Abstract
DNA methylation plays an important role in carcinogenesis and aberrant methylation patterns have been found in many tumors. Methylation is regulated by DNA methyltransferases (DNMT), catalyzing DNA methylation. Therefore inhibition of DNMT is an interesting target for anticancer treatment. RX-3117 (fluorocyclopentenylcytosine) is a novel demethylating antimetabolite that is currently being studied in clinical trials in metastatic bladder and pancreatic cancers. The active nucleotide of RX-3117 is incorporated into DNA leading to downregulation of DNMT1, the maintenance DNA methylation enzyme. Since DNMT1 is a major target for the activity of RX-3117, DNMT1 may be a potential predictive biomarker. Therefore, DNMT1 protein and mRNA expression was investigated in 19 cancer cell lines, 26 human xenografts (hematological, lung, pancreatic, colon, bladder cancer) and 10 colorectal cancer patients. The DNMT1 mRNA expression showed large variation between cell lines (100-fold) and the 26 xenografts (1100-fold) investigated. The DNMT1 protein was overexpressed in colon tumours from patients compared to non-malignant mucosa from the same patients (P = 0.02).
The DNA methylation in these patients was significantly higher in tumour tissues compared to normal mucosa (P = 0.001). DNMT1 expression in normal white blood cells also showed a large variation.
In conclusion, the large variation in DNMT1 expression may serve as a potential biomarker for demethylating therapy such as with RX-3117.
Key words:
Introduction
DNA methylation is a covalent chemical modification resulting in addition of a methyl group at the carbon 5 position of the cytosine ring in CpG dinucleotides. DNA methylation plays important roles in chromatin structure modulation, genomic stability and transcriptional regulation of specific genes.Citation1,Citation2 In comparison with normal cells, human colon cancer cells show a drastic change in DNA methylation status, generally exhibiting global DNA hypomethylation as well as accompanying region-specific hypermethylation.Citation3,Citation4 DNA methyltransferases (DNMT) are a family of enzymes that catalyse DNA methylation with either S-adenosyl-L-methionine or 5-methyl tetrahydrofolate as the methyl donor. Three active family members have been isolated from mammalian tissues (DNMT1, DNMT3A & DNMT3B). DNMT1 is believed to be the "maintenance" enzyme, required to maintain the methylation pattern during DNA replication and has an affinity for hemi-methylated DNA strands.Citation5 DNMT3A/B are known as the de novo methyltransferases each with preferred target sites, different from DNMT1.Citation6,Citation7 DNA methylation has been demonstrated in several instances to show different patterns in malignant tissues compared to normal tissue, partially due to methylation mutations. Mouse models have indicated that DNMT3a may act as a tumour suppressor gene.Citation8 In addition, DNMT1 is crucial for maintenance methylation; inactivation through gene deletion stimulates tumour development.Citation9 Overexpression of DNMTs has also been reported in numerous cell lines supporting the hypothesis that there is an important role for DNMT in cellular transformation, while it has a direct role during tumour initiation and progression.Citation10
RX-3117 (fluorocyclopentenylcytosine) is a new demethylating antimetabolite that is currently being studied in clinical trials in metastatic bladder and metastatic pancreatic cancers.Citation11–13 An active nucleotide of RX-3117 has been shown to be incorporated into both RNA and DNA,Citation14 with the latter possibly being responsible for the downregulation of DNMT1, leading to hypomethylation of DNA. Since DNMT1 is suspected as being a major target for the activity of RX-3117, DNMT1 may also be a potential predictive biomarker for activity of this drug. Therefore, we have characterized the DNMT expression in normal epithelium and in colon cancer, the % global methylation in colon cancer, as well as DNMT1 expression in a panel of 17 cell lines and 26 xenografts from human tumors.
