ABSTRACT
Microbial communities in alpine environments >7,500 m.a.s.l. have not been well studied using modern cultivation-independent sequencing approaches due to the challenges and danger associated with reaching such high elevations. For this reason, we know little about the microorganisms found in sediments on Earth’s tallest mountains, how they reach these surfaces, and how they survive and remain active at such extreme elevations. Here, we explore the microbial diversity recovered from three sediment samples collected from the South Col (~7,900 m.a.s.l.) of Sagarmatha (Mount Everest) using both culturing and next generation sequencing approaches (16S rRNA gene, internal transcribed spacer [ITS] region, and 18S rRNA gene sequencing). Both approaches detected very low diversity of bacteria, protists, and fungi that included a combination of cosmopolitan taxa and specialized microorganisms often found at high elevations like those of the genera Modestobacter and Naganishia. Though we managed to grow viable cultures of many of these taxa, it remains likely that few, if any, can be active in situ at the South Col. Instead, these high-elevation surfaces may act as deep-freeze collection zones of organisms deposited from the atmosphere or left by climbers scaling the Earth’s highest mountain.
Introduction
High-elevation microbial communities must survive a variety of challenging conditions (D. Singh et al. Citation2012; Merino et al. Citation2019). With an increase in elevation comes a corresponding shift in altitude-dependent environmental conditions, including colder temperatures, lower atmospheric pressures, lower oxygen availability, and lower water activity (Merino et al. Citation2019). Sustaining the levels of metabolic activity required to be active becomes more challenging as elevation increases (Margesin et al. Citation2009; Margesin and Miteva Citation2011) and, as a result, microbial diversity tends to decrease correspondingly (Bryant et al. Citation2008; L. Zhang et al. Citation2009; Margesin and Miteva Citation2011). For example, in the Transantarctic Mountains, microbial communities in higher elevation soils are significantly less diverse, have lower biomass, and even grow more slowly than those in lower elevation soils (Dragone et al. Citation2022). The identity and function of the microorganisms can also change as elevation increases. In the Himalayas, fungi dominate sediment communities at elevations from 3,000 to 3,500 m (Petrovič, Gunde-Cimerman, and Zalar Citation2000; Margesin and Miteva Citation2011), archaeal ammonia oxidizers between 4,000 and 5,400 m, and bacterial ammonia oxidizers up to 6,500 m (L. Zhang et al. Citation2009). In addition, phototrophic microbes are thought to be missing in dry high-elevation soils, but active photosynthetic communities have been detected in periglacial soils above 5,500 m in the Himalayas (S. K. Schmidt et al. Citation2011) and above 6,000 m in soils receiving water vapor from fumaroles (Solon et al. Citation2018).
Analysis of samples that have been collected from the highest alpine environments around the world (>7,000 m) suggests that these environments are typically dominated by a small number of polyextremophilic taxa (Lynch et al. Citation2012; Merino et al. Citation2019). Psychrophiles, like the fungus Naganishia friedmannii, which has been shown to remain active in alpine environments above 6,000 m (S. K. Schmidt et al. Citation2017), or the bacterial genus Geodermatophilus, whose type strain was isolated in 1970 from sediment collected at 8,400 m.a.s.l. on Everest (Ishiguro and Fletcher Citation1975), are common. However, not all of the microorganisms identified above 7,000 m are highly adapted to such an extreme environment. Surface snow samples collected at ~8,000 m on the north side of Everest were found to contain high biomass communities of Proteobacteria and Actinobacteria, including the genera Actobacterium, Acinetobacter, and Kocuria (Liu et al. Citation2007), which can be found in temperate and tropical soils around the world (Delgado-Baquerizo et al. Citation2018). Though Liu et al. (Citation2007) suggested that these detected organisms were deposited by snowfall or eolian transport and are probably not active or even viable in situ, their findings highlight that many questions still remain about the microbial communities of the world’s highest alpine environments.
Mt. Everest (Sagarmatha, Chomolungma), located on the China–Nepal border within the Mahalangur Himal subrange of the Himalayas, is the world’s highest mountain. With an elevation of 8,849 m.a.s.l., Everest’s high-elevation environments likely represent some of the most “extreme” terrestrial surfaces on Earth. Though we know that diverse and active microbial communities have been described in many lower elevation environments on the mountain (L. Zhang et al. Citation2009; Margesin and Miteva Citation2011) and bacteria have been isolated from samples collected at 8,400 m (Ishiguro Citation1969; Ishiguro and Fletcher Citation1975), questions still remain about the habitability of the mountain’s highest elevations, in particular of exposed sediment surfaces >7,500 m on the mountain where microbial communities have never been studied using next-generation sequencing approaches. We expect that many microorganisms may never be active in such surface environments, restricted by the extremely challenging conditions experienced at such high elevations.
To better understand the impact that elevation has on microbial habitability and community structure, we analyzed three samples collected from the South Col (~7,900 m.a.s.l) of Everest. At the South Col, a ridge between Everest’s summit and the neighboring Lhotse, temperatures regularly reach −33°C with an air pressure one-third that at sea level (~380 hPA at time of collection; Moore and Semple Citation2004; Matthews et al. Citation2020). Additionally, measurements taken by a weather station installed at the South Col in 2019 have recorded a maximum wind speed of 66.5 m/s and a maximum insolation of 1,500 W/m2, which is greater than the values expected from the top of the atmosphere due to reflectance (Matthews et al. Citation2020). Using a combination of marker gene sequencing and more traditional culture-based methods, we add to previous microbiology research on Mt. Everest by asking two questions: Can we find evidence of microorganisms on terrestrial surfaces >7,000 m and, if so, are these organisms viable and could any be active and able to survive at such extreme elevations?
Materials and methods
Sample collection
Field sampling was performed as part of the National Geographic and Rolex Perpetual Planet Everest Expedition in April to May 2019. A total of three surface sediment samples (0–5 cm depth) were collected in sterile polyethylene bags using aseptic techniques. Sediments were collected about 170 m SE of Camp IV (27.9728°N, 86.9315°E, 7,944 m.a.s.l.; ).
Figure 1. The South Col of Mount Everest (27.9728°N, 86.9315°E). (A) Map displaying the sampling location along the Everest climbing route. Samples were collected ~170 m southeast of Camp IV from an exposed surface environment at 7,944 m.a.s.l. (B) View from sampling location back toward Camp IV. (C) The surface at the collection site was made up of fragments of variable grain size, with the smallest fragments <0.5 mm in diameter and the largest fragments ~2-5 cm in diameter. Photo credit: L. Baker Perry/National Geographic.

The three samples were sent to the University of Colorado Boulder, Colorado. Though the samples remained sealed after collection and throughout the entire shipping process, they did not remain frozen throughout the shipment. After arriving at the University of Colorado, Boulder the samples were refrozen and remained in storage at −70°C until they could be processed.
Cultivation-independent analysis
Samples were removed from the freezer and particles <1 mm in size were manually separated from the larger fragments using a sterilized sediment sieve. DNA was then extracted from 0.5 g of these finest particles in a laminar flow hood using the Qiagen DNeasy Powersoil Kit following the manufacturer’s recommendations. An extraction blank was included to test for any possible contamination introduced during the DNA extraction.
The DNA aliquots extracted from each of the three sediment samples and the associated extraction blank were used for a variety of polymerase chain reaction (PCR) amplifications to assess the archaeal/bacterial, fungal, and microeukaryotic communities. The primer sets that were used for PCR amplification included a primer pair that targets the hypervariable V4 region of the archaeal and bacterial 16S rRNA gene (515 F: 5′-GTGCCAGCMGCCGCGGTAA-3′ and 806-R: 5′-GGACTACHVGGGTWTCTAAT-3′; Walters et al. Citation2016), a primer pair that targets the internal transcribed spacer of the fungal rRNA operon (ITS1-F: 5′-CTTGGTCATTTAGAGGAAGTAA-3′ and ITS2-R: 5′-GCTGCGTTCTTCATCGATGC-3′; Bellemain et al. Citation2010), and a primer pair that targets the hypervariable V9 region of the 18S rRNA gene (1391-F: 5′-GTACACACCGCCCGTC-3′ and EukBR: 5′-TGATCCTTCTGCAGGTTCACCTAC-3′). One no-template PCR control was run with each set of amplifications. All three primer sets included the appropriate Illumina adapters and unique 12 bp barcode sequences to permit multiplexed sequencing (Caporaso et al. Citation2012). PCRs were performed in 25 µL reaction volumes. PCR amplification using the ITS1-F/ITS2-R and 1391-F/EukBR primer sets was performed using GoTaq Hot Start PCR Master Mix and PCR amplification using the 515 F/806-R pair used the Platinum II Hot-Start PCR Master Mix (2X). Cycling parameters for the ITS1F/ITS2R primer sets consisted of an initial denaturation step at 94°C for 3 minutes, followed by thirty-five cycles of denaturation at 94°C (45 seconds), annealing at 50°C (60 seconds), extension at 70°C (90 seconds), and a final extension step at 72°C for 10 minutes. Cycling parameters for the 515 F/806 R primer set consisted of an initial denaturation step at 94°C for 2 minutes, followed by thirty-five cycles of denaturation at 94°C (15 seconds), annealing at 60°C (15 seconds), extension at 68°C (60 seconds), and a final extension step at 72°C for 10 minutes. Cycling parameters for the 1391-F/EukBR primer set consisted of an initial denaturation step at 94°C for 3 minutes, followed by thirty-five cycles of denaturation at 94°C (45 seconds), annealing at 57°C (60 seconds), extension at 72°C (90 seconds), and a final extension step at 72°C for 10 minutes. Amplified product was visualized on a gel and then cleaned and normalized to equimolar concentrations using SequalPrep Normalization Plates. Sequencing was performed by the University of Colorado Boulder’s next-generation sequencing facility using the Illumina MiSeq platform. The V2 2 × 150 bp Illumina sequencing kit was used for the 16S rRNA gene and the 18S rRNA gene sequencing and the 2 × 250 bp paired-end Illumina sequencing kits were used for the internal transcribed spacer (ITS) region amplicons. Raw sequencing data can be accessed in the National Center for Biotechnology Information Sequence Read Archive, project accession number PRJNA882470.
