiple test lines for different analytes on the same strip or by creating an assembly of several strips around a common sample application pad. Several commercial LFAs have been used in the clinical world for biomarkers such as alpha-fetoprotein (AFP), carcinoma embryonic antigen (CEA), prostate-specific antigen (PSA), virus detection [Citation40] and fecal occult blood (FOB). Both to automate and integrate LFAs with medical analyses, smartphone sensing has gained interest in the last decade [Citation41,Citation42] leading to the development of different POC systems for various diseases. Smartphone sensing technology measures colorimetric or fluorescence-based signals with an appropriate software or an app using colored molecules or fluorophores, respectively [Citation43,Citation44]. Thus, a term ‘mobile health (mHealth)’ which refers to the data sharing between patients and physicians emerges, including the use of websites and/or applications to specify and/or monitor a particular health condition [Citation45]. The aim of mHealth is to achieve better personalized healthcare with providing an immediate resource for clinical decision by doctors and pharmacists [Citation46].
3.4. DNA-based protein detection
Partly based on the immunoassay principle other platforms have emerged whereby protein binding by antibodies is combined with DNA-based detection using the advantages of signal amplification. In so-called proximity extension assays (PEA), a pair of antigen-specific antibodies are conjugated with complimentary oligonucleotides that produce a PCR target sequence when both antibodies bind to target protein in close proximity. After amplification, the DNA barcode is quantified by standard microfluidic qPCR using specialized equipment. The advantage of this approach is that it limits cross-reactivity as only matched DNA reporter pairs will be amplified. This allows high specificity and a higher degree of multiplexing in comparison to other multiplex assays (most of them limited to maximum of 10-plex assays). The PEA has been used to analyze a number of potential diagnostic or prognostic biomarkers in areas such as cancer and inflammation research [Citation47]. Commercial PEAs facilitate multiplex assays enabling simultaneous detection of 92 analytes (in total >1100 pre-selected analytes are experimentally available) in only 1 µL of sample [Citation47] ().
4. Mass spectrometry
Mass spectrometry is a powerful technique for detailed molecular characterization of biomolecules, including proteins, metabolites, lipids, and carbohydrates. Rather than just detection, mass spectrometry can reveal unique chemical fragments of such biomolecules, including post-translational protein modifications and specific metabolic isoforms of specific biochemical interactions [Citation48,Citation49].
Mass spectrometry (MS) is well suited for detection of proteins and proteoforms. Yet, because it also detects many other analytes, a thorough sample clean-up of clinical samples is required to remove contaminating salts and endogenous non-protein biological components. Furthermore, the huge dynamic range of proteins present in biological samples is a challenge for mass spectrometry analysis. For cultured cells and liquid biopsies such as urine and saliva, this difference is only 6–9 orders of magnitude whereas in plasma and tissue samples the dynamic range can be greater than 12, with serum albumin representing over 50% of the protein content. To overcome this, either abundant proteins are removed or target biomarkers enriched to enhance their detection [Citation50]. In the former approach, samples (typically plasma or serum) are immunodepleted on columns that capture albumin, endogenous immunoglobulins, transferrin, fibrinogen, and apolipoproteins [Citation51]. Proteins of interest do not bind to the columns and are recovered in the flow-through. In the latter case, protein enrichment may be carried out by subcellular fractionation using ultracentrifugation [Citation52] or affinity chromatography with activity-based chemical probes to isolate specific protein classes based on structure and/or function (e.g. glycosylation, ubiquitination, kinases, phosphoproteins, metalloproteins) [Citation53,Citation54]. Immunoprecipitation, immunocapture, and capture by anti-peptide antibodies (SISCAPA) and mass spectrometric immunoassay (MSIA) are alternative methods dedicated to the enrichment of specific peptide or protein targets as these techniques provide the highest selectivity and sensitivity for low-abundant proteins [Citation55,Citation56] (). Precise and absolute quantification can be reached by the inclusion of stable isotope standards. Most ideally heavy labeled endogenous proteins are used for absolute and relative quantification of proteins of interest. However, for practical and financial reasons this is in many cases impossible, a second best solution is recombinant heavy labeled protein. This gives an identical concentration of the enzymatic generated peptides of the protein, although the folding and possible modifications present in the endogeneous protein are absent in the recombinant protein. A third and most commonly used option is the use of heavy labeled peptides. Because these peptides of one protein are chemically synthesized separately, differences can occur because of inaccuracies in determining the amount of peptide synthesized, different suppression of ions in the mass spectrometer cannot be corrected accurately for differences in behavior among the heavy labeled peptides and variances in recovery of the separate peptides from stock solutions can occur. Despite the shortcomings of stable isotope labeled peptides, it is the most practical solution to perform the quantification of proteins with mass spectrometry. All these steps mentioned above are designed to assist identification of the protein of interest by proteomics-based mass spectrometry, but may increase sample preparation complexity and hence experimental variation.
Table 2. Multiplex protein detection mass spectrometric techniques.
Once the appropriate protein sample has been prepared, mass spectrometry can be employed to analyze intact proteins (‘top-down’ proteomics) or as peptides following proteolysis (‘bottom-up’ proteomics). In ‘top down’ proteomics, proteins are analyses as intact molecules rather than as smaller peptide fragments [Citation83]. Analytical coverage of an intact protein still provides a strong challenge, although mass spectrometers capable of ‘top-down’ proteomics (MALDI MS with in-source decay, Orbitrap-MSn, UHR-QToF MS, Fourier transform mass spectrometry-MSn) are able to analyze an increasing number of intact proteins in one run [Citation84]. Consequently, enrichment and affinity methods as described above play an essential role prior to intact protein analysis by MS. Although it circumvents the need for protease digesting proteins to peptides, ‘top down’ proteomics still has to be improved for broader applications in biomarker discovery and validation, and only limited examples of clinical applications are published [Citation85] ( and ).
‘Bottom-up’ proteomics is the mainstay of biomarker research. However, sample processing methods in the translation from discovery to validation have a substantial impact on reproducibility, particularly when considering analysis of multiple experiments [Citation86]. Whichever biological sources and extraction methods are used for validation studies, protein samples are likely to require clean-up (removal of detergents and other endogenous biomolecules) and necessitate concentration (e.g. urine) prior to proteolytic digestion [Citation87]. Irrespective of the method used (protein precipitation with organic solvents/acids, dialysis, C1 or C4 reverse-phase cartridges, size exclusion chromatography and molecular weight cutoff spin columns), all additional steps risk lowering biomarker yield and increase potential for experimental variation. So far, most of these methods have not been universally standardized, validated, and automated for clinical use ( and ).
4.1. Mass spectrometry for biomarker validation – targeted proteomics (SRM, MRM, and PRM)
Targeted proteomics experiments are classically performed using triple quadrupole mass spectrometers in selected reaction monitoring (SRM) or multiple reaction monitoring (MRM) modes [Citation88,Citation89] (). SRM/MRM approaches have been widely applied for small molecule analysis and only recently emerged as a powerful analytical tool that can be used in ‘bottom-up’ proteomics, due to its wide dynamic range, relative and absolute quantification, and multiplexing capability. For each target protein, signature (or proteotypic) peptides are monitored using highly selective Liquid chromatography (LC)-MS/MS analysis. Particularly, the first and third quadrupoles (Q1 and Q3) of the instrument work as mass filters (typically with a window of ± 1 Da), successively selecting a series of signature peptide ions (Q1) and their corresponding fragments (Q3). Each precursor ion and its fragment ions are called transitions. The intermediate quadrupole (Q2) serves as a collision cell. Due to their multiplexing and scheduling capabilities (adjustment of transition monitoring at specific LC elution times), SRM experiments are especially adapted to large-scale evaluation studies of protein biomarker candidates [Citation90]. Parallel reaction monitoring (PRM) is a similar acquisition mode specific to Orbitrap-type mass spectrometers. In this case peptide ions of interest are selected in a quadrupole in the same way as for triple quadrupole mass spectrometers, but then fragmentation occurs in a linear ion trap before re-routing all targeted product ions to the high-resolution Orbitrap simultaneously rather than sequentially. This enables higher sensitivity and more accurate quantification of the product ions.