Materials and methods
Cell lines and xenografts
Two cell line panels were used to determine the expression of DNMT1, DNMT3A and DNMT3B, and a third panel only for DNMT1. One cell line panel consisted of several variants of Caco-2 and WiDr colon cancer cellsCitation15 grown in standard RPMI 1640 medium containing 2.3 μM folic acid (a supra-physiological concentration referred to as high folate; HF) or in a customized RPMI 1640 medium containing 1 nM folic acid (referred to as low folate; LF). Growth medium was supplemented with 10% dialyzed fetal bovine serum (Gibco, Grand Island, NY, USA), 1 M HEPES, 100 units/ml penicillin and 100 μg/ml streptomycin (all from Invitrogen). The second panel consisted of the cell lines that were earlier reported for the screening of their sensitivity to RX-3117,Citation12 while the third panel consisted of a mix of cell lines from different histological sources (all obtained from ATCC, Manassas, VA, USA). Panels 2 and 3 were routinely maintained in a humidified atmosphere of 5% CO2-95% air and were cultured in either RPMI-1640 (Invitrogen Ltd., Paisley, UK) or DMEM (Sigma-Aldrich, Denmark) medium.
Twenty-six human tumour xenografts (partially patient-derived, partially from cell lines) were established in nude mice as described earlier.Citation16 Tumours were maintained subcutaneously in female nude mice (nu/nu) from about 9-10 weeks old either at the Charles Rivers Discovery Research Services (Seattle, WA, USA) or at Champions Oncology (Hackensack, NJ, USA). Both companies comply with the recommendations of the Guide for Care and use of Laboratory Animals, and are accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International. Tumours were harvested before becoming lethal to the mice and forming metastases. They were between 50-200 mg in size, snap-frozen and transported on dry ice to Amsterdam. From the tumours, RNA was isolated to determine the DNMT1 expression, while from several xenograft tissues cross-sections were prepared for IHC, which was performed as described for the human tumours.
Patient characteristics and tissue handling
Paired samples of primary colorectal cancer and corresponding non-cancerous mucosa were obtained from 10 patients undergoing elective surgery at the Department of Surgery, Naestved County Hospital (Denmark). Tissue was collected from 5 females and 5 males; their mean age was 70 ± 4.6 years (range 56 to 85). The site of the primary tumour was colon ascendens for one, coecum for one, sigmoideum for one, colon descendens for one, rectum for four, colon transversum for one and one from flexura dextra. The primary histological stage (Dukes) was two stages A, 5 stages B and 3 stages C. The tissue samples were snap frozen. Tissue samples taken as far as possible from the tumour site at the margins of resection were designated as “normal”. Visibly necrotic tissue in the tumour samples was manually removed. None of the patients received postoperative treatment, such as radiotherapy or chemotherapy. Information on patient age, gender and tumour stage were obtained from the pathology report. For immunohistochemical (IHC) analysis, normal and neoplastic formalin fixed, paraffin-embedded tissues were obtained from the pathology archives at Naestved County Hospital (Denmark). All of the cases were histologically confirmed by pathologists. Ethics approval was received from The Danish National Committee on biomedical research Ethics. Informed consent was obtained from each patient.
DNA extraction and hydrolysis
DNA was isolated from cells using the QIAmp DNA Mini Kit (Qiagen, Valencia, CA) following the manufacturer’s protocol. DNA hydrolysis was performed as described previously.Citation14 Briefly, 1 μg of genomic DNA was first denatured (45 °C for 120 min) before being degraded by 2U of nuclease P1 (Sigma-Aldrich, Steinheim, Germany). Further breakdown was performed with 0.002U of venom phosphodiesterase I (Sigma-Aldrich, Steinheim, Germany) (37 °C 60 min) and dephosphorylation with 0.5 U alkaline phosphatase (Sigma-Aldrich, Steinheim, Germany)(37 °C 60 min).
RNA Isolation and cDNA Synthesis
Total RNA was isolated from cell extracts using the RNeasy FFPE kit (Qiagen) and from fresh frozen tissue using the RecoverAllTM Total Nucleic Acid Isolation kit (Ambion, Austin, TX, USA). The purity and amount of total RNA were estimated spectrophotometrically by measuring the absorbance at 260 and 280 nm. RNA was reverse transcribed by the QscriptTM SuperMix (Quanta Bioscience, Gaithersburg, USA) and MultiScribe MuLV Reverse Transcriptase (MBI, Vilnius, Lithuania) with random primers (High Capacity cDNA Reverse Transcription Kit, ABI, Foster City, USA), respectively.