Bioinformatics
16S rRNA gene sequences, ITS region sequences, and 18S rRNA gene sequences from the soil extractions were processed using DADA2 pipeline v3.8 (Callahan et al. Citation2016). Sequences were quality filtered and clustered into amplicon sequence variants (ASVs). Taxonomic information was assigned to ASVs using a naive Bayesian classifier method (Wang et al. Citation2007) that takes the set of ASVs generated and compares them to a training set of reference sequences: the 16S rRNA bacterial and archaeal SILVA database v132 (Quast et al. Citation2013; Yilmaz et al. Citation2014), the UNITE database v8.3 for fungi (Nilsson et al. Citation2019), and SILVA 18S SSU rRNA v132 Ref NR 99 database for eukaryotes, which includes fungi (Quast et al. Citation2013; Yilmaz et al. Citation2014). A minimum bootstrapping threshold required to return a taxonomic classification of 50 percent similarity was used for analysis of the 16S rRNA sequences and 18S rRNA sequences and a minimum bootstrapping threshold of 85 percent was used for ITS region sequences.
Downstream analysis of the DADA2 processed data was performed in R v4.0.5 (R Core Team Citation2017) using the package “mctoolsr” (https://github.com/leffj/mctoolsr/) (Leff Citation2016). For the 16S rRNA sequencing, ASVs associated with chloroplasts, mitochondria, and eukaryotes (sixty-two ASVs) were removed. ASVs that made up at least 1 percent of the extraction blank or no-template PCR control and were at least 1 percent of the samples on average were discarded as likely lab contaminants (ASV-17). After this filtering, a total of fifty-seven bacterial ASVs remained. No archaeal sequences were identified from the next-generation sequencing.
For the fungal ITS region sequencing data, ASVs that could not be classified to the phylum level (only classified as “fungi”) were removed prior to downstream analysis (three ASVs). No reads were identified in the extraction blank. The thirteen reads identified in the no-template PCR control were associated with the second most abundant ASV in the samples (ASV-2, Naganishia) that are often dominant members of high-altitude, low-temperature environments (S. K. Schmidt et al. Citation2017). Therefore, these reads were likely not a result of laboratory contamination but are instead most likely derived from the soil samples and represent “tag switching” events (Schnell, Bohmann, and Gilbert Citation2015). After this filtering, sixteen fungal ASVs remained.
For downstream analysis of the 18S rRNA gene sequences, we note that two ASVs were associated with human DNA. Only one of these ASVs was identified in the blanks, with 90 percent of the human reads coming from the samples themselves. Based on this, we do not believe that these were contaminated during processing and were likely in the samples at collection. For downstream analysis, these human ASVs were removed. Additionally, two ASVs in the blank samples were not found in any of the three South Col samples and were removed from the data set. After this filtering, fourteen eukaryotic ASVs remained.
Culturing study/culture identification
To determine whether any microorganisms in these samples were viable, the three sediment samples were plated on seven different types of solid culture media in conditions designed to promote growth. Antarctic bacterial medium (P. Singh, Singh, and Roy Citation2016), modified nutrient broth agar (Pulschen et al. Citation2017), variable nine-salt solution (Egan, Kjelleberg, and Holmström Citation2015), and a modified variable nine-salt solution without the nine-salt solution (Egan, Kjelleberg, and Holmström Citation2015) were chosen because they were designed for the culturing bacteria from oligotrophic environments. Antartic bacterial medium and modified nutrient broth agar were designed to target organisms found in nutrient-poor, cold, and dry Antarctic sediments (P. Singh, Singh, and Roy Citation2016; Pulschen et al. Citation2017) and have been used successfully to grow Proteobacteria, Actinobacteria, Firmicutes, and the fungus Naganishia (Pulschen et al. Citation2015; Dragone et al. Citation2021), which were the most abundant taxa identified through our 16S rRNA gene and ITS region sequencing effort (). Samples were also plated on three nutrient-rich media variants, Luria-Bertani agar (Atlas Citation2004) and R2A agar at pH 7 and pH 9 (Reasoner and Geldreich Citation1985), which were chosen to target the copiotroph taxa identified in our next-generation sequencing approach that we felt may be missed if we just used oligotrophic media (, Data S1). All media were created following the methods described in the cited literature, but more information about the specific pH and conditions can be found in Dataset S3.
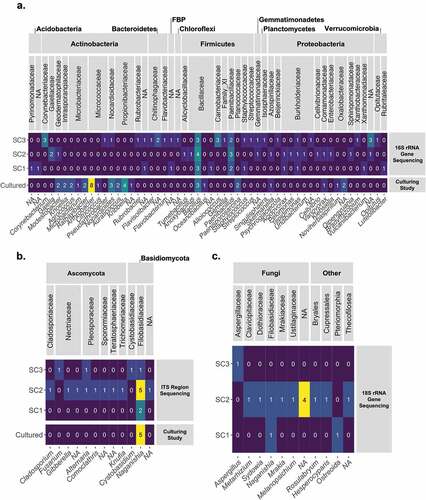
Figure 2. The microganisms identified in the three South Col samples. (A) The bacterial genera identified through the 16S rRNA gene sequencing and the culturing study. (B) The fungal families identified through the ITS sequencing and the culturing study. (C) The identity of the microeukaryotic ASVs identified through the 18S rRNA gene sequencing. For all heat maps, the number of ASVs (or cultures) assigned with each taxonomic identification is displayed in the corresponding box and colored based on the magnitude. “NA” indicates that a taxonomic classification at that level is not avaliable.
One gram of each sample was vortexed and then shaken for 5 minutes in 1 mL of a 7.2 pH phosphate-buffered solution and 60 µL of the resulting soil slurry was pipetted onto each plate (surface area of each plate: 58 cm2) and spread across the plates using flame-sterilized cell spreaders. Two blank plates inoculated with 60 µL PBS were prepared for each media type and handled in an identical manner to the twenty-eight plates inoculated with the soil slurries. Plates of each different media type were incubated at 4°C with regular nocturnal/diurnal light cycles. Plated samples and the uninoculated control plates remained under these conditions for four weeks. All unique colonies that grew were isolated using a streak plate isolation technique. To ensure purity, cultures from the first isolation plate were streaked for isolation two subsequent times.
DNA from each of the isolated colonies was extracted using the Qiagen DNeasy Ultraclean 96 Microbial Kit following the manufacturer’s recommendations. A total of six extraction blanks were included to control for contamination introduced during the DNA extraction.
The DNA aliquots extracted from each of the isolated cultures and associated extraction blanks were PCR amplified in duplicate using a primer pair that targets the archaeal and bacterial 16S rRNA gene (S_27 F: 5′-GTGCCAGCMGCCGCGGTAA-3′ and S_1429 r: 5′-GGACTACHVGGGTWTCTAAT-3′) and using the primer pair (ITS1-F: 5′-CTTGGTCATTTAGAGGAAGTAA-3′ and ITS4-R: 5′-GCTGCGTTCTTCATCGATGC-3′) that targets the ITS of the fungal rRNA operon. Three no-template PCR controls were run for each set of amplifications. PCRs were performed in 25 µL reaction volumes. PCR amplification of the ITS region of the rRNA operon was performed using GoTaq Hot Start PCR Master Mix in 25, while PCR amplification of the 16S rRNA gene was performed using Platinum II Hot-Start PCR Master Mix (2X). Cycling parameters for the ITS1F/ITS4R primer sets consisted of an initial denaturation step at 94°C for 3 minutes, followed by thirty-five cycles of denaturation at 94°C (45 seconds), annealing at 50°C (60 seconds), extension at 70°C (90 seconds), and a final extension step at 72°C for 10 minutes. Cycling parameters for the S_27 F/S_1429_R primer set consisted of an initial denaturation step at 94°C for 2 minutes, followed by thirty-five cycles of denaturation at 94°C (15 seconds), annealing at 60°C (15 seconds), extension at 68°C (60 seconds), and a final extension step at 72°C for 10 minutes. Amplified PCR product from all of the cultured isolates, the six extraction blanks, and the three no-template PCR controls were sequenced by GENEWIZ Sanger sequencing service. Fungal DNA was sequenced using the ITS1_F primer and the bacterial/archaeal DNA was sequenced using the S_27 F primer to capture the full-length sequence of the ITS rRNA operon and 16S rRNA gene, respectively.
Sanger sequence data from the isolates were processed following the “swabs to genomes” workflow described in Dunitz et al. (Citation2015). No sequences from any of the six extraction blanks or three no-template controls were recovered. Briefly, sequences were first filtered based on their length, with only sequences >700 bp kept for identifications based on the 16S rRNA gene and only those >200 bp kept for identifications based on the ITS region. These remaining sequences were cleaned with SeqTrace v0.9.1 (Stucky Citation2012), after which thirty-eight 16S rRNA sequences and six ITS sequences remained. Taxonomy was assigned to all sequences using the RDP classifier v2.10.2 (Wang et al. Citation2007) and the basic local alignment search tool (Altschul et al. Citation1990). Recovered sequences for all fungal and bacterial isolates, as well as their likely taxonomic classification, are reported in Dataset S3.
Geodermatophilaceae phylogenetic tree creation
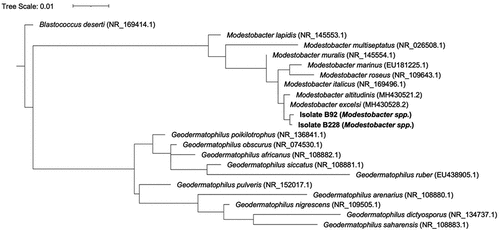
The phylogenetic relationship of our two Modestobacter isolates to previously described Modestobacter taxa was determined from 850 bp sequences of the 16S rRNA gene via maximum likelihood with RaxML (Stamatakis Citation2014). Sequences of 850 bp in length were extracted from partial or complete sequences accessed through the National Center for Biotechnology Information Nucleotide Database, including eight species of Modestobacter and ten species of Geodermatophilus that have been isolated from soils and sediments in cold, dry environments around the world, including the G. obscurus that was previously isolated from a high-elevation surfaces on Mount Everest by Ishiguro and Fletcher (Citation1975). Other sequences included M. lapidis (Trujillo et al. Citation2015), M. multiseptatus (Mevs et al. Citation2000), M. muralis (Trujillo et al. Citation2015), M. marinus (Xiao et al. Citation2011), M. roseus (Qin et al. Citation2013), M. italicus (Montero-Calasanz et al. Citation2019), M. altitudinis (Golinska et al. Citation2020), M. excelsi (Golinska et al. Citation2020), G. poikilotrophi (Montero-Calasanz et al. Citation2014), G. africanus (Montero-Calasanz, Göker, Pötter et al. Citation2013a), G. siccatus (CitationMontero-Calasanz, Göker, Pötter et al. 2013b), G. ruber (Y. Zhang et al. Citation2011), G. pulveris (Hezbri et al. Citation2016), G. arenaris (Montero-Calasanz et al. Citation2012), G. nigrescens (Nie et al. Citation2012), and G. dictyosporus (Montero-Calasanz et al. Citation2015), G. saharensis (Montero-Calasanz, Göker, Rohde et al. Citation2013). First, sequences were aligned using MUSCLE v3.8.31 (Edgar Citation2004). Aligned reads were used to construct a tree with RaxML v8.0.0 (raxmlHPC -f a -m GTRGAMMA -p 12345 -x 12345 -number 100), including Blastococcus deserti (NR_169414.1) as an outgroup for tree rooting (Stamatakis Citation2014). The tree was visualized and annotated using iTOL v6.3.2 (Letunic and Bork Citation2016).