In targeted proteomics experiments absolute quantification of candidate biomarkers is generally obtained using labeled peptide standards incorporating 15 N and/or 13 C isotopes [Citation91]. These stable isotope labeled (SIL) (also referred as AQUA peptides) standards exhibit identical chemical and physical properties to the unlabeled target protein or its surrogate peptides, but they can be distinguished by their mass difference. According to the isotope dilution principle, SRM/MRM/PRM-based quantification can be performed at the peptide level by comparing transition signals from the isotope-labeled and the unlabeled version of a given proteotypic peptide. Different types of internal standards can be used, such as SIL peptides [Citation92], winged SIL peptides [Citation93], SIL peptide concatemers (QconCAT) [Citation94], SIL protein fragments (QPrEST) [Citation95] or full-length SIL proteins (PSAQ or absolute SILAC) [Citation96,Citation97]. Several comparative studies have shown that the quantification performance of targeted proteomics assays can be greatly influenced by the type and quality of the chosen standard [Citation96,Citation98,Citation99]. In this context, the proteomics community has prepared specific recommendations to ensure the development of reliable and reproducible targeted proteomics assays, in a ‘fit-for-purpose’ approach [Citation100,Citation101]. Importantly, to promote the use of targeted proteomics assays, the Clinical Proteomic Tumor Analysis Consortium (CPTAC) has launched an Assay Portal to serve as an open-source repository of well-characterized targeted proteomic assays [Citation102].
4.2. Data-dependent analysis (DDA) and data-independent analysis (DIA)
Historically most ‘bottom-up’ MS approaches use commercially available mass spectrometers (ion traps, Q-TOFs, IMS-TOF MS, Orbitraps, FT-MS, LC-MALDI MS/MS) for Data-Dependent Acquisition (DDA) compilation of proteomics data, particularly for discovery-based strategies. In the DDA strategy, as peptides pass through an LC column and enter the mass spectrometer, the instrument scans all ions but then chooses a subset of those, typically the most abundant ones (MS1 spectra), as precursor ions for further fragmentation usually by Collision-Induced Dissociation (CID). The fragmented ions obtained are then acquired to generate MS/MS data (MS2 spectra), which is used to identify peptides by reconstructing the peptide’s amino acid sequence by database searching. Clinical applications of DDA workflows have proven to be robust and easy to implement in the clinical setting [Citation103,Citation104]. Nonetheless, DDA strategy presents some disadvantages such as the speed of data acquisition, relative low reproducibility compared to immunoassay, absence of important information due to the low-abundant peptides that may not be available for fragmentation and lack of standardization of the quantitative strategy.
To overcome these limitations, a Data-Independent Acquisition (DIA) strategy was developed by Smith group [Citation105] and later refined by the Aebersold’s group in 2012 utilizing triple-TOF mass spectrometers, in which all theoretical spectra within sequential mass windows (known as SWATH-MS) are acquired [Citation106–Citation108]. A similar acquisition approach is possible on Orbitrap mass spectrometers, called WiSIM DIA [Citation109] and on ion-mobility (IMS) mass spectrometers (MSE) [Citation110,Citation111]. The idea behind DIA is to record all MS2 spectra from all precursor ions that fall in a specified mass-to-charge ratio (m/z) range, in a systematic and unbiased manner, leading to the entire set of peptide precursors of a sample. SWATH-MS technique is particularly innovative for discovery purposes due to its proposed data extraction strategy as it combines faster DIA with targeted data extraction. In each SWATH-MS measurement, data acquisition relies on consecutive and established m/z ranges of precursor ions that are isolated in order to be subjected to fragmentation. Through a rapid and recursive scan of these sequential m/z windows, the total precursor ion m/z range of previously digested proteins is covered and fragmentation spectra of precursor ions are obtained.
The DIA data obtained constitutes a complete digital map where ionized ions and specific peptides can be identified and/or quantified by applying targeted data extraction against a previously identified peptide list (also called library). These reference spectral libraries should contain all prior known information regarding peptide components of proteins in a study (e.g. retention time, precursor m/z or MS2 spectra) in order to be extracted from the DIA data. The peptide MS2 assay libraries can be generated locally through DDA analysis, obtained from community data repositories or even a combination of both. Thus, using an in-house generated library or combining it with a publicly accessible one, the overall information should ideally have high degree of protein coverage. The major repository of the peptide library assay [Citation112], covering ~51% of human proteome (>10,000 proteins) is accessible to the scientific community for SWATH-MS analysis in human samples. Other public libraries have been made available by SWATH atlas (www.swathatlas.org). However, at this point, it seems conceptual that a locally generated in-house library is favored as it can be used for several SWATH-MS measurements, with the same LC conditions used to elute the unknown samples. This allows the retention times of eluted peptides to be within a similar range of those in the library. The quality and coverage of the library are of crucial importance for SWATH methodology performance. There are several detailed protocols [Citation113,Citation114] that have been successfully used to build a high-quality library for targeted analysis of SWATH-MS data, supported by DIA analysis software tools. An in-house library spectrum can be expanded at any time just with the addition of more information from DDA acquisition. To build more robust libraries, several sample processing strategies can be applied to reduce proteome complexity before LC-MS analysis, such as protein or peptide fractionation, PTMs or protein-specific enrichment, and depletion of most abundant proteins [Citation114]. Thus, it must be taken into consideration that these procedures increase substantially the sample processing which may compromise analytical reproducibility.
DIA methodologies take advantage of high reproducibility and sensitivity of well-known target methods such as SRM, MRM, and PRM, with the increased proteome coverage normally seen in DDA analysis. In addition to its versatility and due to its capacity of fragmentation of the entire sample, the SWATH-MS technique is useful in characterization and/or quantification of low abundance proteins within a proteome, such as PTMs or other proteins that DDA (through shotgun analysis) approach cannot reach. Furthermore, SWATH-MS is a label-free methodology which is important for biomarker discovery but has yet to make an impact on clinical applications. Despite all of the advances achieved in the last years, DIA still has limitations in sensitivity as a tool for routine clinical diagnosis [Citation107]. The definition of the best operating procedures, the standardization of protocols for sample preparation and acquisition between laboratories, as well as the need for appropriate and inexpensive internal standards will possibly globalize the method but the application of DIA strategies in clinic or large-scale studies is not reached yet at this stage. Currently, efforts have been made in order to achieve these goals and this will contribute to the increase of both databases repositories (spectral libraries and digital maps) ().
5. Alternative and other emerging technologies
5.1. Alternative protein binders
An impressive number of antibodies (monoclonal, polyclonal) have been produced and are commercially available for binding specific proteins in biomarker assays. However, through post-translational modification such as truncation, phosphorylation, acetylation, glycosylation, many proteoforms can be produced from a single gene product [Citation115]. In the case where a higher specificity for a particular proteoform is needed in a biomarker assay, specific affinity matrices are required and can be too time-consuming, inefficient, or impossible to generate the right antibodies for each candidate biomarker to be validated. To that end, alternative binders have become available to replace or supplement antibodies. Firstly, affimers are small and stable recombinant proteins that are based on a stable protein scaffold, derived from the cysteine protease inhibitor family of cystatins. They contain two variable binding loops of nine amino acids each, which can be replaced with alternative randomized sequences to generate affimer libraries of ~1010 clones. These can be screened for affimers with specific properties such as high specificity and/or low dissociation rate [Citation116]. Of interest, affimers are heat and pH-stable, expanding their application as protein affinity matrices in assays. Secondly, aptamers are small molecules that can be synthetically generated using combinatorial chemistry, thus generating libraries that can be screened for protein-specific binding. Interestingly, aptamers are bound by proteins through a conformational binding pocket, resembling drug target and distinguishing from the binding of surface epitopes by antibodies and affimers. SomaLogic developed aptamer-based multiplex biomarker assays, measuring more than 1,300 protein analytes in a small volume of biological matrices in a single run. The detection technology uses Slow Off-rate Modified aptamers with dual nature for high protein affinity, with defined three-dimensional structure and unique nucleotide sequence which is recognizable by specific DNA hybridization probes [Citation117,Citation118]. Thirdly, less specific affinity matrices were developed to enrich specific protein classes based on their physical-chemical properties, such as lectins that have a higher affinity for glycosylated proteins [Citation119]. However, in most cases, an additional high resolution of specific detection method is needed which could be provided by mass spectrometry.