Measurement of DNA content of 5-methylcytosine (5-metC) and deoxyuridine (dU)
DNA methylation analysis was performed as described earlier.Citation14 Briefly, 20 microliters of hydrolysed DNA solution was used for chromatographic analysis. Mixtures of the standard bases 2′-deoxyadenosine (dA), 2′-deoxyguanosine (dG), 2′-deoxythymidine (dT), 2′-deoxycytidine (dC), and 2′-deoxyuridine (dU) at 100 μM and 5-methylcytosine (5-metC) at 5 μM (Sigma-Aldrich, Steinheim, Germany) were used as standards to prepare the calibration curves. Nucleoside quantification was monitored with reverse phase-high performance liquid chromatography (Shimadzu, Columbia, MD, USA) coupled to a PerkinElmer Sciex API 3000 triple-quadropole mass spectrometer (Thornhill, Ontario, Canada). Chromatography was performed on a Prodigy 5 ODS-2, 150 x 3.2 mm column (Phenomenex, Torrance, CA, USA). Two buffers, 0.1% formic acid (Merck, The Netherlands) in water and 50% methanol (Merck, The Netherlands) in water (containing 0.1% formic acid), were used, with a linear gradient increase of 19.3%/min at a flow rate of 0.25 ml/min.
Quantitative RT-PCR
The design of primer sequences and cycling conditions were developed as described.Citation17 PCR was performed using LightCycler-FastStart DNA Master PLUS SYBR Green I kit (Roche Molecular Biochemicals, Mannheim, Germany) with the following primers/probes:
DNMT1, forward-gttcttcctcctggagaatgtcaa; reverse-gggccacgccgtact; probe-ttgtctccttcaagcgctccatggtc
DNMT3A, forward-cctgtgggagcctcaatgtta; reverse-ttcttgcagttttggcacattc; probe-cctggaacaccccctcttcgttgg
DNMT3B, forward-gactcgaagacgcacagctg; reverse-ctcggtctttgccgttgttatag; probe-agccacctctgactactgccccgc
sz-actin, forward-tcacccacactgtgcccatctacga; reverse-cagcggaaccgctcattgccaaagg; probe-atgccctcccccatgccatcctgcgt
Cycling conditions were as follows: denaturation (95 °C for 5 min), followed by 40 cycles (95 °C for 10 s, 60 °C for 15 s, and 72 °C for 10 s). Gene expression values (relative mRNA levels) are expressed as ratios between DNMT and an internal reference gene (β-actin). The analysis was performed using the LightCycler Relative Quantification Software (Roche, Mannheim, Germany).
IHC staining of DNMT in human colon tissue and DNMT western blotting
Immunohistochemistry (IHC) was performed as previously describedCitation18 with primary monoclonal antibodies against DNMT1a (Santa Cruz Biotechnology -10219, Lot N-16, Dilution 1:250), DNMT3A (Cell Signaling Technology-3598, Lot H-295, dilution 1:250) and DNMT 3B (Santa Cruz Biotechnology-130740, Lot Q-25, dilution 1:250). Extracted protein from the cell line A549 (NSCLC) was used a qualitative positive control and purified RNA/DNA free water was used as a negative control in each IHC staining. Lymphocytes, in which DNMT immunoreactivity was previously demonstrated, served as positive controls in all sections. All IHC stained slides were interpreted by an investigator unaware of data. Nuclear DNMT immunoreactivity was assessed as staining intensity (0 = negative; 1= mild; 2 = moderate; 3 = high). IHC for DNMT1 was also performed on 5 μm cross-sections of xenografts using a comparable preparation of the slides.
