Abstract
To investigate the mechanism of trifluorothymidine (TFT)-induced DNA damage, we developed an enzymatic method for the synthesis of single-strand oligonucleotides containing TFT-monophosphate residues. Sixteen-mer oligonucleotides and 14-mer 5′-phosphorylated oligonucleotides were annealed to the template of 25-mer, so as to empty one nucleotide site. TFT-triphosphate was incorporated into the site by DNA polymerase and then ligated to 5′-phosphorylated oligonucleotides by DNA ligase. The synthesized 31-mer oligonucleotides containing TFT residues were isolated from the 25-mer complementary template by denaturing polyacrylamide electrophoresis. Using these single-strand oligonucleotides containing TFT residues, the cleavage of TFT residues from DNA, using mismatch uracil-DNA glycosylase (MUG) of E.coli origin, was compared with that of 5-fluorouracil (5FU) and 5-bromodeoxyuridine (BrdU). The TFT/A pair was not cleaved by MUG, while the other pairs, namely, 5FU/A, 5FU/G, BrdU/A, BrdU/G, and TFT/G, were easily cleaved from each synthesized DNA. Thus, this method is useful for obtaining some site-specifically modified oligonucleotides.
INTRODUCTION
Trifluorothymidine (TFT), which is a component of the oral drug formulation TAS-102, is known to inhibit thymidylate synthase and to be incorporated into the DNA, similar to 5-fluorouracil (5FU) or 5-fluoro-2′-deoxyuridine (FdUrd).
The efficacy of TAS-102 as an oral chemotherapeutic agent for metastatic gastrointestinal cancer is currently under investigation in phase I/II clinical trials.[ Citation 1 ] We demonstrated that short-term high-dose exposure to TFT increased its incorporation into DNA and resulted in potent antitumor activity against human cancer cells, both those sensitive and resistant to 5FU.[ Citation 2 , Citation 3 ] Therefore, it is necessary to investigate the mode of action of TFT against DNA replication and repair enzymes, as compared with that of 5FU and FdUrd. The incorporation of commercially available TFT into DNA using phosphoramidite chemistry[ Citation 4 , Citation 5 ] has been reported. However, after a trityl-on oligonucleotide was synthesized in the solid phase, the trifluoromethyl at the 5-position in the base converted to nitrile under the standard deprotection condition (concd NH4OH at 55°C for 16 hours)[ Citation 6 ] . These results indicated that the conversion was due to the known reactivity and instability of the trifluoromethyl group of 5-trifluoromethyluracil toward nucleophiles under basic conditions. Holzberger et al.[ Citation 7 ] reported that when trifluorothymidine triphosphate (TFT-TP) was synthesized, its nucleotide could be incorporated into the DNA by some DNA polymerases. In order to examine the mode of action of several analogues incorporated into the DNA, it is necessary to use single-strand oligonucleotide site-specific incorporation of these analogues. Therefore, the present study describes the establishment of a method for the generation of single-strand oligonucleotides containing TFT residues for investigating of the effect of TFT against DNA repair and replication enzymes.
MATERIALS AND METHODS
Reagents
5-Trifluorothymidine-5′-triphosphate (TFT-TP) was synthesized by contract research with Moravek Biochemical Inc. (USA). [Gamma-32P]adenosine 5′-triphosphate, tetra (triethylammonium) salt (111 TBq/mmol, NEG502A) was purchased from Perkin Elmer Company (USA).
Enzymes and Oligonucleotides
Sequenase, a version 2.0 DNA polymerase was purchased from USB Corporation (USA). Pfu DNA polymerase was purchased from Fermentas life Sciences (USA). Klenow fragment (3′ → 5′ exonuclease minus and 5′ → 3′ exonuclease minus) was purchased from New England BioLabs (USA). mismatch uracil-DNA glycosylase (MUG) was purchased from Trevigen Inc. (USA). Oligonucleotides containing 5FU, BrdU, U, and other normal bases were purchased from Sigma Chemical Co., as gel-purified grade chemicals (Japan).