5.2. Alternative protein separation
Capillary electrophoresis (CE) is a collective term representing a number of electrokinetic separation techniques performed in narrow bore capillaries or microchips. Capillary zone electrophoresis (CZE) is widely used for the separation of charged species based on differences in their charge density. CE offers an outstanding separation efficiency for peptides and small proteins, being complementary with liquid chromatographic (LC) separations, both in ‘top-down’ and ‘bottom-up’ proteomics. Interfacing CE to MS has matured into a robust clinical investigational tool in several disease areas [Citation120], offering fast separations with good analytical sensitivities in protein biomarker analysis [Citation121]. The most popular interface of CZE-MS coupling is via electrospray ionization (ESI). Both sheath-flow and sheath-less interface designs are employed. To circumvent significant sheath liquid-mediated sample dilution and the limited sample loading capacity of CE, tapered emitters operating in the nanospray regime not only support lower flow rates of the sheath liquid, but also contribute to better desolvation, enhanced sensitivity, and better salt tolerance [Citation122]. Miniaturization to a single microchip (MCE) with the use of an electrophoretic step prior to biomarker detection may lead to an attractive clinical diagnostic tool [Citation123].
5.3. Multi-omics analysis platforms
Optimally, to obtain an in-depth view of biological systems DNA, RNA, proteins and metabolites can be analyzed in one assay. Recently, electrochemical-based biosensors have been developed in which a specific biorecognition element (e.g. antibody, nucleotide strand, or aptamer) bind their biomarker target with high specificity and are combined with sensitive read-out modules [Citation124]. Such biosensors represent an ongoing developing field for fast monitoring and assessment of clinically relevant biomarkers usually from biological fluids. Varying working principles have been put into practice, from platforms containing antibodies or aptamers immobilized on nano-materials for electrochemical biointerfaces [Citation125] to magneto-immunosensors [Citation126] and to formulate multiplexed nanoscale biosensors systems [Citation127] (). As a commercial platform, nanostring developed fluorescent molecular barcodes linked to a specific binder in combinations that are custom-made or pre-designed for particular biomarkers. Following binding to their target, molecular barcode probes are detected digitally through microscopic imaging, thus detecting and counting up to 800 hundred unique biomarkers. Of particular interest, by mixing the binders, different moieties of biomarkers can be detected in one sample, including DNA, RNA, and proteins [Citation128].
5.4. Photonic and plasmonic resonating structure arrays
During the last decade a significant improvement of the analytical performances of POC detection devices was enabled through the development of photonic and plasmonic resonating structure arrays. In a generic way, this signal-to-noise amplification strategy relies on resonance phenomena responsible for a stronger optical energy confinement at specific slots on the substrate. One protein biosensor design focuses on label-free approach based on micro-resonators – ring resonator and crystal cavities arrays – and localized surface plasmon resonance (LSPR) on biofunctionalized metal nanoparticles or nanoantennas [Citation129–Citation133]. The other main signal enhancement strategy focuses on the use of a metal nanostructured substrate and near-infrared fluorophore labels in immunoassays [Citation134,Citation135]. The excitation field enhancement generated at the nanoscale gaps between the metal structures results in an increase of the optical transition rate and thus a stronger emission [Citation136]. In comparison to more conventional LFA-based nitrocellulose, this approach provides a high signal-to-noise ratio, larger dynamic range – typically up to 6 orders of magnitude – and reduced fluorophore auto-fluorescence [Citation137,Citation138].
5.5. Miniaturized sensors
The current trend of miniaturization of immunosensors using micro and nanotechnology promises to increase the sensitivity and multiplexing for the analysis of proteins and other biomarkers, while providing cost-effective alternatives to established assays [Citation139]. Nanostructured electrodes can provide limits of detection at sub-nanomolar concentrations by the use of redox labels with the advantage of using simple portable instrumentation [Citation140–Citation142]. Nanoparticles can be used to increase the original concentration of the analytes using magnetic nanoparticles [Citation143], as well as to increase the transduced signal by optical (plasmonic or quantum dots) [Citation144–Citation146] or electrochemical means (by redox reactions or by changing the conductivity) [Citation147]. However, usually these approaches make the assays more complex due to the use of labels. Other transduction methods based on nanotechnology are evolving to develop label-free biosensors using optical, electrochemical, and mass transduction. Micro cantilevers can be functionalized for the detection of very low concentrations of proteins (fM) [Citation148]. Plasmonic immunosensors based on the localized surface plasmon resonance (LSPR) [Citation146,Citation149] and immunoassays based on potentiometric measurements with immuno-field effect transistors (immunoFETs) [Citation150,Citation151] are promising technologies for label-free sensing offering ultralow sensitivities (fM), cost-effectiveness and the possibility for ultra-high multiplexing combining several sensors with the concept of micro-arrays (immobilization of molecular libraries) [Citation152,Citation153]. These characteristics of transducers like plasmonic biosensors or immunoFETs make them excellent candidates for early detection of cancer-biomarkers and POC diagnostics [Citation154–Citation156]. However, most of the nanotechnology-based devices are still at the stage of ‘proof of concept.’ While some of them have passed the threshold to be commercialized for applications related to DNA, most of them in combination with PCR amplification [Citation157], protein assays still have issues to compete in sensitivity and selectivity with respect to well-stablished methods such as ELISA [Citation158].
5.6. Protein sequencing
The strong improvements of immunoassays, mass spectrometry, and other targeted platforms to analyze proteins in multiplex mode, as discussed above, can still be further improved. In that sense, we may be able to learn from the Next Generation Sequencing field where exponential developments have enabled parallel analysis of single DNA and RNA molecules in multiplex fashion. In particular, parallel single-molecule detection modules are developed for proteins as a nucleotide sequencing equivalent based on fluorescence, tunneling currents, and nanopores [Citation159–Citation161]. Although still early days, such high-throughput single-molecule protein sequencing analyses would revolutionize the proteomics field, providing comprehensive overviews of proteomes, targeted detection of high and low-abundant proteins, and the realization of single-cell proteomics.
6. Expert commentary
6.1. Validation of a method toward application
The validation of candidate protein biomarkers is an imperative component for application in pharmaceutical drug development and clinical sample analysis. Fast, sensitive, and reliable technologies are needed to execute this essential step, including selection of biomarkers for further development to robust assays. The multiplex approach, particularly in parallel read outs is required to reach large numbers of samples for validation in a high-throughput manner.
The sandwich-format immunoassay has been developed into a standardized, robust, and reproducible method and several multiplex versions of the method have emerged. Being a crucial component, the quality of antibodies (or alternative binders) needs to be highly specific and selective, thus being able to bind the target protein with minimal-to-no cross-reactivity and interaction with other proteins. Overall, this part of the process requires more attention in the validation steps. There are web-based resources, such as the Human Protein Atlas or Antibodypedia, that provide researchers with summarized information of available antibodies and their validation status. However, for antibodies and any binders developed for protein quantification it is highly recommended to characterize and confirm binding and cross-reactivity in the specific setting of the aimed method. For this purpose thorough analytical validation of the immunoassay should be performed preferably according to widely acknowledged guidelines such as those of the Clinical Laboratory Standards Institute (CLSI). For monoclonal antibodies, sequence information and epitope mapping information is helpful for standardization [Citation162,Citation163]. Immunoassay technologies show the possibilities for multiplexing with various read out systems. The limiting factor is the accessibility and quality level of the antibodies used. Alternative binders such as affimers or aptamers are expanding the possibilities to specifically bind proteins, by different interaction mechanisms and under conditions with diverse stringency.