Western blotting for DNMT1 in white blood cells from human volunteers was performed as described previously for cell lines.Citation12
Statistics
All data were analysed using the SPSS statistical package, version 17.0 (SPSS Inc., Chicago, IL, USA) for Windows. Differences in mRNA, protein expression and global methylation levels between groups were analysed using the two sided t-test and the Mann-Whitney test. Differences between the mRNA and protein expression of each were determined by the two sided Pearson's Correlation Coefficient. Differences with p values < 0.05 were considered significant.
Results
Expression of DNMT 1, 3A and 3B mRNA in cell lines and xenografts
The expression levels of the DNMT1, DNMT3A and DNMT3B mRNA transcripts were determined by semi-quantitative RT-PCR analysis in a variety of different cell lines. The Caco-2/HF and Caco-2/LF cell lines are derived from human colon epithelial cells and have been grown in a normal culture medium (CaCo2/HF) or a medium containing 1 nM leucovorin (LV) as a replacement for folic acid. Normal medium contains supra-physiological folate levels, hence, can be considered as high folate conditions. Similarly WiDr colon cancer cells were grown in normal medium (WiDr/HF) or adapted to low-folate medium (WiDr/LF). Cells that were adapted to 1 nM LV as only folate source are more comparable to conditions found within a healthy human being (CaCo2/LF and WiDr/LF) (). Comparison with t-test analysis demonstrated that there was no significant difference between DNMT1 mRNA expression in CaCO-2/LF and Caco-2/HF cells, and between WiDr/HF and WiDr/LF cells (p = 0.3). However, DNMT 3A and DNMT 3B mRNA expression was significantly higher in Caco-2/HF cells compared to Caco-2/LF (p = 0.001 and p = 0.02, respectively). DNMT3a expression was higher in WiDr/HF compared to WiDr/LF cells (p< 0.001).
Table 1. Effect of low-folate culture conditions on the expression of DNMT1, DNMT3A and DNMT3B mRNA expression in the colon cancer cell lines CaCo2 and WiDr.
The second cell line panel consists of cells originally used for mechanism of action studies for RX-311712 (). The expression of all three DNMTs was the highest in the U937 leukemia cell line and the lowest in the two non-small cell lung cancer (NSCLC) cell lines. In the AG6000 cells the expression of all three DNMTs was higher than in the parent A2780 ovarian cancer line. In the third cell line panel () the expression of DNMT1 varied from 0.58 to 7.3 (normalized to β-actin), with the LS174T cell line having the lowest expression. The number of cell lines from the various sources (both and ) was too small to establish significant differences.
Figure 1. DNMT1 expression in 8 cell lines from various histological sources normalized to β-actin expression. RaijB is a Burkits lymphoma, RT112 and SW780 are bladder cancer, BT474 and C-33A are cervix carcinoma, LS174T is colon cancer, PANC-1 is pancreatic cancer and HEC-1A is endometrial cancer. The SEM in all assays was less <15%. No statistically significant differences were observed between cell lines with a different pathological origin.
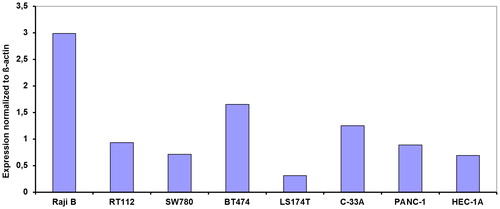
Table 2. Expression of DNMT1, DNMT3A and DNMT3B mRNA expression in 7 separate human cancer cell lines.
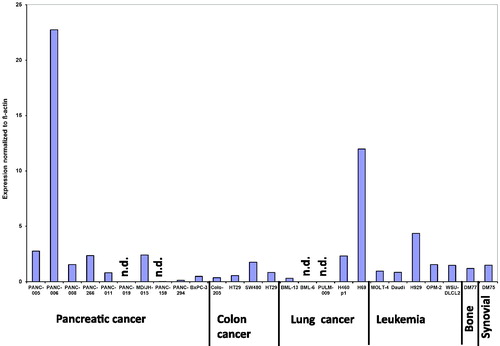
In the xenograft panel () the expression of DNMT1 was 2 times higher in the PANC-006 derived tumour compared to the next tumour in this panel, H69. The H69 tumour had DNMT1 levels 4-times higher than the closest other tumour xenograft H929 (). In three tumours from the tested xenografts panel, DNMT1 expression was at the detection level. Statistical analysis was performed to investigate differences between different tumour types. No statistical significant differences could be determined.