Primer Extension Assay
The 17-base primer was labeled with 32P at its 5′-end and annealed to its complementary M13mp18(+) DNA template.
(5′) 32P- GTAAAACGACGGCCAGT
(3′) CATTTTGCTGCCGGTCACGGTTCGAACGTACGGACGT
The DNA primer extension assay[ Citation 8 , Citation 9 ] was carried out in a total volume of 10 μL, containing 2.5 pmole primer/template, DNA polymerase, and dNTP mixture (containing or not containing FTP-triphosphate), at 37°C for 30 minutes. The reaction was stopped by adding 3.3 μL denaturing loading buffer (95% formamide/10 mM EDTA/0.1%BPB/0.1%XC). The mixture was boiled at 100°C for 3 minutes and cooled on ice. The electrophoresis and the subsequent analysis was performed using the BAS2000 system.
Synthesis of Oligonucleotides Containing TFT
A 16-mer oligonucleotide (DNA(a)) and 14-mer 5′-phosphorylated oligonucleotide (DNA(b)) were annealed with a 25-mer oligonucleotide (DNA(c)) as the template as described below.
DNA(a); 5′-GACTAGTCTGCATGCC-3′
DNA(b); 5′ P-ACGTACGTTGCATG-3′
DNA(c); 5′-GCAACGTACGTAGGCATGCAGACTA-3′
The reaction mixture (2.26 mL) containing 30 nmol of DNA(a), 40 nmol of DNA(b), 40 nmol of DNA(c), 2 μmol of TFT triphosphate (TFT-TP), and 800 U of sequenase in 20 mM Tris-HCl, pH 7.5, 10 mM MgCl2 and 25 mM NaCl was incubated at 37°C for 30 minutes; then, 200 μL of T4 DNA ligase (Nippon Gene Co., Ltd., Japan) and 280 μL of 10 x ligation buffer (500 mM Tris-HCl, 100 mM MgCl2, 200 mM DTT, and 10 mM ATP) were added to the mixture. The reactions were allowed to proceed for 30 minutes at room temperature. The reaction products were concentrated by the addition of butanol and precipitated by ethanol. The DNA crude solution was purified by electrophoresis on 15% polyacrylamide gels containing TBE buffer (100 mM Tris.pH 8.3, 100 mM borate, 2 mM EDTA, 8M urea).
Analysis of Oligonucleotides Containing TFT
The purity of the modified synthetic DNA fragments was checked by polyacrylamide gel electrophoresis and HPLC.
To cleave the phosphodiester bonds, DNA was treated with 5 units of nuclease P1 (Sigma-Aldrich Corp., USA) in 0.05 mM Tris-HCl, pH 6.0, in the presence of 0.5 mM NaCl at 37°C for 1 hour. And then the reaction mixture was treated with alkaline phosphatase (Takara, Japan) in 0.1 mM Tris-HCl, pH 8.0, 0.25 mM NaCl, 1 mM MgCl2 at 37°C for 1 hour. With this treatment, all the nucleotides were dephosphorylated to nucleosides. The resolved nucleosides were analyzed using an HPLC system (Gulliver HPLC system, Jasco Co., Ltd., Japan).
DNA Glycosylase Assay
Oligomers (31-mer) containing a TFT-monophosphate or other deoxyribonucleotide were labeled with 32P at their 5′-end and annealed to their complementary 31-base templates. The DNA glycosylase assay[ Citation 10 ] was carried out in a total volume of 10 μL containing 2.5 pmole primer/template, mismatch uracil-DNA glycosylase (MUG) in 20 mmol/L Tris, 1 mmol/L EDTA, and 50 mmol/L NaCl, the reactions being allowed to proceed at 37°C for 60 minutes. The reactions were stopped, followed by treatment with 100 mM NaOH at 90°C for 15 minutes, in order to cleave the sugar-phosphate backbone at the AP site. The labeled oligonucleotides were precipitated with 3 times their volumes of ethanol at −80°C for 10 hours and dissolved in 10 μL of 50 mM Tris (pH 8.0) and 5 mM MgCl2. This sample was added to 3.3 μL of the denaturing loading buffer (95% formamide/10 mM EDTA/0.1%BPB/0.1%XC), boiled at 100°C for 3 minutes,and then cooled on ice. The reaction products were separated by 15% polyacrylamide sequencing gel electrophoresis and analyzed using the BAS20000 system.