In parallel, high-resolution mass spectrometry has inherent possibility to multiplex many proteins in one run without the use of specific binders. It has the potential to measure proteins at the sub nanogram/mL biofluid or sub ng protein/gram of tissue if appropriate sample preparation is applied. Although mass spectrometry techniques show very good reproducibility, the process is still time-consuming because of the serial approach of protein (or peptide) separation and detection. The less-than impressive sensitivity is caused mostly by ion suppression of peptides that elute simultaneously with the peptides of interest. So inherently low levels of detection can be reached although at the cost of lower throughput. Therefore, breakthroughs in the more efficient loading of ions in the mass spectrometer will increase the sensitivity. Developments such as IMS, FAIMS separation technology integrated online to the mass spectrometer and further improvements in DIA are experimental tools to reach the clinic. Of particular interest to biomarker scientists, mass spectrometry is particularly suited for analysis of post-translational modifications of proteins. These can strongly influence the stability, folding, interaction with other biomolecules, and thus activity of the protein. Mass spectrometry can provide such analysis relatively straightforward in an unbiased manner, in contrast to immunoassays where specific binders have to be developed. Novel experimental MS workflows such as IMS, that provide structural information on proteins, are expected to further increase the depth of protein characterization. Mass spectrometry does lead the way forward in providing the means for multiple, parallel analyte analyses, and in the context of multiple biomarkers the opportunity for increased diagnostic accuracy. The emergence of machine learning and artificial intelligence will further enhance the predictive capability of these complex multi-biomarker data sets to define disease susceptibility or onset [Citation164,Citation165].
We expect that alternative protein binders to supplement or replace antibodies will be strong drivers of clinical protein biomarker assays. Affimers, that are generated as recombinant proteins in stable scaffolds, with a high diversity, can be selected for a particular property as desired (specificity, selectivity, association/dissociation rates, stability) to a much better extent than antibodies. Also of interest, aptamers that mimic small molecules that are bound by particular enzymes will particularly detect a protein subpopulation that has drug target like properties, and as such complements the selection of antibodies based on surface binding affinity. Regarding protein biomarker detection, we view the emergence of molecular barcodes as particularly interesting as this enables the combination of different types of binders in one integrated analysis and the possibility to achieve semi-quantitation of DNA, RNA, and proteins. Finally, we see very high potential if analytical protein platforms could adopt some principles of the exponential developments in next-generation sequencing, for instance by single-molecule sequencing through nanopores, as this would significantly improve throughput and clinical applications. Although examples for DNA are established, the application for proteins is still premature.
Concluding, several analytical platforms have been developed to analyze protein biomarkers in a multiplex manner, some of which have the potential to seriously impact the way we do future biomarker research and clinical implementation and will drive the impact of personalized medicine [Citation166]. Mass spectrometers are for clinical use especially applied in metabolomics, drugs vitamins, antibiotic resistance. The clinical use of mass spectrometers for clinical use of proteins is near future. Especially changes in Medical Device Regulation within Europe will change and accelerate this process of quantifying proteins of interest in a clinical environment (https://ec.europa.eu/health/sites/health/files/docs/20200325_news_md_en.pdf).
Article highlights
High-throughput methods to validate proteins of choice by multiplex methods in a robust, time-efficient, and cost-effective manner are in place to further drive protein biomarker research and implementation in the clinic.
Clinical immunoassays are established as a robust platform for multiplex protein analyses. Recent improvements in protein binding, detection, and miniaturisation have yielded robust assays to semi-quantitate a large number of preselected protein biomarkers in integrated assay panels. Particular attention has to be given to the characterization of protein binders as this drives assay specificity, selectivity, sensitivity, and throughput.
Mass spectrometry has the unique capability to detect proteins and their post-translational modifications in an unbiased manner, reducing the necessity of selective protein binders. New developments in PRM, DIA, SWATH, FAIMS, and IMS will further improve the resolution and throughput of protein analytics. An elaborate understanding and solution of ion suppression will be found to increase the protein sensitivity.
Alternative technologies in immunoassay based on optical label-free technologies will mature to a sensitive, multiplex, and label-free detection of biomarkers.
The concomitant experimental development of nanotechnology-enabled resonator-based biosensors, site-directed bio-functionalization techniques, multi-spectral analytical approaches, and single-molecule protein sequencing will open promising perspectives for high-throughput protein clinical detection systems.
Declaration of interest
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Reviewer disclosures
Peer reviewers on this manuscript have no relevant financial or other relationships to disclose.
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Schork NJ. Personalized medicine: time for one-person trials. Nature. 2015Apr30;520(7549):609–611..
- van Gool AJ, Henry B, Sprengers ED. From biomarker strategies to biomarker activities and back. Drug Discov Today. 2010 Feb;15(3–4):121–126.
- Pegram MD, Lipton A, Hayes DF, et al. Phase II study of receptor-enhanced chemosensitivity using recombinant humanized anti-p185HER2/neu monoclonal antibody plus cisplatin in patients with HER2/neu-overexpressing metastatic breast cancer refractory to chemotherapy treatment. J Clin Oncol. 1998 Aug;16(8):2659–2671.
- Freedman LP, Cockburn IM, Simcoe TS. The economics of reproducibility in preclinical research. PLoS Biol. 2015 Jun;13(6):e1002165.
- van Gool AJ, Bietrix F, Caldenhoven E, et al. Bridging the translational innovation gap through good biomarker practice. Nat Rev Drug Discov. 2017 Sep;16(9):587–588.
- Mischak H, Ioannidis JP, Argiles A, et al. Implementation of proteomic biomarkers: making it work. Eur J Clin Invest. 2012 Sep;42(9):1027–1036.
- Rosenling T, Slim CL, Christin C, et al. The effect of preanalytical factors on stability of the proteome and selected metabolites in cerebrospinal fluid (CSF). J Proteome Res. 2009 Dec;8(12):5511–5522.
- Cristobal A, van den Toorn HWP, van de Wetering M, et al. Personalized proteome profiles of healthy and tumor human colon organoids reveal both individual diversity and basic features of colorectal cancer. Cell Rep. 2017Jan3;18(1):263–274.
- Hornbeck P, Winston SE, Fuller SA. Enzyme-linked immunosorbent assays (ELISA). Curr Protoc Mol Biol. 2001 May. Chapter 11: Unit 11.2.
- Engvall E, Perlmann P. Enzyme-linked immunosorbent assay (ELISA). Quantitative assay of immunoglobulin G. Immunochemistry. 1971 Sep;8(9):871–874.
- Aydin S. A short history, principles, and types of ELISA, and our laboratory experience with peptide/protein analyses using ELISA. Peptides. 2015 Oct;72:4–15.
- Gan SD, Patel KR. Enzyme immunoassay and enzyme-linked immunosorbent assay. J Invest Dermatol. 2013 Sep;133(9):e12.
- Revelen R, D’Arbonneau F, Guillevin L, et al. Comparison of cell-ELISA, flow cytometry and Western blotting for the detection of antiendothelial cell antibodies. Clin Exp Rheumatol. 2002 Jan-Feb;20(1):19–26.
- Czerkinsky CC, Nilsson LA, Nygren H, et al. A solid-phase enzyme-linked immunospot (ELISPOT) assay for enumeration of specific antibody-secreting cells. J Immunol Methods. 1983 Dec 16;65(1–2):109–121.