Figure 2. DNMT1 expression in 26 different xenografts including several patient derived xenografts (PDX) normalized to β-actin expression. DM77 is bone cancer, PANC-005, PANC-006, PANC-008, PANC-011, PANC-019, PANC-159, PANC-294, PANC-266, MD/JH-015, BxPC-3 are pancreatic cancer, DM75 is synovial cancer, PULM-009, BML-13, BML-6, H460 and H69 are lung cancer, Colo-205, HT29 and SW480 are colon cancer and MOLT-4, Daudi, H929, OPM-2 and WSU-DLCL are hematological malignancies. The SEM in all assays was less <15%. There was not statistically significant difference between the tumour types.
Global methylation and DNMT 1, 3A and 3B mRNA expression in normal colorectal and corresponding cancerous tissue
The level of global methylation in colorectal tumour tissue (mean 3.42 ± 0.18) was significantly higher (p = 0.0001) compared to corresponding normal epithelial colon tissue (mean 3.07 ± 0.17). The level of DNMT1 mRNA expression was higher in 5 of 10 colorectal cancers (50%) compared with that in the corresponding non-cancerous tissue (), but the mean in cancerous tissue was not significantly different compared to the level in the corresponding normal colorectal mucosa. However, for DNMT3A and DNMT3B mRNA expression the level was higher in 8 of 10 colorectal cancers (80%) compared with that in the corresponding non-cancerous tissue. The mean expression was significantly higher for both DNMT3A (p = 0.03) and DNMT3B (p = 0.004) mRNA expression.
Table 3. Protein and mRNA expression levels of DNMT1, 3A and 3B in normal and corresponding tumour tissue
DNMT 1, 3A and 3B protein levels in white blood cells, normal colorectal and corresponding cancerous tissue, and in xenografts
shows examples of IHC staining for DNMT 1, 3A and 3B in neoplastic colorectal tissue samples and corresponding “normal” colorectal mucosa. The distribution of the DNMT protein expression scores is summarized in . Very high and very low levels of staining for DNMT1A levels were found in the same cancer tissue (results not shown). Unlike DNMT3B that is present in the cytosol and nuclei, DNMT1 and DNMT3A are mainly found in the cytoplasm of tumour cells. (, insert). For comparison we also stained two of the xenografts (the highest and lowest mRNA expression) for DNMT1 () and observed a very intense staining for H69, the highest expressor from the xenografts.
Figure 3. IHC staining of DNA methyltransferase 1, 3A and 3B protein in colorectal cancer (CRC) patients and DNMT1 in two 5 µm xenograft sections (G), selected for high and low DNMT1 mRNA expression. Positive immunoreactivity for DNMT1, 3A and 3B was found in CRC tissue (A, C, E) and corresponding normal tissue (B, D, F). Unlike DNMT3B that is present in the cytosol and nuclei (E insert), DNMT1 and DNMT3A are mainly well distributed only in the cytoplasm of tumour cells. DAB staining shows DNMT1 expression in brown. Pictures were taken with 40x lens magnification.
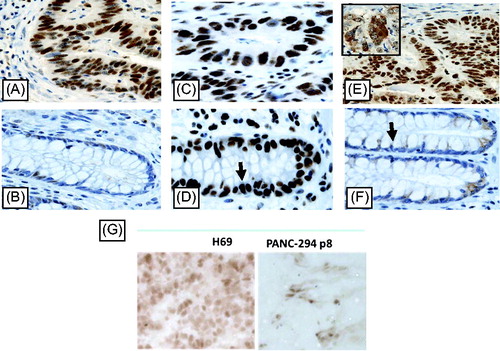
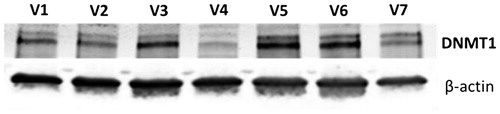
Figure 4. DNMT1 expression in white blood cells from 7 human volunteers (V1-V7) as determined with western blotting equal amounts of protein were used for each sample. β-actin was used as a loading control.