Geochemical analysis
Sediment pH was determined according to the method described in King et al. (Citation2010). Specifically, 5 g of soil and 5 g of deionized water were placed in a 15-mL conical tube and shaken for 2 hours at 200 rpm. pH was then measured with an Orion Star A211 Benchtop pH meter. Total water content of samples was measured following a method described in Vimercati, Darcy, and Schmidt (Citation2019). In summary, 5 g of sediment of each sample were placed in sterile glass tubes. The tubes were left open to dry at 60°C in an oven for 48 hours. Water content was measured as the percentage difference between the wet and dry sample.
Other geochemical measurements were performed on freeze-dried and crushed aliquots of the three samples. Total nitrogen (TN) content measurements were measured by the Arikaree Laboratory at the University of Colorado Boulder using a Shimadzu TOC-L/TNM-L TOC/TN analyzer. Total organic carbon (TOC) content was measured by Activation Laboratories Ltd.. Lithogeochemistry analysis was also performed by Activation Laboratories Ltd. Mineral composition of the samples was performed via lithium borate fusion/inductively couple plasma-optical emission spectroscopy and the composition of trace elements was measured via lithium borate fusion/inductively coupled plasma-mass spectrometry.
Results
Environmental properties of South Col sediment
The properties of these sediments create a unique lithic environment for microbial communities, most notably defined by its extremely high elevation (>7,900 m). Unsurprising, given the high elevation, the sediments were dry (water content <1.0 percent for all samples, mean = 0.31 percent water content) and contained low measured concentrations of organic carbon (average = 0.06 percent g/g) and no detectable nitrogen. Sediment pH was an average of 9.4 across the three samples, suggesting a basic environment. Though DNA extraction and culturing were only performed on the smallest sediment fragments (<1 mm diameter), all three samples were made up of fragments of variable grain side, with the largest sediment fragments ~2-5 cm in diameter () showing the rocky nature of the substrate.
Additional analysis of the three samples that were collected from the South Col show that the mineralogical composition (weight percentage) was on average 59.1 percent SiO2, 15.7 percent Al2O3, 6.4 percent Fe2O3, 5.2 percent CaO, 2.6 percent MgO, 3.5 percent K2O, and 2.1 percent Na2O, with P2O5, TiO2, and MnO each making up <1 percent (). These “Everest series greenschists,” which were described by Searle (Citation1999) in more detail, are a type of metamorphic rock formed under the lowest temperatures and pressures experienced by the Himalayan region. These rocks also contained several trace elements, the most abundant of which included barium (average = 596 ppm), strontium (average = 393 ppm), zirconium (average = 221 ppm), rubidium (average = 181 ppm), zinc (average = 100 ppm), cerium (average = 98 ppm), chromium (average = 87 ppm), vanadium (average = 80 ppm), and lanthanum (average = 51 ppm). For more details on mineralogical and elemental composition of samples see Dataset S1.
Table 1. Minerological composition of the three South Col sediments.
Cultivation-independent marker gene sequencing
All three samples yielded enough PCR-amplifiable DNA to characterize the bacterial and eukaryotic taxa using marker gene sequencing (see Dataset S2). 16S rRNA gene reads averaged 4,699 reads/sample (3,101–6,708), ITS region reads averaged 9,556 reads/sample (661–25,284 reads), and 18S rRNA gene reads averaged 14,450 reads/sample (480–28,173 reads). As expected, the number of taxa identified per sample (microbial richness) was low, with average richness of twenty-three 16S ASVs (fifteen to thirty-three), six ITS ASVs (two to fourteen), and five 18S ASVs (one to twelve). No archaeal sequences were identified using these methods.
The thirty-six bacterial ASVs identified by cultivation-independent marker gene sequencing included taxa of the phyla Firmicutes (sixteen ASVs), Proteobacteria (sixteen ASVs), Actinobacteria (ten ASVs), Bacteroidetes (four ASV), Chloroflexi (two ASVs), Acidobacteria (two ASVs), Verrucomicrobia (two ASVs), Planctomycetes (one ASV), Abditibacteriota (one ASV), and Gemmatimonadota (one ASV). In total, thirty-one families were represented, the most common of which were Bacillaceae (six ASVs), Burkholderiaceae (six ASVs), Carnobacteriaceae (three ASVs), Corynebacteriaceae (three ASVs), Chitinophagaceae (one ASV), and Paenibacillaceae (three ASVs). All other families were represented by only one ASV ().
All sixteen of the fungal ASVs identified through the next-generation marker gene sequencing effort were of either the phyla Ascomycota (nine ASVs) or Basidiomycota (seven ASVs). The most common fungal family was Filobasidiaceae (six ASVs), all of which were identified to the genus Naganishia. The seven other fungal families included Nectriaceae (three ASVs), Pleosporaceae (two ASVs), Teratosphaeriaceae (one ASV), Sporormiaceae (one ASV), Cladosporiaceae (one ASV), Trichomeriaceae (one ASV), and one Basidiomycota ASV not identified to a fungal family ().
The 18S rRNA gene sequencing effort identified fourteen ASVs. Most of these sequences (ten ASVs) were fungal () and 77 percent of all reads recovered from the three samples were one ASV that had 100 percent identity over the very short read of 129 bp with members of the genera Piskurozyma and Naganishia. Nonfungal ASVs were identified as being of the genera Rosulabryum (one ASV) and Crassostrea (one ASV), the order Ostreoida (one ASV), and an unidentified ASV from the phylum Cercozoa (one ASV; ).
Culture dependent methods
Growth of at least one colony occurred on all plates that were inoculated with samples. No growth occurred on any of the fourteen control plates. One hundred eighty-six bacterial colonies and twenty-nine fungal colonies were streaked for isolation after the four-week incubation. Sanger sequencing revealed that a total of thirty-eight unique bacterial isolates and six fungal isolates were grown from the three sediment samples (Dataset S3).
All bacterial cultures grown were identified as being of the phyla Actinobacteria (twenty-seven isolates), Firmicutes (six isolates), or Proteobacteria (five isolates). Eleven different families were identified, including Micrococcaceae (eight isolates), Propionibacteriaceae (seven), Microbacteriaceae (five), Bacillaceae (four), Oxalobacteraceae (three), Nocardioidaceae (three), Planococcaceae (two), Intrasporangiaceae (two), Geodermatophilaceae (two), Azospirillaceae (one), and Comamonadaceae (one; ).
All of the fungal isolates were identified as being of the cold-adapted yeast genus Naganishia (), including three isolates most closely related to N. vishniacii and two other isolates most closely related to N. albidosimilis and N. adeliensis.
Discussion
The environmental properties of the South Col sediments we analyzed in this study are not unique to Everest. Many high alpine environments experience limited water and nutrients and high pH, among other elevation-influenced properties (Merino et al. Citation2019), and still have diverse microbial communities (D. Singh et al. Citation2012; Yuan et al. Citation2014; Vimercati, Darcy, and Schmidt Citation2019). Our three samples, however, are notable for the extremely high elevation from which they were collected (>7,900 m). There are only fifteen mountains in the world, including Mt. Everest, with peaks higher in elevation than the South Col. To our knowledge, these samples represent the highest elevation sediment environment to be explored for microorganisms using cultivation-independent next-generation sequencing methods. Organisms found at such high elevations, if any, must be able to cope with extremely high insolation (Matthews et al. Citation2020), cold temperatures, low atmospheric pressures, and low oxygen availability, among other corresponding environmental conditions (Merino et al. Citation2019).
Though the three samples South Col sediment samples were collected using aseptic techniques and sterile sampling material and remained sealed from the moment of collection until they were processed, the samples did not remain frozen through the entire shipping process. For this reason, our analyses focused on the diversity of organisms identified in the samples and not on their abundance.
Taxonomic diversity of bacteria
Both the Illumina marker gene sequencing and culture-based identifications yielded similar microbial diversity in the South Col sediments, though the next-generation sequencing approach identified a greater diversity of microorganisms than the culturing study (ten phyla versus three phyla; ). Both techniques revealed that the most common bacterial phyla were Firmicutes, Proteobacteria, and Actinobacteria. At finer taxonomic levels, Illumina sequencing identified a greater diversity of Firmicutes, whereas the culturing methods identified a greater diversity of Actinobacteria, though both methods identified many of the same taxa (e.g., Geodermatophilaceae, Nocardioidaceae). These differences are not surprising, because culturing methods are inherently selective. Many of the media we used for our study were designed for culturing of organisms in cold soil environments, where Actinobacteria often are the dominant taxa (Babalola et al. Citation2009; D. J. Smith et al. Citation2012; Dragone et al. Citation2021).
Both methods captured a multitude of taxa with closely related sequences that have been retrieved from other cold, dry, lithic environments. The dominant phyla identified in our South Col sediments were Actinobacteria, Firmicutes, and Proteobacteria, taxonomic groups that often dominate microbial communities in Antarctic soils (Cowan et al. Citation2014). We see few Acidobacteria and Bacteroidetes, phyla that are common to Arctic tundra (Nemergut et al. Citation2005). At a finer taxonomic resolution, the families Nocardioidaceae, Planococcaceae, Oxalobacteraceae, Geodermatophilaceae, and Chitinophagaceae have closely related sequences that have previously been retrieved from the Transantarctic Mountains and the McMurdo Dry Valleys (J. J. Smith et al. Citation2006; Dragone et al. Citation2021, Citation2022), as well as from cryoconite holes on Antarctic glaciers (Sommers et al. Citation2018). We also see taxa that have been identified in other high alpine sediment environments around the world, including near the top of Kilimanjaro (Vimercati, Darcy, and Schmidt Citation2019) and the Puna de Atacama Volcanic Zone (Solon et al. Citation2018). Other genera, like Auraticoccus and Kribbella, have been found on rocks (Cheema et al. Citation2020), in deserts (Saygin et al. Citation2019), and on the walls of stone monuments and catacombs (Alonso-Vega et al. Citation2011).