A 31-mer 32P labeled drug-containing or normal oligomer (DNA(1)) and 31-mer oligomer (DNA(2) or DNA(3)) were also prepared:
DNA(1); *5′-GACTAGTCTGCATGCCXACGTACGTTGCATG-3′
where X is U, T(DNA(1)[T]), TFT(DNA(1)[TFT]), 5BrU or 5FU.
DNA(2); 5′-CATGCAACGTACGTAGGCATGCAGACTAGTC -3′
DNA(3); 5′-CATGCAACGTACGTGGGCATGCAGACTAGTC -3′
Electrophoresis and Autoradiography
Products from the DNA synthesis and ligation reactions were analyzed by electrophoresis on 15% polyacrylamide gels containing TBE buffer (100 mM Tris.pH 8.3, 100 mM borate, 2 mM EDTA, 8 M urea) at 1500V for 2–3 hours.
Autoradiography was performed for 2–10 hours using the Fujix BAS 2000 system (Fuji Photo-Film, Tokyo, Japan).
RESULTS
Analysis of Synthesized TFT-Containing Oligonucleotide
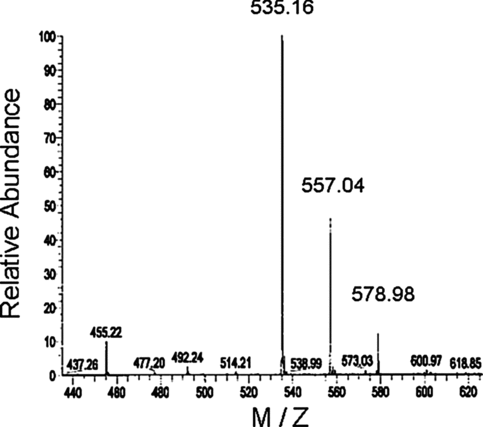
The synthesized trifluorothymidine-triphosphate (TFT-TP) was analyzed by mass spectrometry (). TFT-TP was detected as the peak corresponding to 535.16 m/z.
Incorporation TFT into DNA by Several DNA Polymerases
DNA primer extension assay was used to measure the activity of the incorporated TFT-TP into DNA by several commercial available DNA polymerases ().
FIGURE 2 Incorporation TFT into DNA by several DNA polymerases. Actions of TFT and BrdU on DNA primer extension in vitro. The ability of pfu DNA polymerase (pfu, 2.5 U/10 μL), Sequenase DNA polymerase (Sequenase, 13 U/10 μL), and Klenow fragment DNA polymerase (Klenow, 2.0 U/10 μL) to extend the 5′-32P-labeled primer/M13mp19(+) DNA (5 μg/mL) was evaluated in the presence of dATP,dGTP, and dCTP with or without dTTP, with TFT-TP, and with BrdUTP. Lanes 1–4, dideoxynucleotide sequencing of the M13mp19(+) DNA template with ddGTP, ddATP, ddTTP, and ddCTP, respectively, are indicated.
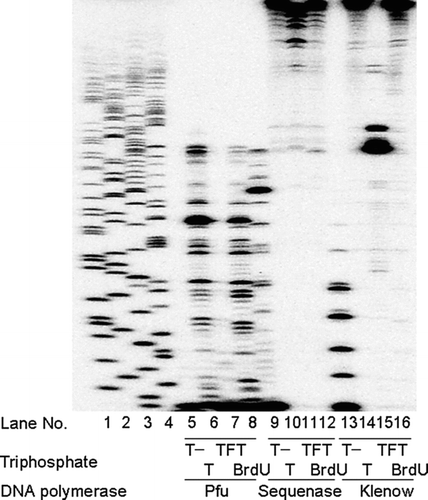
When dTTP was omitted from the mixture (T-reaction), the primer was elongated by sequenase DNA polymerase and klenow fragment DNA polymerase, which paused at the T site, but not by pfu DNA polymerase (, lanes 5, 10, and 13). These results suggest the error-prone property of these enzymes.[ Citation 11 ] Sequenase DNA polymerase and klenow fragment DNA polymerase were able to incorporate TFT-TP into DNA, but pfu DNA polymerase showed little activity in this situation.