The expression of DNMT1 and DNMT3B was significantly higher in cancer tissue, compared to normal tissue (p = 0.02 and p= 0.0005, respectively), however, there was no significant difference in protein expression of DNMT3A between cancer and normal colonic tissue (p = 0.5). To determine whether there was a correlation between mRNA and protein expression, the Pearson correlation coefficient was determined. No correlation between the mRNA and protein levels were found (r values ranging from – 0.16 to 0.40), ().
Since DNMT1 is down-regulated by RX-3117 we aimed to determine whether RX-3117 treatment would indeed lead to down-regulation in patients. As a first step for this purpose we measured DNMT1 in white blood cells, to be used as a surrogate tissue. The expression of DNMT1 was easily detectable in white blood cells from 7 volunteers () indicating that there would be no problem in detection when white blood cells would be used as surrogate tissue.
Discussion
This study shows that in colon tumours there is a higher expression of DNMTs in the tumour compared to normal tissues. This was most evident for DNMT3A and DNMT3B when mRNA expression was evaluated, but using IHC the expression level was higher for DNMT1 and DNMT3B. This may be related to a different splicing pattern.Citation19 Overexpression of DNMTs was reported earlier in several human cancers including colorectal cancer,Citation20 although the extent of difference was initially reported to be 13-fold, but later reports are in the same range as our study. Interestingly, the difference in expression in xenografts evaluated by mRNA expression or protein was comparable, indicating that the use of a proper antibody would enable to evaluate such a difference. Since DNMT expression is related to cell cycle progression, it can be postulated that the observed difference between normal and tumour tissues might be a consequence of a higher proportion of proliferating cells in the tumour compared to normal tissue.Citation21 However, another explanation for the difference in expression might be related to a different ubiquitination and subsequent protein degradation.Citation22,Citation23 The latter might also explain the lack of correlation between protein and mRNA expression. Alternative splicing might play a role as well as mentioned above.
In the cell lines, we also found a large difference in DNMT expression, which is in line with data observed in various types of tumours. The difference in DNMT expression is, however, not reflected in a difference in global methylation, neither in this panel of colon tumours, nor in the panel of cell lines, for which we published global methylation earlier.Citation14 However, in some of the fast growing tumours and cell lines (e.g. H69 and U937) we also found a high DNMT expression, which might be in line with the association of DNMT1 with proliferation.
Folate depletion may affect the level of S-adenosyl-methionine, the methyl donor for DNA methylation, and therefore DNA methylation and DNMT1 levels.Citation24 For that purpose and since cancer patients may have a relative folate deficiency, we evaluated the DNMT expression in colon cancer cell lines adapted to folate depleted medium. Some small difference were observed, especially for DNMT3A and DNMT3B, but the overall global methylation was not different either.Citation14
Several novel anticancer drugs have been developed to target DNMTs,Citation23 with the aim to directly inhibit tumour growth or to modulate DNA methylation to enhance the efficacy of other drugs. For RX-3117 it was recently demonstrated that it is not only incorporated into RNA and DNA, but it can also inhibit DNMT1. It is not clear yet, which mechanism is responsible for this anti-tumour effect. One possibility to elucidate the mechanism of action in patients would be to determine the effect of RX-3117 on DNMT1 expression. Therefore we wondered whether DNMT expression can be used as a biomarker for these drugs. For this purpose we also measured the expression in white blood cells, which was readily detectable despite being resting cells. This offers the possibility to follow the expression of DNMT1 during treatment. It is expected that the expression should go down and it recommended to include this expression as a marker for activity of drugs such as the novel RX-3117.
In conclusion, we found a large variation of DNMT expression in cell lines, xenografts and tumour from patients. It is expected that this variation may play a role in the efficacy of drugs targeting DNMTs.
Acknowledgements
Rexahn Pharmaceuticals provided grant support for this study.