Of special note is our identification of members of the family Geodermatophilaceae, including Geodermatophilus obscurus, which was originally isolated from a sediment sample taken at 8,400 m.a.s.l. by the American Everest expedition of 1963 and later described by Ishiguro and Fletcher (Citation1975). Though G. obscurus was not identified in our samples, we did identify taxa of the related genus Modestobacter (), with one cultured isolate identified as being most closely related to M. roseus (Qin et al. Citation2013). Organisms of the genus Modestobacter are often identified in other cold, dry soils and sediments (Mevs et al. Citation2000; Busarakam et al. Citation2016; Golinska et al. Citation2020) and, like the closely related genus Geodermatophilus (Montero-Calasanz et al. Citation2017), have recently been found to have many psychrophilic and oligotrophic adaptations (Golinska et al. Citation2020; Tarlachkov et al. Citation2020). Genomic analysis of Modestobacter taxa (see Golinska et al. [Citation2020] for detailed summary) has found genes related to cold-shock regulation (e.g., Csp protein family; Essoussi et al. Citation2010), pathways related to desiccation tolerance via the uptake of trehalose (Reina-Bueno et al. Citation2012), and multiple copies of the uvrD genes that have previously been linked to increased ultraviolet (UV) tolerance (Normand et al. Citation2012). Of additional note is the presence of genes that allow for the use of carbon monoxide and other trace gases as a source of carbon and energy (e.g., coxD, coxE, coxG; Lorite et al. Citation2000). These potential functional attributes suggest adaptations that might allow for survival in environments like the South Col.
Figure 3. Modestobacter and Geodermatophilus phylogenetic tree. Phylogenetic relationships displayed are based on partial 16S rRNA gene sequences of at least 800 bp in length. The twenty sequences represented in this tree include two isolates recovered during the culturing study, eight representatives of the genus Modestobacter, and ten representatives of the genus Geodermatophilus, including G. obscurus, the organism isolated from a sediment sample taken at 8,400 m.a.s.l. by the American Everest expedition of 1963. The two cultured Geodermatophilaceae isolates were most closely related to M. excelsi and M. altitudinus, taxa most often associated with cold, dry, high alpine environments like soils of the Atacama Desert and Antarctica. The tree is rooted by Blastococcus deserti, a representative of another Geodermatophilaceae family.
Along with Geodermatophilaceae and the other cold-associated organisms described previously, the diversity of bacteria we identified in the South Col samples included more cosmopolitan species that are not characteristic of high alpine or cold soil/sediment environments. Many taxa, including those of the families Paenibacillaceae and Bacillaceae, are endospore-forming species (Montes et al. Citation2004; Mandic-Mulec, Stefanic, and van Elsas Citation2015). Bradyrhizobium is a nitrogen-fixing genus that is most often found in symbiotic association with legumes, though it is also a common soil taxon (Klepa et al. Citation2021). Alloiococcus, Staphylococcus, and Streptococcus are genera that include human pathogens (Aguirre and Collins Citation1992; Becher et al. Citation2009; Palmieri, Varaldo, and Facinelli Citation2011), though the latter two often are often found in certain soil environments (Delgado-Baquerizo et al. Citation2018).
Taxonomic diversity of eukaryotes
Both the Illumina marker gene sequencing of the ITS region and culture-based identifications yielded similar microbial diversity in the South Col and showed that most of the diversity of these samples was made up of the fungal family Filobasidiaceae. More specifically, the majority of the fungal ASVs identified in the marker gene sequencing and all of the fungi that grew in culture were of the genus Naganishia. This is not surprising, because Naganishia is a genus of cold-adapted poly-extremophile yeasts that are commonly found in cold and dry ecosystems, including in Antarctica (Duarte et al. Citation2018; Nizovoy et al. Citation2021), on Kilimanjaro (Vimercati, Darcy, and Schmidt Citation2019), and in high-elevation sediments of the hyperarid Puna de Atacama (Pulschen et al. Citation2015; S. K. Schmidt et al. Citation2018). In these other environments, Naganishia is thought to survive by periodically coming out of dormancy during brief periods of favorable conditions (S. K. Schmidt et al. Citation2017). This genus of cold-adapted poly-extremophilic yeasts is commonly found in a variety of extreme cold, dry environments and is adapted to be resistant to high doses of UV radiation, can grow at temperatures below 0°C, and can survive and even grow during extreme daily freeze–thaw cycles (Vimercati et al. Citation2016; S. K. Schmidt et al. Citation2017).
The non-Naganishia fungal taxa identified in the samples are mostly cosmopolitan, many of which are commonly found in association with plants (Gibberella, Alternaria, Cladosporium; Zanne et al. Citation2020). One taxon, Knufia, belongs to a genus that includes extremotolerant fungi often found on rocks (Breitenbach et al. Citation2018).
The 18S rRNA gene sequencing results generally supported the patterns described from the ITS data. Though many of the other ASVs were not classified past the phylum level, this sequencing effort revealed a similar mix of cosmopolitan taxa and extremotolerant taxa like that revealed by the ITS region sequencing. The genus Sporisorium, for example, includes plant pathogens and mutualists (Ghareeb et al. Citation2011) and the genus Mrakia includes cryotolerant and xerophilic species (Thomas-Hall et al. Citation2010; Turchetti et al. Citation2011). Most notable, 77 percent of all 18S rRNA gene reads were one ASV that was a 100 percent match to several different yeasts in the Filoblasidiales, including numerous Naganishia like N. vishniacii and the genus Piskurozyma, a fungus closely related to Naganishia. Given the very short reads from the Illumina sequencing run (129 bp), we cannot definitively assign a specific taxonomy to this ASV at this time, but given results from the ITS and long-read 18S data from cultures, it is very likely that this ASV is a member of the genus Naganishia.
Implications for microbial survival in extremely high-altitude environments
Our results add to previous work describing the microbial communities on Mount Everest and in the Himalayas (Freeman et al. Citation2009; L. Zhang et al. Citation2009; S. K. Schmidt et al. Citation2011). Previous studies have found that Himalayan microbial communities are dominated by psychrophilic fungi at elevations from 3,000 to 5,300 m (Petrovič, Gunde-Cimerman, and Zalar Citation2000; Margesin and Miteva Citation2011). At higher elevations between 4,000 m and 6,500 m, communities of ammonia oxidizers were dominated by archaeal taxa below 5,400 m and bacteria taxa above (L. Zhang et al. Citation2009), whereas even algae and chytrids were found in soils wet by snowmelt above 5,500 m (Freeman et al. Citation2009; S. K. Schmidt et al. Citation2011). Across all studies, microbial diversity decreased with increasing elevation. Our three sediments from the South Col contained extremely low microbial diversity, much more like the microbial diversity found in the dry tephra “soils” on the volcanic peaks of the inner Atacama Desert (Costello et al. Citation2009; S. K. Schmidt et al. Citation2018; Solon et al. Citation2018) than the diversity found at lower elevations on the mountain. This is not surprising given that microorganisms at such an extreme elevation would have to be able to cope with correspondingly high UV radiation, extreme cold temperatures, low atmospheric pressures, and lower oxygen availability, among other corresponding environmental conditions (Merino et al. Citation2019).
We expect that most of the organisms we identified may have been seeded by eolian transport from other less extreme terrestrial surfaces. Wind-driven transport of microorganisms and DNA is well described in a variety of environments (Aalismail et al. Citation2019; Souza et al. Citation2019) and has been shown to lead to the detection of organisms in environments where they cannot grow (Archer et al. Citation2019; Maki et al. Citation2019). This includes on Mount Everest, where Liu et al. (Citation2007) found many cosmopolitan bacterial taxa in snow at 8,000 m. Our identification of cosmopolitan taxa and foreign plant and animal DNA (e.g., Rosulabryum and Crassostrea DNA; ) in these samples supports this theory. In fact, the most dominant prokaryotic phyla (Proteobacteria, Firmicutes, and Actinobacteria) and fungal phyla (Ascomycota) identified in our samples are the same phyla that typically dominate microbial sequences recovered by bioaerosol studies (Aalismail et al. Citation2019; Archer et al. Citation2019; Souza et al. Citation2019; Ruiz-Gil et al. Citation2020) and include some of the taxa Liu et al. (Citation2007) suggested could not be active at such high elevations on Everest. Even the most highly cold-adapted genera (e.g., Modestobacter, Naganishia, or Ishiguro’s Geodermatophilus as described in Ishiguro and Fletcher [Citation1975] and Ishiguro and Wolfe [Citation1970]) are likely to have been seeded from lower elevation source populations. Representatives of these genera have been detected, and even isolated, from air samples collected around the world (J. J. Smith et al. Citation2006; Amato et al. Citation2007; S. K. Schmidt et al. Citation2017).
The prospect should also be raised that all, or most, organisms found at the South Col in this study rarely, if ever, grow there. The organisms we isolated and grew likely only came out of dormancy due to the less extreme, consistent conditions of the laboratory setting. Though the intense insolation does indicate that melt occurs at the South Col, consistent with reports from climbers, it remains unclear whether this free water would be enough to support brief periods of microbial growth, given that air temperatures rarely rise above −10°C at such high elevations on Mount Everest (Matthews et al. Citation2020; Potocki et al. Citation2022). However, previous work at high elevations in the Atacama has shown that even when air temperatures are below −10°C (at 2 m height), surface soil temperatures can reach +5°C due to the intense insolation (D. Schmidt Citation1999; Schubert Citation2014). Organisms like Naganishia sp. may be able to take advantage of even these short periods of water availability beause they can grow during repeated freeze–thaw cycles and at constant temperatures as low as −6°C (Vimercati et al. Citation2016). Obviously, it would be hard to test this idea in situ, but more work on the unique microbes isolated in the present study could illuminate the potential for organisms to function during brief thaw periods at extreme elevations. It also very likely that the South Col and other extremely high-elevation environments may just be deep-freeze collection points for deposited organisms (either from the atmosphere or by transiting humans). Given the challenge, and danger, of performing in situ tests at such extreme high-elevation sites, we cannot say whether the brief periods when free water would be available are enough to support microbial growth. Perhaps extended monitoring by the weather station installed at this site where these samples were collected will shed some light on this mystery in the future (Matthews et al. Citation2020; Potocki et al. Citation2022).
Though our culturing study could not re-create the extreme conditions at the South Col, our results show that many of the organisms, including the most ubiquitous fungal and bacterial taxa identified, are in fact viable and could be grown at 4°C. Though the average air temperature from 1991 to 2022 at the South Col was −22.6°C, well below our culturing conditions, air temperatures in the Mount Everest region have been increasing at a rate of ~0.33°C per decade (Kang et al. Citation2022), and a record high of −1.4°C was measured at the South Col in July 2022 (National Geographic Society Citation2022). Organisms that are not active in situ now may become active in the future if the current rate of warming continues (described in Kang et al. Citation2022).