- Tighe PJ, Ryder RR, Todd I, et al. ELISA in the multiplex era: potentials and pitfalls. Proteomics Clin Appl. 2015 Apr;9(3–4):406–422.
- Ma C, Fan R, Ahmad H, et al. A clinical microchip for evaluation of single immune cells reveals high functional heterogeneity in phenotypically similar T cells. Nat Med. 2011 Jun;17(6):738–743.
- Huang RP. Cytokine protein arrays. Methods Mol Biol. 2004;264:215–231.
- Fulton RJ, McDade RL, Smith PL, et al. Advanced multiplexed analysis with the FlowMetrix system. Clin Chem. 1997 Sep;43(9):1749–1756.
- Appleyard DC, Chapin SC, Srinivas RL, et al. Bar-coded hydrogel microparticles for protein detection: synthesis, assay and scanning. Nat Protoc. 2011 Oct 20;6(11):1761–1774.
- Hemmila I, Holttinen S, Pettersson K, et al. Double-label time-resolved immunofluorometry of lutropin and follitropin in serum. Clin Chem. 1987 Dec;33(12):2281–2283.
- Ogata A, Tagoh H, Lee T, et al. A new highly sensitive immunoassay for cytokines by dissociation-enhanced lanthanide fluoroimmunoassay (DELFIA). J Immunol Methods. 1992 Apr 8;148(1–2):15–22.
- Gilbert M, Livingston R, Felberg J, et al. Multiplex single molecule counting technology used to generate interleukin 4, interleukin 6, and interleukin 10 reference limits. Anal Biochem. 2016 Jun 15;503:11–20.
- Rissin DM, Kan CW, Song L, et al. Multiplexed single molecule immunoassays. Lab Chip. 2013 Aug 7;13(15):2902–2911.
- Gold L, Ayers D, Bertino J, et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS One. 2010 Dec 7;5(12):e15004.
- Ngo D, Sinha S, Shen D, et al. Aptamer-based proteomic profiling reveals novel candidate biomarkers and pathways in cardiovascular disease. Circulation. 2016 Jul 26;134(4):270–285.
- Moody MD, Van Arsdell SW, Murphy KP, et al. Array-based ELISAs for high-throughput analysis of human cytokines. Biotechniques. 2001 Jul;31(1):186–90, 192–4.
- Leng SX, McElhaney JE, Walston JD, et al. and multiplex technologies for cytokine measurement in inflammation and aging research. J Gerontol A Biol Sci Med Sci. 2008 Aug;63(8):879–884.
- Esmaeili R, Zhang M, Sternberg MR, et al. The Quansys multiplex immunoassay for serum ferritin, C-reactive protein, and alpha-1-acid glycoprotein showed good comparability with reference-type assays but not for soluble transferrin receptor and retinol-binding protein. PLoS One. 2019;14(4):e0215782.
- Roda A, Mirasoli M, Michelini E, et al. Progress in chemical luminescence-based biosensors: A critical review [review]. Biosens Bioelectron. 2016 Feb;76:164–179.
- Miao WJ. Electrogenerated chemiluminescence and its biorelated applications [Review]. Chem Rev. 2008 Jul;108(7):2506–2553.
- Sek S, Vacek J, Dorcak V. Electrochemistry of peptides [review]. Curr Opin Electrochem. 2019 Apr;14:166–172.
- Ding CF, Zhang W, Wang W, et al. Amplification strategies using electrochemiluminescence biosensors for the detection of DNA, bioactive molecules and cancer biomarkers [review]. Trac-Trends Anal Chem. 2015 Feb;65:137–150.
- Toedter G, Hayden K, Wagner C, et al. Simultaneous detection of eight analytes in human serum by two commercially available platforms for multiplex cytokine analysis. Clin Vaccine Immunol. 2008 Jan;15(1):42–48.
- Gao WY, Saqib M, Qi LM, et al. Recent advances in electrochemiluminescence devices for point-of-care testing [review]. Curr Opin Electrochem. 2017 Jun;3(1):4–10.
- Wu MR, Lai QY, Ju Q, et al. Paper-based fluorogenic devices for in vitro diagnostics [review]. Biosens Bioelectron. 2018 Apr;102:256–266.
- Bouffier L, Arbault S, Kuhn A, et al. Generation of electrochemiluminescence at bipolar electrodes: concepts and applications [Review]. Anal Bioanal Chem. 2016 Oct;408(25):7003–7011.
- Davies P, Carlisle D. Five days that shook the NHS. Health Serv J. 2008 Jul;3;Suppl:4-7. DOI:10.1016/0006-291x(75)90498-2.
- Dzantiev BB, Byzova NA, Urusov AE, et al. Immunochromatographic methods in food analysis. Trends Analyt Chem. 2014 March 01;55:81–93.
- Mao X, Wang W, Du T-E. Rapid quantitative immunochromatographic strip for multiple proteins test. Sens Actuators B Chem. 2013 Sep 01;186:315–320.
- Bocoum FY, Ouedraogo H, Tarnagda G, et al. Evaluation of the diagnostic performance and operational characteristics of four rapid immunochromatographic syphilis tests in Burkina Faso. Afr Health Sci. 2015 Jun;15(2):360–367.
- Majumder S, Deen MJ. Smartphone sensors for health monitoring and diagnosis. Sensors (Basel). 2019 May 9;19(9):2164.
- Zhang D, Liu Q. Biosensors and bioelectronics on smartphone for portable biochemical detection. Biosens Bioelectron. 2016 Jan;15(75):273–284.
- Li Z, Wang Y, Wang J, et al. Rapid and sensitive detection of protein biomarker using a portable fluorescence biosensor based on quantum dots and a lateral flow test strip. Anal Chem. 2010 Aug 15;82(16):7008–7014.
- Guler E, Yilmaz Sengel T, Gumus ZP, et al. Mobile phone sensing of cocaine in a lateral flow assay combined with a biomimetic material. Anal Chem. 2017 Sep 19;89(18):9629–9632.
- Martinez-Perez B, de la Torre-diez I, Lopez-Coronado M. Mobile health applications for the most prevalent conditions by the World Health Organization: review and analysis. J Med Internet Res. 2013 Jun 14;15(6):e120.
- Oh H, Rizo C, Enkin M, et al. What is eHealth (3): a systematic review of published definitions. J Med Internet Res. 2005 Feb 24;7(1):e1.
- Lundberg M, Eriksson A, Tran B, et al. Homogeneous antibody-based proximity extension assays provide sensitive and specific detection of low-abundant proteins in human blood. Nucleic Acids Res. 2011 Aug;39(15):e102.
- Aebersold R, Mann M. Mass-spectrometric exploration of proteome structure and function. Nature. 2016Sep15;537(7620):347–355.
- Doll S, Gnad F, Mann M. The case for proteomics and phospho-proteomics in personalized cancer medicine. Proteomics Clin Appl. 2019 Mar;13(2):e1800113.
- Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002 Nov;1(11):845–867.
- Wu C, Duan J, Liu T, et al. Contributions of immunoaffinity chromatography to deep proteome profiling of human biofluids. J Chromatogr B Analyt Technol Biomed Life Sci. 2016 May 15;1021:57–68.
- Mulvey CM, Breckels LM, Geladaki A, et al. Using hyperLOPIT to perform high-resolution mapping of the spatial proteome. Nat Protoc. 2017 Jun;12(6):1110–1135.
- Lemeer S, Zorgiebel C, Ruprecht B, et al. Comparing immobilized kinase inhibitors and covalent ATP probes for proteomic profiling of kinase expression and drug selectivity. J Proteome Res. 2013 Apr 5;12(4):1723–1731.
- Sutton CW. The role of targeted chemical proteomics in pharmacology. Br J Pharmacol. 2012 May;166(2):457–475.
- Ten HS, Boulon S, Ahmad Y, et al. Mass spectrometry-based immuno-precipitation proteomics - the user’s guide. Proteomics. 2011 Mar;11(6):1153–1159.