Enzymatic Synthesis of Oligonucleotides Containing TFT
In order to investigate the actions of TFT on the DNA replication and repair enzymes, 31-mer oligonucleotides containing TFT were synthesized enzymatically (). First, for the DNA polymerase reaction step, we used sequenase DNA polymerase, as shown in . TFT-TP was completely incorporated into the nucleotide site (T-site) between the 16-mer oligonucleotides and 5′-phosphorylated 14-mer oligonucleotides within 10 minutes. This reaction does not allow elongation of more than one nucleotide site, because the pre-annealed 5′-phosphorylated 14-mer oligonucleotides hinder the next elongation.
FIGURE 3 Enzymatic reaction of DNA polymerase and DNA ligase. Sixteen-mer oligonucleotides were labeled with 32P and aliquots (0.05 pmol/μL) were added to the reaction mixture for monitoring the enzymatic reaction. Except adding the 32P labeled oligonucleotides, the same experiments were performed as described in synthesis oligonucleotides containing TFT section of Materials and Methods).
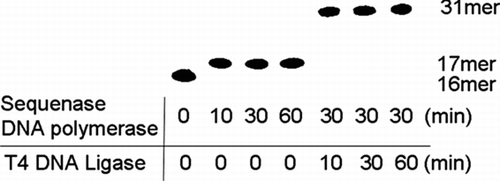
Second, in the DNA ligase reaction step, if TFT were not incorporated into the T-site or were incorporated in more than one nucleotide site, then DNA ligase would not be able to connect its oligomers with the 5′-phosphorylated 14-mer oligonucleotides. Therefore, the synthesized 31-mer oligomer appears to be site-specifically incorporated into the TFT-monophosphate residues.
Third, we purified these oligonucleotides by denaturing polyacrylamide electrophoresis. At this step, we did not use ethidium bromide to stain the DNA, because the band of the 31-mer oligomer was immediately detected by direct UV irradiation from a UV lamp.
Analysis of the Synthesized Oligonucleotides Containing TFT
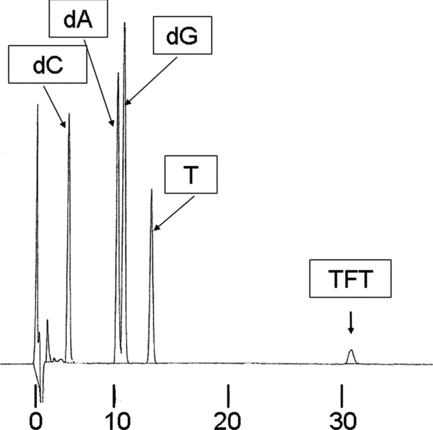
The oligonucleotides were digested by nuclease P1 and alkaline phosphatase and were confirmed to show the TFT peak in the HPLC ().
FIGURE 4 Analysis of oligonucleotide containing TFT using high-performance liquid chromatography. Oligonucleotides containing TFT were resolved into nucleosides by the treatment with phosphodiesterase and alkaline phosphatase as described in Material and Methods. Aliquots (50 μL) of the treated samples were applied to a Chemcosorb 300-5C18 column (4.6 i.d. × 250 mm, Chemco Corp., Ltd.) under the following chromatographic conditions: mobile phase, 2% acetonitrile containing 0.01% trifluoroacetic acid; flow rate, 1.ml/min;monitoring wavelength, 254 nm.