Evidence of human-associated contamination of Mount Everest
The microorganisms identified in these South Col sediments included taxa that are often associated with human contamination. Many of the sequences recovered could be deposited from mountaineers through coughing, nasal discharge, and associated sputum (Grice and Segre Citation2011; Wade Citation2013; Ibironke et al. Citation2020). Staphylococcus, for example, is one of the most common skin and nose bacteria, and Streptococcus is the dominant genus in the human mouth (Otto Citation2010; Gevers et al. Citation2012; Ursell et al. Citation2013; Escapa et al. Citation2018). Though these genera are widespread in nature and are not always associated with humans (e.g., Ko et al. Citation2011; Zhao et al. Citation2015), the sequences that were identified as being most closely related to our taxa based on analysis using blastn are human associated. For example, the Staphylococcus sequence identified in our samples shares a 100 percent identity with the skin colonizers S. hominis and S. epidermis (Nagase et al. Citation2002; Otto Citation2009), and the Streptococcus sequence shares a 100 percent identity with oral cavity colonizer S. oralis (Kennedy et al. Citation2000). Because none of these human-associated taxa were identified in our extraction blanks and no-template PCR controls and none grew on our control plates in our culturing study, it remains likely that these organisms were from the collected sediments.
Mount Everest is heavily trafficked by climbers and guides each year, prompting concerns that humans have irrevocably changed the high alpine environment. Our study provides evidence that climbing activity may increase the transport of human-associated microorganisms onto the mountain’s highest surfaces. However, we expect that at such high elevations these microorganisms are not active. We predict that if we sampled in the more human-utilized areas on the mountain we may find even more microbial evidence of human impact on the environment, as has been well described at lower elevations in the Himalayas (Rashid and Romshoo Citation2013; Ahmad et al. Citation2021; Rather et al. Citation2022) and around the world (Sinton, Finlay, and Hannah Citation1998; Joergensen and Emmerling Citation2006; Harwood et al. Citation2014).
Conclusions
The South Col of Everest represents a unique surface environment, most notably for being >7,900 m. Despite the extreme elevation and corresponding high UV radiation, cold temperatures, low atmospheric pressures, lower oxygen availability, we have identified bacterial and fungal DNA sequences and have grown bacterial and fungal cultures from sediments collected from the South Col. The mix of cold-adapted and cosmopolitan species identified using our methods suggests that most of these organisms are inactive in situ and may have been seeded by eolian transport from other less extreme terrestrial surfaces. Others may have been transported to the South Col by the activity of climbers. Indeed, our data suggest that the South Col and other extremely high-elevation environments may be deep-freeze collection points for deposited organisms, including human-borne contaminants that may never leave once they arrive.
Data and materials availability
All data used in this study are available in the main text or the supplementary material with the raw sequence data available in the National Center for Biotechnology Information (NCBI) Sequence Read Database, project accession number PRJNA882470, BioSample accessions SAMN30937384, SAMN30937385, SAMN30937386.
Supplemental Material
Download Zip (71.2 KB)Acknowledgments
The 2019 National Geographic and Rolex Everest Expedition was organized by National Geographic Society and supported by Rolex as part of its Perpetual Planet initiative. This research was conducted in partnership with Tribhuvan University, with approval from all relevant agencies of the Government of Nepal (Letter numbers 075/076, 576, and 2351, issued by the Department of National Parks and Wildlife Conservation of the Government of Nepal). We also thank the communities of the Khumbu region, our field support team, and Shangri-La Nepal Trek. We also thank Jessica Henley, Matthew Gebert, Caihong Vanderburgh, and Claire Mastrangelo for help with the laboratory work; the University of Colorado Boulder Next Generation Sequencing Facility and GENEWIZ for help with the sequencing effort; and The Arikaree Laboratory at CU Boulder and Activation Laboratories for assistance with the geochemical analyses. Finally, we thank Sam Guilford for his help creating the map in .
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15230430.2023.2164999.
Additional information
Funding
References
- Aalismail, N. A., D. K. Ngugi, R. Díaz-Rúa, I. Alam, M. Cusack, and C. M. Duarte. 2019. Functional metagenomic analysis of dust-associated microbiomes above the Red Sea. Scientific Reports 9:13741. doi:10.1038/s41598-019-50194-0.
- Aguirre, M., and M. D. Collins. 1992. Phylogenetic analysis of Alloiococcus otitis gen. nov., sp. nov., an organism from human middle ear fluid. International Journal of Systematic and Evolutionary Microbiology 42:79–17. doi:10.1099/00207713-42-1-79.
- Ahmad, T., G. Gupta, A. Sharma, B. Kaur, M. A. El-Sheikh, and M. N. Alyemeni. 2021. Metagenomic analysis exploring taxonomic and functional diversity of bacterial communities of a Himalayan urban freshwater lake. PLOS ONE 16:e0248116. doi:10.1371/journal.pone.0248116.
- Alonso-Vega, P., L. Carro, E. Martínez-Molina, and M. E. Trujillo. 2011. Auraticoccus monumenti gen. nov., sp. nov., an Actinomycete isolated from a deteriorated sandstone monument. International Journal of Systematic and Evolutionary Microbiology 61:1098–103. doi:10.1099/ijs.0.024257-0.
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. Journal of Molecular Biology 215:403–10. doi:10.1016/S0022-2836(05)80360-2.
- Amato, P., M. Parazols, M. Sancelme, P. Laj, G. Mailhot, and A. Delort. 2007. Microorganisms isolated from the water phase of tropospheric clouds at the Puy de Dôme: Major groups and growth abilities at low temperatures. FEMS Microbiology Ecology 59:242–54. doi:10.1111/j.1574-6941.2006.00199.x.
- Archer, S. D. J., K. C. Lee, T. Caruso, T. Maki, C. K. Lee, S. C. Cary, D. A. Cowan, F. T. Maestre, and S. B. Pointing. 2019. Airborne microbial transport limitation to isolated Antarctic soil habitats. Nature Microbiology 4:925–32. doi:10.1038/s41564-019-0370-4.
- Atlas, R. M. 2004. Handbook of microbiological media. Boca Raton, FL: CRC press.
- Babalola, O. O., B. M. Kirby, M. Le Roes-Hill, A. E. Cook, S. C. Cary, S. G. Burton, and D. A. Cowan. 2009. Phylogenetic analysis of Actinobacterial populations associated with Antarctic Dry Valley mineral soils. Environmental Microbiology 11:566–76. doi:10.1111/j.1462-2920.2008.01809.x.
- Becher, D., K. Hempel, S. Sievers, D. Zühlke, J. Pané-Farré, A. Otto, S. Fuchs, D. Albrecht, J. Bernhardt, S. Engelmann, et al. 2009. A proteomic view of an important human pathogen – Towards the quantification of the entire Staphylococcus aureus proteome. PLOS ONE 4:e8176. doi:10.1371/journal.pone.0008176.
- Bellemain, E., T. Carlsen, C. Brochmann, E. Coissac, P. Taberlet, and H. Kauserud. 2010. ITS as an environmental DNA barcode for fungi: an in silico approach reveals potential PCR biases. BMC Microbiology 10:189. doi:10.1186/1471-2180-10-189.
- Breitenbach, R., D. Silbernagl, J. Toepel, H. Sturm, W. J. Broughton, G. L. Sassaki, and A. A. Gorbushina. 2018. Corrosive extracellular polysaccharides of the rock-inhabiting model fungus Knufia petricola. Extremophiles 22:165–75. doi:10.1007/s00792-017-0984-5.
- Bryant, J. A., C. Lamanna, H. Morlon, A. J. Kerkhoff, B. J. Enquist, and J. L. Green. 2008. Microbes on mountainsides: Contrasting elevational patterns of bacterial and plant diversity. Proceedings of the National Academy of Sciences 105:11505–11. doi:10.1073/pnas.0801920105.
- Busarakam, K., A. T. Bull, M. E. Trujillo, R. Riesco, V. Sangal, G. P. van Wezel, and M. Goodfellow. 2016. Modestobacter caceresii sp. nov., novel Actinobacteria with an insight into their adaptive mechanisms for survival in extreme hyper-arid Atacama Desert soils. Systematic and Applied Microbiology 39:243–51. doi:10.1016/j.syapm.2016.03.007.
- Callahan, B. J., P. J. McMurdie, M. J. Rosen, A. W. Han, A. A. Johnson, and S. P. Holmes. 2016. DADA2: High-resolution sample inference from Illumina amplicon data. Nature Methods 13:581–83. doi:10.1038/nmeth.3869.
- Caporaso, J. G., C. L. Lauber, W. A. Walters, D. Berg-Lyons, J. Huntley, N. Fierer, S. M. Owens, J. Betley, L. Fraser, M. Bauer, et al. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. The ISME Journal 6:1621–24. doi:10.1038/ismej.2012.8.
- Cheema, M. T., J. Ye, F. Li, Q. Lu, M. Abbas, I. Sajid, D. Huang, S. Liu, and C. Sun. 2020. Auraticoccus cholistanensis sp. nov., an Actinomycete isolated from soil of the Cholistan Desert, and emended description of the genus Auraticoccus. International Journal of Systematic and Evolutionary Microbiology 70:3179–85. doi:10.1099/ijsem.0.004152.
- Costello, E. K., S. R. P. Halloy, S. C. Reed, P. Sowell, and S. K. Schmidt. 2009. Fumarole-supported islands of biodiversity within a hyperarid, high-elevation landscape on Socompa Volcano, Puna de Atacama, Andes. Applied and Environmental Microbiology 75:735–47. doi:10.1128/AEM.01469-08.
- Cowan, D. A., T. P. Makhalanyane, P. G. Dennis, and D. W. Hopkins. 2014. Microbial ecology and biogeochemistry of continental Antarctic soils. Frontiers in Microbiology 5:154. doi:10.3389/fmicb.2014.00154.
- Delgado-Baquerizo, M., A. M. Oliverio, T. E. Brewer, A. Benavent-González, D. J. Eldridge, R. D. Bardgett, F. T. Maestre, B. K. Singh, and N. Fierer. 2018. A global atlas of the dominant bacteria found in soil. Science 359:320–25. doi:10.1126/science.aap9516.
- Dragone, N. B., M. A. Diaz, I. D. Hogg, W. B. Lyons, W. A. Jackson, D. H. Wall, B. J. Adams, and N. Fierer. 2021. Exploring the boundaries of microbial habitability in soil. Journal of Geophysical Research: Biogeosciences 126:e2020JG006052. doi:10.1029/2020JG006052.