References
- Calcagno, D. Q.; Cardoso Smith, M. A.; Burbano, R. R. Cancer Type-Specific Epigenetic Changes: Gastric Cancer Methods Mol Biol. 2015; 1238:79–101
- Baylin, S. B. Mechanisms Underlying Epigenetically Mediated Gene Silencing in Cancer. Semin. Cancer Biol. 2002, 12(5), 331–337.
- Jones, P. A. The DNA Methylation Paradox. Trends Genet. 1999, 15(1), 34–37.
- Robertson, K. D.; Jones, P. A. DNA Methylation: Past, Present and Future Directions. Carcinogenesis 2000, 21(3), 461–467.
- Bestor, T. H.; Ingram, V. M. Two DNA Methyltransferases from Murine Erythroleukemia Cells: Purification, Sequence Specificity, and Mode of Interaction with DNA. Proc. Natl. Acad. Sci. U. S. A. 1983. 80(18):5559–63
- Okano, M.; Xie, S.; Li, E. Cloning and Characterization of a Family of Novel Mammalian DNA (Cytosine-5) Methyltransferases. Nat. Genet. 1998, 19, 219–220.
- Xie, S.; Wang, Z.; Okano, M.; Nogami, M.; Li, Y.; He, W. W.; Okumura, K.; Li, E. Cloning, Expression and Chromosome Locations of the Human DNMT3 Gene Family. Gene 1999, 236(1), 87–95.
- Peters, S. L.; Hlady, R. A.; Opavska, J.; Klinkebiel, D.; Pirruccello, S. J.; Talmon, G. A.; Sharp, J. G.; Wu, L.; Jaenisch, R.; Simpson, M. A.; Karpf, A. R.; Opavsky, R. Tumor Suppressor Functions of Dnmt3a and Dnmt3b in the Prevention of Malignant Mouse Lymphopoiesis. Leukemia 2014, 28, 1138–1142.
- Peters, S. L.; Hlady, R. A.; Opavska, J.; Klinkebiel, D.; Novakova, S.; Smith, L. M.; Lewis, R. E.; Karpf, A. R.; Simpson, M. A.; Wu, L.; Opavsky, R. Essential Role for Dnmt1 in the Prevention and Maintenance of MYC-Induced T-Cell Lymphomas. Mol. Cell. Biol. 2013, 33(21), 4321–4333.
- Zhang, W.; Xu, J. DNA Methyltransferases and Their Roles in Tumorigenesis. Biomark. Res. 2017, 5(1), 1.
- Choi, W. J.; Chung, H.-J.; Chandra, G.; Alexander, V.; Zhao, L. X.; Lee, H. W.; Nayak, A.; Majik, M. S.; Kim, H. O.; Kim, J.-H.; Lee, Y. B.; Ahn, C. H.; Lee, S. K.; Jeong, L. S. Fluorocyclopentenyl-Cytosine with Broad Spectrum and Potent Antitumor Activity. J. Med. Chem. 2012, 55(9), 4521–4525.
- Peters, G. J.; Smid, K.; Vecchi, L.; Kathmann, I.; Sarkisjan, D.; Honeywell, R. J.; Losekoot, N.; Ohne, O.; Orbach, A.; Blaugrund, E.; Jeong, L. S.; Lee, Y. B.; Ahn, C. H.; Kim, D. J. Metabolism, Mechanism of Action and Sensitivity Profile of Fluorocyclopentenylcytosine (RX-3117; TV-1360). Invest New Drugs 2013, 31, 1444–1457.
- Rasco, D.; Peterson, C.; Benaim, E.; Merchan, R. RX3117, an Oral Antimetabolite Nucleoside Shows Activity in Subjects with Pancreatic Cancer. Preliminary Results of Stage 1 of the Phase 1a/2b Study. J. Clin. Oncol. 2017, 35(4_suppl), 445.