Excision Activity by E. coli Mismatch Uracil-DNA Glycosylase of TFT-Containing Oligonucleotides
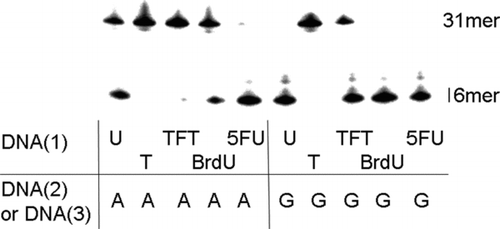
In order to investigate the cleavage activity of TFT residues by E. coli mismatch uracil-DNA glycosylase (MUG), single-strand oligonucleotides containing TFT monophosphate residues and complementary oligonucleotides (TFT/A pair or TFT/G pair) were prepared (). MUG (4 unit) excised not only the U/G pair, but also the U/A pair following incubation at 37°C for 1 hour. These results were consistent with a previous report.[ Citation 12 ] Under these conditions, all of the 5FU/A, 5FU/G, and BrdU/G, BrdU/A, and TFT/G pairs were removed, but not the TFT/A pair. These studies indicate that MUG does not have the ability to recognize and remove TFT paired to adenine in the double-strand DNA.
FIGURE 5 Cleavage by E. coli mismatch uracil-DNA glycosylase (MUG) of oligonucleotides containing several 5-substituted pyrimidine nucleotide residues. Lanes 1–5 show cleavage of a double-strand oligonucleotide with several 5-substituted pyrimidine nucleotide residues paired to the adenine base, and lanes 6–10 show the same sequence with the analogues paired to the guanine base. The oligonulcleotides (0.125 pmol/μl) were incubated with 4.0 units of MUG at 37°C for 60 minutes.
DISCUSSION
5-Fluorouracil (5FU) an antitumor pyrimidine, has been widely used for the treatment of gastric, colorectal, head and neck and breast cancers. Like 5FU, TFT is believed to exert its cytocidal effect by inhibiting TS.[ Citation 13 ] However, we found that when tumors were exposed to high concentrations of TFT for short periods of time, the drug was incorporated into the DNA and exerted its cytocidal activity by causing DNA fragmentation rather than by inhibiting TS.[ Citation 2 , Citation 3 , Citation 14 ] Therefore, we considered it necessary to prepare oligonucleotides to investigate with the mode of action of trifluorothymidine (TFT) against DNA replication and repair enzymes as compared to 5FU.
We succeeded in synthesizing single-strand oligonucleotides containing TFT-monophosphate residues using DNA polymerase and DNA ligase, but failed to synthesize these oligonucleotides using the principle of phosphoramidite chemistry[ Citation 6 ] (data not shown). We used a simple and rapid method in the present study, making use of enzymatic reactions under physiological conditions.
E. coli mismatch uracil-DNA glycosylase (MUG) can remove 5FU/A, 5FU/G, and 5-bromodeoxyuridine (BrdU)/G, BrdU/A,and TFT/G pairs, but not the TFT/A pair.
MUG belongs to family 2 of the uracil-DNA glycosylase superfamily.[ Citation 15 ] Thymine DNA glycosylase (TDG) also belongs to the same family. This family of MUG and TDG displays quite distinct properties, being insensitive to inhibition by UGI and inactive on ss-uracil-DNA, and lacking the conserved catalytic residues and specificity determinants of the Family-1 UDGs.[ Citation 16 ]
When TFT is mispaired with guanine, MUG is able to excise the TFT from the DNA, similar to BrdU and 5FU. Unlike BrdU and 5FU, however, MUG was not able to recognize and remove TFT paired to adenine in the double-strand DNA.
In vivo or ex vivo experiments using live cell cultures are very important, and we plan to continue studying this mechanism. We believe that investigation of the reactivity of several analogues incorporated into the DNA with other DNA repair enzymes is a useful approach to identify new therapeutic targets.
REFERENCES
- Overman , M. J. , Kopetz , S. , Varadhachary , G. , Fukushima , M. , Kuwata , K. , Mita , A. , Wolff , R. A. , Hoff , P. , Xiong , H. and Abbruzzese , J. L. 2008 . Phase I clinical study of three times a day oral administration of TAS-102 in patients with solid tumors . Cancer Invest. , 26 : 794 – 799 .