- Dragone, N. B., J. B. Henley, H. Holland-Moritz, M. Diaz, I. D. Hogg, W. B. Lyons, D. H. Wall, B. J. Adams, and N. Fierer. 2022. Elevational constraints on the composition and genomic attributes of soil microbial communities in Antarctica. mSystems 7:e01330–21. doi:10.1128/msystems.01330-21.
- Duarte, A. W. F., J. A. Dos Santos, M. V. Vianna, J. M. F. Vieira, V. H. Mallagutti, F. J. Inforsato, L. C. P. Wentzel, L. D. Lario, A. Rodrigues, F. C. Pagnocca, et al. 2018. Cold-adapted enzymes produced by fungi from terrestrial and marine Antarctic environments. Critical Reviews in Biotechnology 38:600–19. doi:10.1080/07388551.2017.1379468.
- Dunitz, M. I., J. M. Lang, G. Jospin, A. E. Darling, J. A. Eisen, and D. A. Coil. 2015. Swabs to genomes: A comprehensive workflow. PeerJ 3:e960. doi:10.7717/peerj.960.
- Edgar, R. C. 2004. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32:1792–97. doi:10.1093/nar/gkh340.
- Egan, S., S. Kjelleberg, and C. Holmström. 2015. Pseudoalteromonas ulvae sp. nov., a bacterium with antifouling activities isolated from the surface of a marine alga. International Journal of Systematic and Evolutionary Microbiology 51:1499–504. doi:10.1099/00207713-51-4-1499.
- Escapa, I. F., T. Chen, Y. Huang, P. Gajare, F. E. Dewhirst, and K. P. Lemon. 2018. New insights into human nostril microbiome from the Expanded Human Oral Microbiome Database (EHOMD): A resource for the microbiome of the human aerodigestive tract. Msystems 3:e00187–18. doi:10.1128/mSystems.00187-18.
- Essoussi, I., F. Ghodhbane-Gtari, H. Amairi, H. Sghaier, A. Jaouani, L. Brusetti, D. Daffonchio, A. Boudabous, and M. Gtari. 2010. Esterase as an enzymatic signature of Geodermatophilaceae adaptability to Sahara Desert stones and monuments. Journal of Applied Microbiology 108:1723–32. doi:10.1111/j.1365-2672.2009.04580.x.
- Freeman, K. R., A. P. Martin, D. Karki, R. C. Lynch, M. S. Mitter, A. F. Meyer, J. E. Longcore, D. R. Simmons, and S. K. Schmidt. 2009. Evidence that chytrids dominate fungal communities in high-elevation soils. Proceedings of the National Academy of Sciences 106:18315–20. doi:10.1073/pnas.0907303106.
- Gevers, D., R. Knight, J. F. Petrosino, K. Huang, A. L. McGuire, B. W. Birren, K. E. Nelson, O. White, B. A. Methé, and C. Huttenhower. 2012. The Human Microbiome Project: A community resource for the healthy human microbiome. PLOS Biology 10:e1001377. doi:10.1371/journal.pbio.1001377.
- Ghareeb, H., A. Becker, T. Iven, I. Feussner, and J. Schirawski. 2011. Sporisorium reilianum infection changes inflorescence and branching architectures of maize. Plant Physiology 156:2037–52. doi:10.1104/pp.111.179499.
- Golinska, P., M. Montero-Calasanz, M. Świecimska, A. Yaramis, J. M. Igual, A. T. Bull, and M. Goodfellow. 2020. Modestobacter excelsi sp. nov., a novel Actinobacterium isolated from a high altitude Atacama Desert soil. Systematic and Applied Microbiology 43:126051. doi:10.1016/j.syapm.2019.126051.
- Grice, E. A., and J. A. Segre. 2011. The skin microbiome. Nature Reviews. Microbiology 9:244–53. doi:10.1038/nrmicro2537.
- Harwood, V., C. Staley, B. D. Badgley, K. Borges, and A. Korajkic. 2014. Microbial source tracking markers for detection of fecal contamination in environmental waters: Relationships between pathogens and human health outcomes. FEMS Microbiology Reviews 38:1–40. doi:10.1111/1574-6976.12031.
- Hezbri, K., F. Ghodhbane-Gtari, M. Montero-Calasanz, I. Nouioui, M. Rohde, C. Spröer, P. Schumann, H. Klenk, and M. Gtari. 2016. Geodermatophilus pulveris sp. nov., a gamma-radiation-resistant Actinobacterium isolated from the Sahara Desert. International Journal of Systematic and Evolutionary Microbiology 66:3828–34. doi:10.1099/ijsem.0.001272.
- Ibironke, O., L. R. McGuinness, S. Lu, Y. Wang, S. Hussain, C. P. Weisel, and L. J. Kerkhof. 2020. Species-level evaluation of the human respiratory microbiome. GigaScience 9:giaa038. doi:10.1093/gigascience/giaa038.
- Ishiguro, E. E. 1969. The microbiology of Mount Everest soils. PhD Diss., San Francisco State College.
- Ishiguro, E. E., and D. W. Fletcher. 1975. Characterization of Geodermatophilus strains isolated from high altitude Mount Everest soils. Mikrobiologika 12:99–108.
- Ishiguro, E. E., and R. S. Wolfe. 1970. Control of morphogenesis in Geodermatophilus: Ultrastructural studies. Journal of Bacteriology 104:566–80. doi:10.1128/jb.104.1.566-580.1970.
- Joergensen, R. G., and C. Emmerling. 2006. Methods for evaluating human impact on soil microorganisms based on their activity, biomass, and diversity in agricultural soils. Journal of Plant Nutrition and Soil Science 169:295–309. doi:10.1002/jpln.200521941.
- Kang, S., Q. Zhang, Y. Zhang, W. Guo, Z. Ji, M. Shen, S. Wang, X. Wang, L. Tripathee, Y. Liu, et al. 2022. Warming and thawing in the Mt. Everest region: A review of climate and environmental changes. Earth-Science Reviews 225:103911. doi:10.1016/j.earscirev.2021.103911.
- Kennedy, H. F., D. Morrison, M. E. Kaufmann, M. S. Jackson, J. Bagg, B. E. S. Gibson, C. G. Gemmell, and J. R. Michie. 2000. Origins of Staphylococcus epidermidis and Streptococcus oralis causing bacteraemia in a bone marrow transplant patient. Journal of Medical Microbiology 49:367–70. doi:10.1099/0022-1317-49-4-367.
- King, A. J., K. R. Freeman, K. F. McCormick, R. C. Lynch, C. Lozupone, R. Knight, and S. K. Schmidt. 2010. Biogeography and habitat modelling of high-alpine bacteria. Nature Communications 1:53. doi:10.1038/ncomms1055.
- Klepa, M. S., L. C. F. Helene, G. O’Hara, and M. Hungria. 2021. Bradyrhizobium agreste sp. nov., Bradyrhizobium glycinis sp. nov. and Bradyrhizobium diversitatis sp. Nov., isolated from a biodiversity hotspot of the genus Glycine in Western Australia. International Journal of Systematic and Evolutionary Microbiology 71:004742. doi:10.1099/ijsem.0.004742.
- Ko, K. S., S. Lim, S. Jung, J. M. Yoon, J. Y. Choi, and J. Song. 2011. Sequence type 72 meticillin-resistant Staphylococcus aureus isolates from humans, raw meat and soil in South Korea. Journal of Medical Microbiology 60:442–45. doi:10.1099/jmm.0.026484-0.
- Leff, J. 2016. mctoolsR. v.1.1. GitHub. https://github.com/leffj/mctoolsr/
- Letunic, I., and P. Bork. 2016. Interactive tree of life (ITOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Research 44:W242–W245. doi:10.1093/nar/gkw290.
- Liu, Y. Q., T. D. Yao, S. C. Kang, N. Z. Jiao, Y. H. Zeng, S. J. Huang, and T. W. Luo. 2007. Microbial community structure in major habitats above 6000 m on Mount Everest. Chinese Science Bulletin 52:2350–57. doi:10.1007/s11434-007-0360-4.
- Lorite, M. J., J. Tachil, J. Sanjuán, O. Meyer, and E. J. Bedmar. 2000. Carbon monoxide dehydrogenase activity in Bradyrhizobium japonicum. Applied and Environmental Microbiology 66:1871–76. doi:10.1128/AEM.66.5.1871-1876.2000.
- Lynch, R. C., A. J. King, M. E. Farías, P. Sowell, C. Vitry, and S. K. Schmidt. 2012. The potential for microbial life in the highest-elevation (>6000 m.a.s.l.) mineral soils of the Atacama Region. Journal of Geophysical Research 117:G02028. doi:10.1029/2012JG001961.
- Maki, T., K. C. Lee, K. Kawai, K. Onishi, C. S. Hong, Y. Kurosaki, M. Shinoda, K. Kai, Y. Iwasaka, S. D. J. Archer, et al. 2019. Aeolian dispersal of bacteria associated with desert dust and anthropogenic particles over continental and oceanic surfaces. Journal of Geophysical Research: Atmospheres 124:5579–88. doi:10.1029/2018JD029597.
- Mandic-Mulec, I., P. Stefanic, and J. D. van Elsas. 2015. Ecology of Bacillaceae. Microbiology Spectrum 3:3–2. doi:10.1128/microbiolspec.TBS-0017-2013.
- Margesin, R., M. Jud, D. Tscherko, and F. Schinner. 2009. Microbial communities and activities in alpine and subalpine soils. FEMS Microbiology Ecology 67:208–18. doi:10.1111/j.1574-6941.2008.00620.x.
- Margesin, R., and V. Miteva. 2011. Diversity and ecology of psychrophilic microorganisms. Research in Microbiology 162:346–61. doi:10.1016/j.resmic.2010.12.004.
- Matthews, T., L. B. Perry, I. Koch, D. Aryal, A. Khadka, D. Shrestha, K. Abernathy, A. C. Elmore, A. Seimon, A. Tait, et al. 2020. Going to extremes: Installing the world’s highest weather stations on Mount Everest. Bulletin of the American Meteorological Society 101:E1870–E1890. doi:10.1175/BAMS-D-19-0198.1.
- Merino, N., H. S. Aronson, D. P. Bojanova, J. Feyhl-Buska, M. L. Wong, S. Zhang, and D. Giovannelli. 2019. Living at the extremes: Extremophiles and the limits of life in a planetary context. Frontiers in Microbiology 10:780. doi:10.3389/fmicb.2019.00780.