- Trenchevska O, Nelson RW, Nedelkov D. Mass spectrometric immunoassays in characterization of clinically significant proteoforms. Proteomes. 2016 Mar 17;4(1):13.
- Ducret A, James I, Wilson S, et al. Translation and evaluation of a pre-clinical 5-protein response prediction signature in a breast cancer phase Ib clinical trial. PLoS One. 2019;14(3):e0213892.
- Collins CJ, Chang IJ, Jung S, et al. Rapid multiplexed proteomic screening for primary immunodeficiency disorders from dried blood spots. Front Immunol. 2018;9:2756.
- Fu Q, Kowalski MP, Mastali M, et al. Highly reproducible automated proteomics sample preparation workflow for quantitative mass spectrometry. J Proteome Res. 2018 Jan 5;17(1):420–428.
- Mbasu RJ, Heaney LM, Molloy BJ, et al. Advances in quadrupole and time-of-flight mass spectrometry for peptide MRM based translational research analysis. Proteomics. 2016 Aug;16(15–16):2206–2220.
- Abbatiello SE, Schilling B, Mani DR, et al. Large-scale interlaboratory study to develop, analytically validate and apply highly multiplexed, quantitative peptide assays to measure cancer-relevant proteins in plasma. Mol Cell Proteomics. 2015 Sep;14(9):2357–2374.
- Percy AJ, Yang J, Hardie DB, et al. Precise quantitation of 136 urinary proteins by LC/MRM-MS using stable isotope labeled peptides as internal standards for biomarker discovery and/or verification studies. Methods. 2015 Jun 15;81:24–33.
- Klont F, Pouwels SD, Hermans J, et al. A fully validated liquid chromatography-mass spectrometry method for the quantification of the soluble receptor of advanced glycation end-products (sRAGE) in serum using immunopurification in a 96-well plate format. Talanta. 2018 May 15;182:414–421.
- Klont F, Joosten MR, Ten Hacken NHT, et al. Quantification of the soluble receptor of advanced glycation end-products (sRAGE) by LC-MS after enrichment by strong cation exchange (SCX) solid-phase extraction (SPE) at the protein level. Anal Chim Acta. 2018 Dec 28;1043:45–51.
- De Marchi T, Kuhn E, Dekker LJ, et al. Targeted MS assay predicting tamoxifen resistance in estrogen-receptor-positive breast cancer tissues and sera. J Proteome Res. 2016 Apr 1;15(4):1230–1242.
- Yang X, Naughton SX, Han Z, et al. Mass spectrometric quantitation of tubulin acetylation from pepsin-digested rat brain tissue using a novel stable-isotope standard and capture by anti-peptide antibody (SISCAPA) method. Anal Chem. 2018 Feb 6;90(3):2155–2163.
- Hsiao YC, Chi LM, Chien KY, et al. Development of a multiplexed assay for oral cancer candidate biomarkers using peptide immunoaffinity enrichment and targeted mass spectrometry. Mol Cell Proteomics. 2017 Oct;16(10):1829–1849.
- Chen YT, Chen HW, Wu CF, et al. Development of a multiplexed liquid chromatography multiple-reaction-monitoring mass spectrometry (LC-MRM/MS) method for evaluation of salivary proteins as oral cancer biomarkers. Mol Cell Proteomics. 2017 May;16(5):799–811.
- Duangkumpha K, Stoll T, Phetcharaburanin J, et al. Urine proteomics study reveals potential biomarkers for the differential diagnosis of cholangiocarcinoma and periductal fibrosis. PLoS One. 2019;14(8):e0221024.
- Mun S, Lee J, Park A, et al. Proteomics approach for the discovery of rheumatoid arthritis biomarkers using mass spectrometry. Int J Mol Sci. 2019 Sep 5;20(18). DOI:10.3390/ijms20184368.
- Van Raemdonck GA, Osbak KK, Van Ostade X, et al. Needle lost in the haystack: multiple reaction monitoring fails to detect Treponema pallidum candidate protein biomarkers in plasma and urine samples from individuals with syphilis. F1000Res. 2018;7:336.
- Chen J, Tang MS, Xu LC, et al. Proteomic analysis of biomarkers predicting the response to triple therapy in patients with rheumatoid arthritis. Biomed Pharmacother. 2019 Aug;116:109026.
- Zhang Q, Salzler R, Dore A, et al. Multiplex immuno-liquid chromatography-mass spectrometry-parallel reaction monitoring (LC-MS-PRM) quantitation of CD8A, CD4, LAG3, PD1, PD-L1, and PD-L2 in frozen human tissues. J Proteome Res. 2018 Nov 2;17(11):3932–3940.
- Martinez-Garcia E, Lesur A, Devis L, et al. Development of a sequential workflow based on LC-PRM for the verification of endometrial cancer protein biomarkers in uterine aspirate samples. Oncotarget. 2016 Aug 16;7(33):53102–53115.
- Henderson CM, Bollinger JG, Becker JO, et al. Quantification by nano liquid chromatography parallel reaction monitoring mass spectrometry of human apolipoprotein A-I, apolipoprotein B, and hemoglobin A1c in dried blood spots. Proteomics Clin Appl. 2017 Jul;11(7–8):1600103.
- Guzel C, Govorukhina NI, Wisman GBA, et al. Proteomic alterations in early stage cervical cancer. Oncotarget. 2018 Apr 6;9(26):18128–18147.
- van der Ende EL, Meeter LH, Stingl C, et al. Novel CSF biomarkers in genetic frontotemporal dementia identified by proteomics. Ann Clin Transl Neurol. 2019 Apr;6(4):698–707.
- Skillback T, Mattsson N, Hansson K, et al. A novel quantification-driven proteomic strategy identifies an endogenous peptide of pleiotrophin as a new biomarker of Alzheimer’s disease. Sci Rep. 2017 Oct 17;7(1):13333.
- Theodorescu D, Wittke S, Ross MM, et al. Discovery and validation of new protein biomarkers for urothelial cancer: a prospective analysis. Lancet Oncol. 2006 Mar;7(3):230–240.
- Rodriguez-Ortiz ME, Pontillo C, Rodriguez M, et al. Novel urinary biomarkers for improved prediction of progressive egfr loss in early chronic kidney disease stages and in high risk individuals without chronic kidney disease. Sci Rep. 2018 Oct 29;8(1):15940.
- van den Broek I, Nouta J, Razavi M, et al. Quantification of serum apolipoproteins A-I and B-100 in clinical samples using an automated SISCAPA-MALDI-TOF-MS workflow. Methods. 2015 Jun 15;81:74–85.
- Popp R, Li H, LeBlanc A, et al. Immuno-matrix-assisted laser desorption/ionization assays for quantifying AKT1 and AKT2 in breast and colorectal cancer cell lines and tumors. Anal Chem. 2017 Oct 3;89(19):10592–10600.
- Tran JC, Zamdborg L, Ahlf DR, et al. Mapping intact protein isoforms in discovery mode using top-down proteomics. Nature. 2011 Oct 30;480(7376):254–258.
- Schmit PO, Vialaret J, Wessels H, et al. Towards a routine application of top-down approaches for label-free discovery workflows. J Proteomics. 2018 Mar;20(175):12–26.
- Tegtmeyer LC, Rust S, van Scherpenzeel M, et al. Multiple phenotypes in phosphoglucomutase 1 deficiency. N Engl J Med. 2014 Feb 6;370(6):533–542.
- Blum BC, Mousavi F, Emili A. Single-platform ‘multi-omic’ profiling: unified mass spectrometry and computational workflows for integrative proteomics-metabolomics analysis. Mol Omics. 2018 Oct 8;14(5):307–319.
- Rogers JC, Bomgarden RD. Sample preparation for mass spectrometry-based proteomics; from proteomes to peptides. Adv Exp Med Biol. 2016;919:43–62.