- Murakami , Y. , Kazuno , H. , Emura , T. , Tsujimoto , H. , Suzuki , N. and Fukushima , M. 2000 . Different mechanisms of acquired resistance to fluorinated pyrimidines in human colorectal cancer cells . Int. J. Oncol. , 17 : 277 – 283 .
- Emura , T. , Suzuki , N. , Yamaguchi , M. , Ohshimo , H. and Fukushima , M . 2004 . A novel combination antimetabolite, TAS-102, exhibits antitumor activity in FU-resistant human cancer cells through a mechanism involving FTD incorporation in DNA . Int. J. Oncol. , 25 : 571 – 578 .
- Gmeiner , W. H. , Pon , R. T. and Lown , J. W. 1991 . Synthesis, annealing properties, fluorine-19 NMR characterization, and detection limits of a trifluorothymidine-labeled antisense oligodeoxyribonucleotide 21 mer . J. Org. Chem. , 56 3602–3608
- Cook , P. D. and Sanghvi , Y. S. 1992 . Nuclease resistant,pyrimidine modified oligonucleotides that detect and modulate gene expression. US Patent 5,614,617 . Chem. Abstr. , 117 : 111996 1992
- Markley , J. C. , Chirakul , P. , Sologub , D. and Sigurdsson , S. T. 2001 . Incorporation of 2′-deoxy-5-(trifluoromethyl)uridine and 5-Cyano-2′-deoxyuridine into DNA . Bioorg. Med. Chem. Lett. , 11 : 2453 – 2455 .
- Holzberger , B. and Marx , A. 2009 . Enzymatic synthesis of perfluoroalkylated DNA . Bioorgan. Med. Chem. , 17 : 3653 – 3658 .
- Mikita , T. and Beardsley , G. P. 1988 . Functional consequences of the arabinosylcytosine structural lesion in DNA . Biochemistry , 27 : 4698 – 4705 .
- Huang , P. , Chubb , S. , Hertel , L. W. , Grindey , G. B. and Plunkett , W. 1991 . Action of 2′,2′-difluorodeoxycytidine on DNA synthesis . Cancer Res. , 51 : 6110 – 6117 .
- Neddermann , P. and Jiricny , J. 1993 . The purification of a mismatch-specific thymine-DNA glycosylase from HeLa cells . J. Biol. Chem. , 268 : 21218 – 21224 .
- Kunkel , T. A. , Sabatino , R. D. and Bambara , R. A. 1987 . Exonucleolytic proofreading by calf thymus DNA polymerase delta . Proc. Natl. Acad. Sci. USA , 84 : 4865 – 4869 .
- Liu , P. , Theruvathu , J. A. , Darwanto , A. , Lao , V. V. , Pascal , T. , Goddard , W. 3rd. and Sowers , L. C. 2008 . Mechanisms of base selection by the Escherichia coli mispaired uracil glycosylase . J. Biol. Chem. , 283 : 8829 – 8836 .
- Peters , G. J. , van der Wilt , C. L. , van Triest , B. , Codacci-Pisanelli , G. , Johnston , P. G. , van Groeningen , C. J. and Pinedo , H. M. 1995 . Thymidylate synthase and drug resistance . Eur. J. Cancer , 31A : 1299 – 1305 .
- Emura , T. , Nakagawa , F. , Fujioka , A. , Ohshimo , H. , Yokogawa , T. , Okabe , H. and Kitazato , K. 2004 . An optimal dosing schedule for a novel combination antimetabolite, TAS-102, based on its intracellular metabolism and its incorporation into DNA . Int. J. Mol. Med. , 13 : 249 – 255 .
- Pearl , L. H. 2000 . Structure and function in the uracil-DNA glycosylase superfamily . Mutat. Res. , 460 : 165 – 181 .
- Barrett , T. E. , Savva , R. , Panayotou , G. , Barlow , T. , Brown , T. , Jiricny , J. and Pearl , LH. 1998 . Crystal structure of a G:T/U mismatch-specific DNA glycosylase: mismatch recognition by complementary-strand interactions . Cell , 92 : 117 – 129 .