- Mevs, U., E. Stackebrandt, P. Schumann, C. A. Gallikowski, and P. Hirsch. 2000. Modestobacter multiseptatus gen. nov., sp. nov., a budding Actinomycete from soils of the Asgard Range (Transantarctic Mountains). International Journal of Systematic and Evolutionary Microbiology 50:337–46. doi:10.1099/00207713-50-1-337.
- Montero-Calasanz, M. C., M. Göker, G. Pötter, M. Rohde, C. Spröer, P. Schumann, A. A. Gorbushina, and H. P. Klenk. 2012. Geodermatophilus arenarius sp. Nov., a xerophilic Actinomycete isolated from Saharan Desert sand in Chad. Extremophiles 16:903–09. doi:10.1007/s00792-012-0486-4.
- Montero-Calasanz, M. C., M. Göker, G. Pötter, M. Rohde, C. Spröer, P. Schumann, A. A. Gorbushina, and H. Klenk. 2013a. Geodermatophilus africanus sp. nov., a halotolerant Actinomycete isolated from Saharan Desert sand. Antonie van Leeuwenhoek 104:207–16. doi:10.1007/s10482-013-9939-8.
- Montero-Calasanz, M. C., M. Göker, G. Pötter, M. Rohde, C. Spröer, P. Schumann, A. A. Gorbushina, and H. P. Klenk. 2013b. Geodermatophilus arenarius sp. Nov., a xerophilic Actinomycete isolated from Saharan Desert sand in Chad. Archives of Microbiology 195:153–59. doi:10.1007/s00203-012-0860-8.
- Montero-Calasanz, M. C., M. Göker, M. Rohde, P. Schumann, G. Pötter, C. Spröer, A. A. Gorbushina, and H. Klenk. 2013. Geodermatophilus siccatus sp. nov., isolated from arid sand of the Saharan Desert in Chad. Antonie van Leeuwenhoek 103:449–56. doi:10.1007/s10482-012-9824-x.
- Montero-Calasanz, M. C., K. Hezbri, M. Göker, H. Sghaier, M. Rohde, C. Spröer, P. Schumann, and H. Klenk. 2015. Description of gamma radiation-resistant Geodermatophilus dictyosporus sp. nov. to accommodate the not validly named Geodermatophilus obscurus subsp. dictyosporus (Luedemann, 1968). Extremophiles 19:77–85. doi:10.1007/s00792-014-0708-z.
- Montero-Calasanz, M. C., B. Hofner, M. Göker, M. Rohde, C. Spröer, K. Hezbri, M. Gtari, P. Schumann, and H. Klenk. 2014. Geodermatophilus poikilotrophi sp. nov.: A multitolerant Actinomycete isolated from dolomitic marble. BioMed Research International 2014:e914767. doi:10.1155/2014/914767.
- Montero-Calasanz, M. C., J. P. Meier-Kolthoff, D. Zhang, A. Yaramis, M. Rohde, T. Woyke, N. C. Kyrpides, P. Schumann, W. Li, and M. Göker. 2017. Genome-scale data call for a taxonomic rearrangement of Geodermatophilaceae. Frontiers in Microbiology 8:2501. doi:10.3389/fmicb.2017.02501.
- Montero-Calasanz, M. C., A. Yaramis, I. Nouioui, J. M. Igual, C. Spröer, J. F. Castro, P. Schumann, H. Klenk, and C. Urzì. 2019. Modestobacter italicus sp. nov., isolated from Carrara marble quarry and emended descriptions of the genus Modestobacter and the species Modestobacter marinus, Modestobacter multiseptatus, Modestobacter roseus and Modestobacter versicolor. International Journal of Systematic and Evolutionary Microbiology 69:1537–45. doi:10.1099/ijsem.0.003282.
- Montes, M. J., E. Mercadé, N. Bozal, and J. Guinea. 2004. Paenibacillus antarcticus sp. nov., a novel psychrotolerant organism from the Antarctic environment. International Journal of Systematic and Evolutionary Microbiology 54:1521–26. doi:10.1099/ijs.0.63078-0.
- Moore, G. W. K., and J. L. Semple. 2004. High Himalayan meteorology: Weather at the South Col of Mount Everest. Geophysical Research Letters 31:L18109. doi:10.1029/2004GL020621.
- Nagase, N., A. Sasaki, K. Yamashita, A. Shimizu, Y. Wakita, S. Kitai, and J. Kawano. 2002. Isolation and species distribution of Staphylococci from animal and human skin. Journal of Veterinary Medical Science 64:245–50. doi:10.1292/jvms.64.245.
- National Geographic Society. 2022. Weather data from Mt. Everest. Online Resource. https://www.nationalgeographic.org/projects/perpetual-planet/everest/weather-data/
- Nemergut, D. R., E. K. Costello, A. F. Meyer, M. Y. Pescador, M. N. Weintraub, and S. K. Schmidt. 2005. Structure and function of alpine and Arctic soil microbial communities. Research in Microbiology 156:775–84. doi:10.1016/j.resmic.2005.03.004.
- Nie, G., H. Ming, S. Li, E. Zhou, J. Cheng, T. Yu, J. Zhang, H. Feng, S. Tang, and W. Li. 2012. Geodermatophilus nigrescens sp. nov., isolated from a dry-hot valley. Antonie van Leeuwenhoek 101:811–17. doi:10.1007/s10482-012-9696-0.
- Nilsson, R. H., K. Larsson, A. F. S. Taylor, J. Bengtsson-Palme, T. S. Jeppesen, D. Schigel, P. Kennedy, K. Picard, F. O. Glöckner, L. Tedersoo, et al. 2019. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Research 47:D259–D264. doi:10.1093/nar/gky1022.
- Nizovoy, P., N. Bellora, S. Haridas, H. Sun, C. Daum, K. Barry, I. V. Grigoriev, D. Libkind, L. B. Connell, and M. Moliné. 2021. Unique genomic traits for cold adaptation in Naganishia vishniacii, a polyextremophile yeast isolated from Antarctica. FEMS Yeast Research 21:foaa056. doi:10.1093/femsyr/foaa056.
- Normand, P., J. Gury, P. Pujic, B. Chouaia, E. Crotti, L. Brusetti, D. Daffonchio, B. Vacherie, V. Barbe, C. Medigue, et al. 2012. Genome sequence of radiation-resistant Modestobacter marinus strain BC501, a representative Actinobacterium that thrives on calcareous stone surfaces. Journal of Bacteriology 2012:4773–74. https://journals.asm.org/doi/abs/10.1128/JB.01029-12
- Otto, M. 2009. Staphylococcus epidermidis—the ‘accidental’ pathogen. Nature Reviews. Microbiology 7:555–67. doi:10.1038/nrmicro2182.
- Otto, M. 2010. Staphylococcus colonization of the skin and antimicrobial peptides. Expert Review of Dermatology 5:183–95. doi:10.1586/edm.10.6.
- Palmieri, C., P. Varaldo, and B. Facinelli. 2011. Streptococcus suis, an emerging drug-resistant animal and human pathogen. Frontiers in Microbiology 2:235. doi:10.3389/fmicb.2011.00235.
- Petrovič, U., N. Gunde-Cimerman, and P. Zalar. 2000. Xerotolerant mycobiota from high altitude Annapurna soil, Nepal. FEMS Microbiology Letters 182:339–42. doi:10.1111/j.1574-6968.2000.tb08918.x.
- Potocki, M., P. A. Mayewski, T. Matthews, L. B. Perry, M. Schwikowski, A. M. Tait, E. Korotkikh, H. Clifford, S. Kang, T. C. Sherpa, et al. 2022. Mt. Everest’s highest glacier is a sentinel for accelerating ice loss. njp Climate and Atmospheric Science 5:1–8. doi:10.1038/s41612-022-00230-0.
- Pulschen, A. A., A. G. Bendia, A. D. Fricker, V. H. Pellizari, D. Galante, and F. Rodrigues. 2017. Isolation of uncultured bacteria from Antarctica using long incubation periods and low nutritional media. Frontiers in Microbiology 8:1346. doi:10.3389/fmicb.2017.01346.
- Pulschen, A. A., F. Rodrigues, R. T. D. Duarte, G. G. Araujo, I. F. Santiago, I. G. Paulino-Lima, C. A. Rosa, M. J. Kato, V. H. Pellizari, and D. Galante. 2015. UV-resistant yeasts isolated from a high-altitude volcanic area on the Atacama Desert as eukaryotic models for astrobiology. MicrobiologyOpen 4:574–488. doi:10.1002/mbo3.262.
- Qin, S., G. Bian, Y. Zhang, K. Xing, C. Cao, C. Liu, C. Dai, W. Li, and J. Jiang. 2013. Modestobacter roseus sp. nov., an endophytic Actinomycete isolated from the coastal halophyte Salicornia europaea linn., and emended description of the genus Modestobacter. International Journal of Systematic and Evolutionary Microbiology 63:2197–202. doi:10.1099/ijs.0.044412-0.
- Quast, C., E. Pruesse, P. Yilmaz, J. Gerken, T. Schweer, P. Yarza, J. Peplies, and F. O. Glöckner. 2013. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Research 41:D590–D596. doi:10.1093/nar/gks1219.
- Rashid, I., and S. A. Romshoo. 2013. Impact of anthropogenic activities on water quality of Lidder River in Kashmir Himalayas. Environmental Monitoring and Assessment 185:4705–19. doi:10.1007/s10661-012-2898-0.
- Rather, R. A., H. Bano, S. A. Padder, T. R. Baba, S. Ara, F. A. Lone, and S. Nazir. 2022. Impact of anthropogenic pressure on physico-chemical characteristics of forest soils of Kashmir Himalaya. Bulletin of Environmental Contamination and Toxicology 2022:1–10. doi:10.1007/s00128-022-03458-x.
- R Core Team. 2017. R: A language and environment for statistical computing. Vienna, AT: R Foundation for Statistical Computing. https://www.R-project.org/
- Reasoner, D. J., and E. E. Geldreich. 1985. A new medium for the enumeration and subculture of bacteria from potable water. Applied and Environmental Microbiology 49:1–7. doi:10.1128/AEM.49.1.1-7.1985.
- Reina-Bueno, M., M. Argandoña, J. J. Nieto, A. Hidalgo-García, F. Iglesias-Guerra, M. J. Delgado, and C. Vargas. 2012. Role of trehalose in heat and desiccation tolerance in the soil Bacterium rhizobium etli. BMC Microbiology 12:207. doi:10.1186/1471-2180-12-207.
- Ruiz-Gil, T., J. J. Acuña, S. Fujiyoshi, D. Tanaka, J. Noda, F. Maruyama, and M. A. Jorquera. 2020. Airborne bacterial communities of outdoor environments and their associated influencing factors. Environment International 145:106156. doi:10.1016/j.envint.2020.106156.