- Lange V, Picotti P, Domon B, et al. Selected reaction monitoring for quantitative proteomics: a tutorial. Mol Syst Biol. 2008;4(1):222.
- Gallien S, Domon B. Detection and quantification of proteins in clinical samples using high resolution mass spectrometry. Methods. 2015 Jun 15;81:15–23.
- Picotti P, Aebersold R. Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nat Methods. 2012 May 30;9(6):555–566.
- Brun V, Masselon C, Garin J, et al. Isotope dilution strategies for absolute quantitative proteomics. J Proteomics. 2009 Jul 21;72(5):740–749.
- Gerber SA, Rush J, Stemman O, et al. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc Natl Acad Sci U S A. 2003 Jun 10;100(12):6940–6945.
- Kushnir MM, Rockwood AL, Roberts WL, et al. Measurement of thyroglobulin by liquid chromatography-tandem mass spectrometry in serum and plasma in the presence of antithyroglobulin autoantibodies. Clin Chem. 2013 Jun;59(6):982–990.
- Beynon RJ, Doherty MK, Pratt JM, et al. Multiplexed absolute quantification in proteomics using artificial QCAT proteins of concatenated signature peptides. Nat Methods. 2005 Aug;2(8):587–589.
- Zeiler M, Straube WL, Lundberg E, et al. A protein epitope signature tag (PrEST) library allows SILAC-based absolute quantification and multiplexed determination of protein copy numbers in cell lines. Mol Cell Proteomics. 2012 Mar;11(3):O111 009613.
- Brun V, Dupuis A, Adrait A, et al. Isotope-labeled protein standards: toward absolute quantitative proteomics. Mol Cell Proteomics. 2007 Dec;6(12):2139–2149.
- Hanke S, Besir H, Oesterhelt D, et al. Absolute SILAC for accurate quantitation of proteins in complex mixtures down to the attomole level. J Proteome Res. 2008 Mar;7(3):1118–1130.
- Kuhn E, Whiteaker JR, Mani DR, et al. Interlaboratory evaluation of automated, multiplexed peptide immunoaffinity enrichment coupled to multiple reaction monitoring mass spectrometry for quantifying proteins in plasma. Mol Cell Proteomics. 2012 Jun;11(6):M111 013854.
- Oeckl P, Steinacker P, Otto M. Comparison of internal standard approaches for SRM analysis of alpha-synuclein in cerebrospinal fluid. J Proteome Res. 2018 Jan 5;17(1):516–523.
- Carr SA, Abbatiello SE, Ackermann BL, et al. Targeted peptide measurements in biology and medicine: best practices for mass spectrometry-based assay development using a fit-for-purpose approach. Mol Cell Proteomics. 2014 Mar;13(3):907–917.
- Abbatiello S, Ackermann BL, Borchers C, et al. New guidelines for publication of manuscripts describing development and application of targeted mass spectrometry measurements of peptides and proteins. Mol Cell Proteomics. 2017 Mar;16(3):327–328.
- Whiteaker JR, Halusa GN, Hoofnagle AN, et al. CPTAC assay portal: a repository of targeted proteomic assays. Nat Methods. 2014 Jul;11(7):703–704.
- Kemna EH, Tjalsma H, Podust VN, et al. Mass spectrometry-based hepcidin measurements in serum and urine: analytical aspects and clinical implications. Clin Chem. 2007 Apr;53(4):620–628.
- Eigner U, Holfelder M, Oberdorfer K, et al. Performance of a matrix-assisted laser desorption ionization-time-of-flight mass spectrometry system for the identification of bacterial isolates in the clinical routine laboratory. Clin Lab. 2009;55(7–8):289–296.
- Pasa-Tolic L, Masselon C, Barry RC, et al. Proteomic analyses using an accurate mass and time tag strategy. Biotechniques. 2004 Oct;37(4):621–4, 626–33, 636 passim.
- Gillet LC, Navarro P, Tate S, et al. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol Cell Proteomics. 2012 Jun;11(6):O111 016717.
- Sajic T, Liu Y, Aebersold R. Using data-independent, high-resolution mass spectrometry in protein biomarker research: perspectives and clinical applications. Proteomics Clin Appl. 2015 Apr;9(3–4):307–321.
- Anjo SI, Santa C, Manadas B. SWATH-MS as a tool for biomarker discovery: from basic research to clinical applications. Proteomics. 2017 Feb;17(3–4):1600278.
- Koopmans F, Ho JTC, Smit AB, et al. Comparative analyses of data independent acquisition mass spectrometric approaches: DIA, WiSIM-DIA, and untargeted DIA. Proteomics. 2018 Jan;18(1):1700304.
- Souza GH, Guest PC, Martins-de-Souza D. LC-MS(E), multiplex MS/MS, ion mobility, and label-free quantitation in clinical proteomics. Methods Mol Biol. 2017;1546:57–73.
- Helm S, Baginsky S. MSE for label-free absolute protein quantification in complex proteomes. Methods Mol Biol. 2018;1696:235–247.
- Rosenberger G, Koh CC, Guo T, et al. A repository of assays to quantify 10,000 human proteins by SWATH-MS. Sci Data. 2014;1(1):140031.
- Schubert OT, Gillet LC, Collins BC, et al. Building high-quality assay libraries for targeted analysis of SWATH MS data. Nat Protoc. 2015 Mar;10(3):426–441.
- Wu JX, Song X, Pascovici D, et al. SWATH mass spectrometry performance using extended peptide MS/MS assay libraries. Mol Cell Proteomics. 2016 Jul;15(7):2501–2514.
- Smith LM, Kelleher NL. Consortium for Top Down P. Proteoform: a single term describing protein complexity. Nat Methods. 2013 Mar;10(3):186–187.
- Tiede C, Bedford R, Heseltine SJ, et al. Affimer proteins are versatile and renewable affinity reagents. Elife. 2017 Jun 27;6. DOI:10.7554/eLife.24903.
- Lollo B, Steele F, Gold L. Beyond antibodies: new affinity reagents to unlock the proteome. Proteomics. 2014 Mar;14(6):638–644.
- Gold L, Walker JJ, Wilcox SK, et al. Advances in human proteomics at high scale with the SOMAscan proteomics platform. N Biotechnol. 2012Jun15;29(5):543–549.
- Katrlik J, Svitel J, Gemeiner P, et al. Glycan and lectin microarrays for glycomics and medicinal applications. Med Res Rev. 2010 Mar;30(2):394–418.
- Latosinska A, Siwy J, Mischak H, et al. Peptidomics and proteomics based on CE-MS as a robust tool in clinical application: the past, the present, and the future. Electrophoresis. 2019 Sep;40(18–19):2294–2308.
- Mischak H, Vlahou A, Ioannidis JP. Technical aspects and inter-laboratory variability in native peptide profiling: the CE-MS experience. Clin Biochem. 2013 Apr;46(6):432–443.
- Sun L, Zhu G, Zhang Z, et al. Third-generation electrokinetically pumped sheath-flow nanospray interface with improved stability and sensitivity for automated capillary zone electrophoresis-mass spectrometry analysis of complex proteome digests. J Proteome Res. 2015 May 1;14(5):2312–2321.
- Wuethrich A, Quirino JP. A decade of microchip electrophoresis for clinical diagnostics - A review of 2008-2017. Anal Chim Acta. 2019 Jan 3;1045:42–66.
- Mascini M, Palchetti I, Tombelli S. Nucleic acid and peptide aptamers: fundamentals and bioanalytical aspects. Angew Chem Int Ed Engl. 2012 Feb 6;51(6):1316–1332.
- Zakynthinos E, Pappa N. Inflammatory biomarkers in coronary artery disease. J Cardiol. 2009 Jun;53(3):317–333.