- Honeywell, R. J.; Sarkisjan, D.; Kathmann, I.; Kristensen, M. H.; Peters, G. J. Sensitive Liquid Chromatography Mass Spectrometry (LC-MS) Assay Reveals Novel Insights on DNA Methylation and Incorporation of Gemcitabine, Its Metabolite Difluorodeoxyuridine, Deoxyuridine, and RX-3117 into DNA. Nucleosides, Nucleotides and Nucleic Acids 2016, 35(10–12).
- Lemos, C.; Kathmann, I.; Giovannetti, E.; Belien, J. A.; Scheffer, G. L.; Calhau, C.; Jansen, G.; Peters, G. J. Cellular Folate Status Modulates the Expression of BCRP and MRP Multidrug Transporters in Cancer Cell Lines from Different Origins. Mol.Cancer Ther. 2009, 8, 3, 655–664.
- Yang, M. Y.; Lee, Y. B.; Ahn, C. H.; Kaye, J.; Fine, T.; Kashi, R.; Ohne, O.; Smid, K.; Peters, G. J.; Kim, D. J. A Novel Cytidine Analog, RX-3117, Shows Potent Efficacy in Xenograft Models, Even in Tumors That Are Resistant to Gemcitabine. Anticancer Res. 2014, 34(12), 6951–6959.
- Sigmond, J.; Kroep, J. R.; Loves, W.; Codacci-Pisanelli, G.; Peters, G. J. Quantitative Real Time PCR of Deoxycytidine Kinase MRNA by Light Cycler PCR in Relation to Enzyme Activity and Gemcitabine Sensitivity. Cancer Lett. 2004, 213, 173–179.
- Kristensen, M. H.; Weidinger, M.; Bzorek, M.; Pedersen, P. L.; Mejer, J. Correlation between Thymidylate Synthase Gene Variants, RNA and Protein Levels in Primary Colorectal Adenocarcinomas. J. Int. Med. Res. 2010, 38(2), 484–497.
- Gopalakrishnan, S.; Van Emburgh, B. O.; Shan, J.; Su, Z.; Fields, C. R.; Vieweg, J.; Hamazaki, T.; Schwartz, P. H.; Terada, N.; Robertson, K. D. A Novel DNMT3B Splice Variant Expressed in Tumor and Pluripotent Cells Modulates Genomic DNA Methylation Patterns and Displays Altered DNA Binding. Mol. Cancer Res. 2009, 7(10), 1622–1634.
- Subramaniam, D.; Thombre, R.; Dhar, A.; Anant, S. DNA Methyltransferases: A Novel Target for Prevention and Therapy. Front. Oncol. 2014, 4:80.
- Robertson, K. D.; Keyomarsi, K.; Gonzales, F. A.; Velicescu, M.; Jones, P. A. Differential MRNA Expression of the Human DNA Methyltransferases (DNMTs) 1, 3a and 3b during the G(0)/G(1) to S Phase Transition in Normal and Tumor Cells. Nucleic Acids Res. 2000, 28(10), 2108–2113.
- Du, Z.; Song, J.; Wang, Y.; Zhao, Y.; Guda, K.; Yang, S.; Kao, H. Y.; Xu, Y.; Willis, J.; Markowitz, S. D.; Sedwick, D.; Ewing, R. M.; Wang, Z. DNMT1 Stability Is Regulated by Proteins Coordinating Deubiquitination and Acetylation-Driven Ubiquitination. Sci. Signal. 2010, 3(146), ra80.
- Sarkisjan, D.; Steenbergen, R. D. M.; Cloos, J.; Peters, G. J. Re-Emerging Antimetabolites with Novel Mechanism of Action with Respect to Epigenetic Regulation: Basic Aspects. In Chemotherapy for Leukemia: Novel Drugs and Treatment; Chapter 18 (Ed: T. Ueda). Springer Nature, Singapore Pte Ltd, 2017; pp. 311–326
- Farias, N.; Ho, N.; Butler, S.; Delaney, L.; Morrison, J.; Shahrzad, S.; Coomber, B. L. The Effects of Folic Acid on Global DNA Methylation and Colonosphere Formation in Colon Cancer Cell Lines. J. Nutr. Biochem. 2015, 26(8), 818–826.