- Saygin, H., H. Ay, K. Guven, and N. Sahin. 2019. Kribbella turkmenica sp. nov., isolated from the Karakum Desert. International Journal of Systematic and Evolutionary Microbiology 69:2533–40. doi:10.1099/ijsem.0.003538.
- Schmidt, D. 1999. Das Extremklima Der Nordchilenischen Hochatacama Unter Besonderer Berücksichtigung Der Höhengradienten. Dresdener Geographische Beiträge 4:1–122.
- Schmidt, S. K., E. M. S. Gendron, K. Vincent, A. J. Solon, P. Sommers, Z. R. Schubert, L. Vimercati, D. L. Porazinska, J. L. Darcy, and P. Sowell. 2018. Life at extreme elevations on Atacama volcanoes: The closest thing to Mars on Earth? Antonie van Leeuwenhoek 111:1389–401. doi:10.1007/s10482-018-1066-0.
- Schmidt, S. K., R. C. Lynch, A. J. King, D. Karki, M. S. Robeson, L. Nagy, M. W. Williams, M. S. Mitter, and K. R. Freeman. 2011. Phylogeography of microbial phototrophs in the dry valleys of the high Himalayas and Antarctica. Proceedings of the Royal Society B: Biological Sciences 278:702–08. doi:10.1098/rspb.2010.1254.
- Schmidt, S. K., L. Vimercati, J. L. Darcy, P. Arán, E. M. S. Gendron, A. J. Solon, D. Porazinska, and C. Dorador. 2017. A Naganishia in high places: Functioning populations or dormant cells from the atmosphere?. Mycology 8:153–63. doi:10.1080/21501203.2017.1344154.
- Schnell, I. B., K. Bohmann, and M. T. P. Gilbert. 2015. Tag jumps illuminated – Reducing sequence-to-sample misidentifications in metabarcoding studies. Molecular Ecology Resources 15:1289–303. doi:10.1111/1755-0998.12402.
- Schubert, Z. R. 2014. Dew formation and water availability at high elevation in the Atacama Desert, Chile. Undergraduate Honors Thesis. Paper 192. University of Colorado, Boulder
- Searle, M. P. 1999. Extensional and compressional faults in the Everest–Lhotse massif, Khumbu Himalaya, Nepal. Journal of the Geological Society 156 (2): 227–240. doi:10.1144/gsjgs.156.2.0227.
- Singh, P., S. M. Singh, and U. Roy. 2016. Taxonomic characterization and the bio-potential of bacteria isolated from glacier ice cores in the High Arctic. Journal of Basic Microbiology 56:275–85. doi:10.1002/jobm.201500298.
- Singh, D., K. Takahashi, M. Kim, J. Chun, and J. M. Adams. 2012. A hump-backed trend in bacterial diversity with elevation on Mount Fuji, Japan. Microbial Ecology 63:429–37. doi:10.1007/s00248-011-9900-1.
- Sinton, L. W., R. K. Finlay, and D. J. Hannah. 1998. Distinguishing human from animal faecal contamination in water: A review. New Zealand Journal of Marine and Freshwater Research 32:323–48. doi:10.1080/00288330.1998.9516828.
- Smith, D. J., D. A. Jaffe, M. N. Birmele, D. W. Griffin, A. C. Schuerger, J. Hee, and M. S. Roberts. 2012. Free tropospheric transport of microorganisms from Asia to North America. Microbial Ecology 64:973–85. doi:10.1007/s00248-012-0088-9.
- Smith, J. J., L. A. Tow, W. Stafford, C. Cary, and D. A. Cowan. 2006. Bacterial diversity in three different Antarctic cold desert mineral soils. Microbial Ecology 51:413–21. doi:10.1007/s00248-006-9022-3.
- Solon, A. J., L. Vimercati, J. L. Darcy, P. Arán, D. Porazinska, C. Dorador, M. E. Farías, and S. K. Schmidt. 2018. Microbial communities of high-elevation fumaroles, penitentes, and dry tephra ‘soils’ of the Puna de Atacama volcanic zone. Microbial Ecology 76:340–51. doi:10.1007/s00248-017-1129-1.
- Sommers, P., J. L. Darcy, E. M. S. Gendron, L. F. Stanish, E. A. Bagshaw, D. L. Porazinska, and S. K. Schmidt. 2018. Diversity patterns of microbial eukaryotes mirror those of bacteria in Antarctic cryoconite holes. FEMS Microbiology Ecology 94:fix167. doi:10.1093/femsec/fix167.
- Souza, F. F. C., D. V. Rissi, F. O. Pedrosa, E. M. Souza, V. A. Baura, R. A. Monteiro, E. Balsanelli, L. M. Cruz, R. A. F. Souza, M. O. Andreae, et al. 2019. Uncovering prokaryotic biodiversity within aerosols of the pristine Amazon Forest. Science of the Total Environment 688:83–86. doi:10.1016/j.scitotenv.2019.06.218.
- Stamatakis, A. 2014. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–13. doi:10.1093/bioinformatics/btu033.
- Stucky, B. J. 2012. SeqTrace: A graphical tool for rapidly processing DNA sequencing chromatograms. Journal of Biomolecular Techniques 23:90–93. doi:10.7171/jbt.12-2303-004.
- Tarlachkov, S. V., T. V. Shevchuk, M. C. Montero-Calasanz, and I. P. Starodumova. 2020. Diversity of rhodopsins in cultivated bacteria of the family Geodermatophilaceae associated with non-aquatic environments. Bioinformatics 36:1668–72. doi:10.1093/bioinformatics/btz840.
- Thomas-Hall, S. R., B. Turchetti, P. Buzzini, E. Branda, T. Boekhout, B. Theelen, and K. Watson. 2010. Cold-adapted yeasts from Antarctica and the Italian Alps—description of three novel species: Mrakia robertii sp. nov., Mrakia blollopis sp. nov. and Mrakiella niccombsii sp. nov. Extremophiles 14:47–59. doi:10.1007/s00792-009-0286-7.
- Trujillo, M. E., M. Goodfellow, K. Busarakam, and R. Riesco. 2015. Modestobacter lapidis sp. nov. and Modestobacter muralis sp. nov., isolated from a deteriorated sandstone historic building in Salamanca, Spain. Antonie van Leeuwenhoek 108:311–20. doi:10.1007/s10482-015-0482-7.
- Turchetti, B., S. R. Thomas-Hall, L. B. Connell, E. Branda, P. Buzzini, B. Theelen, W. H. Müller, and T. Boekhout. 2011. Psychrophilic yeasts from Antarctica and European glaciers: Description of Glaciozyma gen. nov., Glaciozyma martinii sp. nov. and Glaciozyma watsonii sp. nov. Extremophiles 15:573. doi:10.1007/s00792-011-0388-x.
- Ursell, L. K., W. Van Treuren, J. L. Metcalf, M. Pirrung, A. Gewirtz, and R. Knight. 2013. Replenishing our defensive microbes. BioEssays 35:810–17. doi:10.1002/bies.201300018.
- Vimercati, L., J. L. Darcy, and S. K. Schmidt. 2019. The disappearing periglacial ecosystem atop Mt. Kilimanjaro supports both cosmopolitan and endemic microbial communities. Scientific Reports 9:10676. doi:10.1038/s41598-019-46521-0.
- Vimercati, L., S. Hamsher, Z. Schubert, and S. K. Schmidt. 2016. Growth of a high-elevation Cryptococcus sp. during extreme freeze-thaw cycles. Extremophiles 20:579–88. doi:10.1007/s00792-016-0844-8.
- Wade, W. G. 2013. Characterisation of the human oral microbiome. Journal of Oral Biosciences 55:143–48. doi:10.1016/j.job.2013.06.001.
- Walters, W., E. R. Hyde, D. Berg-Lyons, G. Ackermann, G. Humphrey, A. Parada, J. A. Gilbert, J. K. Jansson, J. G. Caporaso, J. A. Fuhrman, A. Apprill, and R. Knight. 2016. Improved bacterial 16S rRNA gene (V4 and V4-5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. mSystems 1:e00009-15. doi:10.1128/mSystems.00009-15.
- Wang, Q., G. M. Garrity, J. M. Tiedje, and J. R. Cole. 2007. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Applied and Environmental Microbiology 73:5261–67. doi:10.1128/AEM.00062-07.
- Xiao, J., Y. Luo, J. Xu, S. Xie, and J. Xu. 2011. Modestobacter marinus sp. nov., a psychrotolerant Actinobacterium from deep-sea sediment, and emended description of the genus Modestobacter. International Journal of Systematic and Evolutionary Microbiology 61:1710–14. doi:10.1099/ijs.0.023085-0.
- Yilmaz, P., L. W. Parfrey, P. Yarza, J. Gerken, E. Pruesse, C. Quast, T. Schweer, J. Peplies, W. Ludwig, and F. O. Glöckner. 2014. The SILVA and ‘all-species living tree project (LTP)’ taxonomic frameworks. Nucleic Acids Research 42:D643–D648. doi:10.1093/nar/gkt1209.
- Yuan, Y., G. Si, J. Wang, T. Luo, and G. Zhang. 2014. Bacterial community in alpine grasslands along an altitudinal gradient on the Tibetan Plateau. FEMS Microbiology Ecology 87:121–32. doi:10.1111/1574-6941.12197.
- Zanne, A. E., K. Abarenkov, M. E. Afkhami, C. A. Aguilar-Trigueros, S. Bates, J. M. Bhatnagar, P. E. Busby, N. Christian, W. K. Cornwell, T. W. Crowther, et al. 2020. Fungal functional ecology: Bringing a trait-based approach to plant-associated fungi. Biological Reviews 95:409–33. doi:10.1111/brv.12570.
- Zhang, Y., J. Chen, H. Liu, Y. Zhang, W. Li, and L. Yu. 2011. Geodermatophilus ruber sp. nov., isolated from rhizosphere soil of a medicinal plant. International Journal of Systematic and Evolutionary Microbiology 61:190–93. doi:10.1099/ijs.0.020610-0.
- Zhang, L., M. Wang, J. I. Prosser, Y. Zheng, and J. He. 2009. Altitude ammonia-oxidizing bacteria and archaea in soils of Mount Everest. FEMS Microbiology Ecology 70:208–17. doi:10.1111/j.1574-6941.2009.00775.x.
- Zhao, W., X. Liu, Q. Huang, and P. Cai. 2015. Streptococcus suis sorption on agricultural soils: Role of soil physico-chemical properties. Chemosphere 119:52–58. doi:10.1016/j.chemosphere.2014.05.060.