- de Avila BE, Escamilla-Gomez V, Campuzano S, et al. Disposable amperometric magnetoimmunosensor for the sensitive detection of the cardiac biomarker amino-terminal pro-B-type natriuretic peptide in human serum. Anal Chim Acta. 2013 Jun 19;784:18–24.
- Farzin L, Shamsipur M, Samandari L, et al. Recent advances in designing nanomaterial based biointerfaces for electrochemical biosensing cardiovascular biomarkers. J Pharm Biomed Anal. 2018 Nov 30;161:344–376.
- Tsang HF, Xue VW, Koh SP, et al. NanoString, a novel digital color-coded barcode technology: current and future applications in molecular diagnostics. Expert Rev Mol Diagn. 2017 Jan;17(1):95–103.
- Bruzas I, Unser S, Yazdi S, et al. Ultrasensitive plasmonic platform for label-free detection of membrane-associated species [article]. Anal Chem. 2016 Aug;88(16):7968–7974.
- Ciminelli C, Dell’Olio F, Conteduca D, et al. Integrated photonic and plasmonic resonant devices for label-free biosensing and trapping at the nanoscale. Phys Status Solidi A. 2019;216(3):1800561.
- Acimovic SS, Sipova H, Emilsson G, et al. Superior LSPR substrates based on electromagnetic decoupling for on-a-chip high-throughput label-free biosensing [Article]. Light Sci Appl. 2017 Aug 6(8):e17042.
- Yavas O, Acimovic SS, Garcia-Guirado J, et al. Self-calibrating on-chip localized surface plasmon resonance sensing for quantitative and multiplexed detection of cancer markers in human serum [article]. ACS Sens. 2018 Jul;3(7):1376–1384.
- Wittenberg NJ, Im H, Johnson TW, et al. Facile assembly of micro- and nanoarrays for sensing with natural cell membranes [article]. Acs Nano. 2011 Sep;5(9):7555–7564.
- Liu B, Li YL, Wan H, et al. High performance, multiplexed lung cancer biomarker detection on a plasmonic gold chip [article]. Adv Funct Mater. 2016 Nov;26(44):7994–8002.
- Zhang R, Le BA, Xu W, et al. Magnetic “Squashing” of circulating tumor cells on plasmonic substrates for ultrasensitive NIR fluorescence detection [article]. Small Methods. 2019 Feb;3(2):7.
- Fothergill SM, Joyce C, Xie F. Metal enhanced fluorescence biosensing: from ultra-violet towards second near-infrared window [Review]. Nanoscale. 2018 Dec;10(45):20914–20929.
- Jawad ZAR, Theodorou IG, Jiao LR, et al. Highly sensitive plasmonic detection of the pancreatic cancer biomarker CA 19-9 [Article]. Sci Rep. 2017 Oct;7:7.
- Tabakman SM, Lau L, Robinson JT, et al. Plasmonic substrates for multiplexed protein microarrays with femtomolar sensitivity and broad dynamic range [Article]. Nat Commun. 2011 Sep;2(1):9.
- Ansari AA, Alhoshan M, Alsalhi MS, et al. Prospects of nanotechnology in clinical immunodiagnostics. Lect Notes Electr En. 2010 Jul;10(7):6535–6581.
- Ramachandran R, Chen SM, Kumar GPG, et al. An overview of fabricating nanostructured electrode materials for biosensor applications. Int J Electrochem Sci. 2015;Oct;10(10):8607–8629.
- Arris FA, Benoudjit AM, Sanober F, et al. Characterization of electrochemical transducers for biosensor applications. Multifaceted protocol in biotechnology. Singapore: Springer; 2018:119–137.
- Escamilla-Gómez V, Hernández-Santos D, González-García MB, et al. Simultaneous detection of free and total prostate specific antigen on a screen-printed electrochemical dual sensor. Biosens Bioelectron. 2009;24(8):2678–2683.
- Rocha-Santos TA. Sensors and biosensors based on magnetic nanoparticles. Trends Analyt Chem. 2014;62:28–36.
- Gill R, Zayats M, Willner I. Semiconductor quantum dots for bioanalysis. Angew Chem. 2008;47(40):7602–7625.
- Li J, Wu N. Biosensors based on nanomaterials and nanodevices. CRC Press, Boca Raton; 2014.
- Anker JN, Hall WP, Lyandres O, et al. Biosensing with plasmonic nanosensors. Nat Mater. 2008;7(6):442–453.
- Pei X, Zhang B, Tang J, et al. Sandwich-type immunosensors and immunoassays exploiting nanostructure labels: A review. Anal Chim Acta. 2013;758:1–18.
- Arntz Y, Seelig JD, Lang H, et al. Label-free protein assay based on a nanomechanical cantilever array. Nanotechnology. 2002;14(1):86.
- Hong Y, Huh Y-M, Yoon DS, et al. Nanobiosensors based on localized surface plasmon resonance for biomarker detection. J Nanomater. 2012;2012:111.
- Stern E, Klemic JF, Routenberg DA, et al. Label-free immunodetection with CMOS-compatible semiconducting nanowires. Nature. 2007;445(7127):519.
- de Moraes A, Kubota L. Recent trends in field-effect transistors-based immunosensors. Chemosensors. 2016;4(4):20.
- Syahir A, Usui K, Tomizaki K-Y, et al. Label and label-free detection techniques for protein microarrays. Microarrays. 2015;4(2):228–244.
- Stekel D. Microarrays: making them and using them. Microarray Bioinf. 2003, Cambridge university press, chapter 1;1–18.
- Bellassai N, D’Agata R, Jungbluth V, et al. Surface plasmon resonance for biomarker detection: advances in non-invasive cancer diagnosis. Front Chem. 2019;7:570.
- Doucey M-A, Carrara S. Nanowire sensors in cancer. Trends Biotechnol. 2019;37(1):86–99.
- Vashist S. Point-of-care diagnostics: recent advances and trends. Biosensors (Basel).2017 Dec 18;7(4). pii: E62.
- Popowitch EB, O’Neill SS, Miller MB. Comparison of the biofire filmArray RP, Genmark eSensor RVP, Luminex xTAG RVPv1, and Luminex xTAG RVP fast multiplex assays for detection of respiratory viruses. J Clin Microbiol. 2013;51(5):1528–1533.
- Cohen L, Walt DR. Single-molecule arrays for protein and nucleic acid analysis. Annu Rev Anal Chem (Palo Alto Calif). 2017 Jun 12;10(1):345–363.
- Restrepo-Perez L, Joo C, Dekker C. Paving the way to single-molecule protein sequencing. Nat Nanotechnol. 2018 Sep;13(9):786–796.
- Di Muccio G, Rossini AE, Di Marino D, et al. Insights into protein sequencing with an alpha-Hemolysin nanopore by atomistic simulations. Sci Rep. 2019 Apr 23;9(1):6440.
- Swaminathan J, Boulgakov AA, Hernandez ET, et al. Highly parallel single-molecule identification of proteins in zeptomole-scale mixtures. Nat Biotechnol. 2018 Oct 22;36(11):1076–1082.
- Bradbury A, Pluckthun A. Reproducibility: standardize antibodies used in research. Nature. 2015 Feb 5;518(7537):27–29.
- Bradbury AM, Pluckthun A. Antibodies: validate recombinants once. Nature. 2015 Apr 16;520(7547):295.
- Xu T, Fang Y, Rong A, et al. Flexible combination of multiple diagnostic biomarkers to improve diagnostic accuracy. BMC Med Res Methodol. 2015 Oct;31(15):94.
- Captur G, Heywood WE, Coats C, et al. Identification of a multiplex biomarker panel for hypertrophic cardiomyopathy using quantitative proteomics and machine learning. Mol Cell Proteomics. 2020 Jan;19(1):114–127.
- Van Eyk JE, Snyder MP. Precision medicine: role of proteomics in changing clinical management and care. J Proteome Res. 2019 Jan 4;18(1):1–6.
