Abstract
Sterile alpha motif and histidine-aspartic acid domain containing protein-1 (SAMHD1) is a deoxynucleoside triphosphate (dNTP) hydrolase that controls dNTP pools and detoxifies cancer cells of chemotherapy metabolites. TH6342 is a recently reported small molecule inhibitor of SAMHD1 that interacts with the protein in vitro and non-competitively prevents dimerisation, a prerequisite for catalysis. The binding site of TH6342 on SAMHD1 is currently unknown. In the present study we demonstrate that the N-terminal SAM domain of SAMHD1 is not required for inhibition by TH6342.
Sterile alpha motif and histidine-aspartic acid domain containing protein-1 (SAMHD1) is a deoxynucleoside triphosphate (dNTP) triphosphohydrolase that controls dNTP pools in mammalian cells.[Citation1] This activity also contributes to therapy failure during cancer treatment, as the active metabolites of several chemotherapeutic nucleoside analogues can be hydrolysed by SAMHD1 and thus converted back into their inactive prodrug forms.[Citation2] This is best characterised for the deoxycytidine analogue cytarabine,[Citation3,Citation4] part of standard-of-care therapy for acute myeloid leukaemia. Accordingly, pharmacological approaches to inactivate this enzyme are of great interest,[Citation2] both as basic science tools to dissect dNTP metabolic pathways, and as a strategy to improve the efficacy of several nucleoside-based cancer therapies.
We recently reported a chemical series capable of inhibiting the dNTP hydrolase activity of SAMHD1.[Citation5] We demonstrated that these molecules, represented by TH6342 and analogues, inactivated SAMHD1 in vitro by directly binding to the protein and deterring dimerisation, which is required for forming the catalytically competent homotetramer. However, the molecular determinants of how this series of molecules inhibit SAMHD1, specifically where these ligands bind upon SAMHD1, remained elusive. Structural studies of SAMHD1 typically use a truncated version of the protein lacking the N-terminal SAM domain, as the flexibility of the linker connecting this domain to the catalytic HD domain prevents structural studies of full length wild-type SAMHD1. Thus, in the present study, as a prerequisite for structure-based efforts to identify the TH6342 binding interface, and directly interrogate the involvement of the N-terminal SAM domain upon the inhibitory activity of this chemical series, we investigated whether TH6342 retained inhibitory activity against SAMHD1 lacking the N-terminal SAM domain.
2. Results & discussion
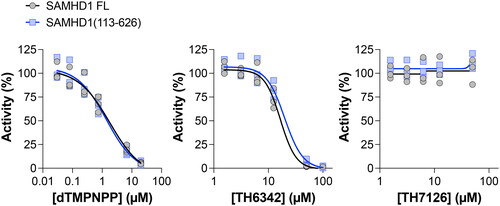
We began by producing recombinant SAMHD1 protein with an N-terminal truncation of the first 112 amino acids, resulting in a SAMHD1 variant – SAMHD1(113-626) – that lacked the SAM domain ( and ). We evaluated the dNTP hydrolase activity of SAMHD1(113-626) together with full-length wild-type SAMHD1 using a previously established and validated enzyme-coupled activity assay.[Citation6] In this assay, the inorganic triphosphate produced by SAMHD1 dNTP hydrolysis is a substrate for the coupled enzyme inorganic pyrophosphatase (PPase), included in excess in the reaction mixture, which then produces inorganic phosphate that can then be measured using an absorbance assay based on malachite green. The dGTP hydrolysis activity of SAMHD1(113-626), evaluated over a range of dGTP concentrations, was comparable to that of the full-length enzyme (). Next we verified that the non-hydrolysable dNTP analogue, 2′-deoxythymidine-5′-[(α,β)imido]triphosphate (dTMPNPP), inhibited SAMHD1(113-626), which was expected as this molecule is a competitive inhibitor occupying the catalytic site and allosteric site 2 (AS2) of SAMHD1 located within the HD domain[Citation7] (). Subsequently, we tested TH6342 and the inactive analogue control from this series, TH7126,[Citation5] in the enzyme-coupled activity assay and observed dose-dependent inhibition of dGTP hydrolysis by TH6342 with comparable inhibition observed between SAMHD1(113-626) and the full-length enzyme (). No inhibition was observed with the inactive control analogue TH7126 ().
Figure 1. SAMHD1(113-626), a truncation lacking the N-terminal SAM domain, retains dNTP hydrolysis activity. (a) Domain schematic of full-length (FL) human SAMHD1 and the truncation removing the N-terminal SAM domain (SAMHD1 113-626). (b) SDS-PAGE analysis of SAMHD1 FL and SAMHD1(113-626). Representative (n = 2) image shown. (c) SAMHD1 FL and SAMHD1(113-626) have comparable dGTP hydrolysis activity in the enzyme-coupled activity assay. Schematic of assay is shown. SAMHD1 FL and SAMHD1(113-626) were incubated with a range of dGTP concentrations for 20 min before the reaction was stopped and dGTP hydrolysis measured. Average values from 3 independent experiments (each performed in quadruplet) shown. Sigmoidal substrate-velocity curves were fitted using the allosteric enzyme kinetic model in Prism 10 (GraphPad Software).
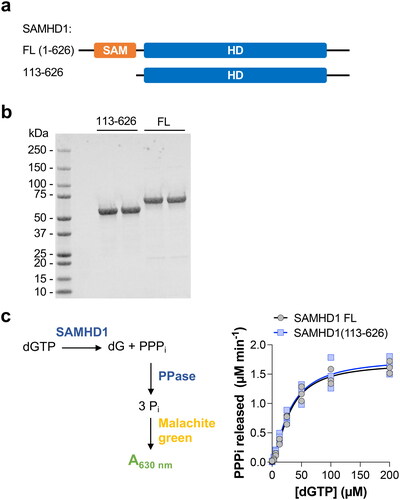
Figure 2. The N-terminal SAM domain of the dNTPase SAMHD1 is not required for inhibition by small molecule TH6342. SAMHD1 full-length (FL) or SAMHD1(113-626), the truncation lacking the N-terminal SAM domain, were incubated with a titration of non-hydrolysable dNTP analogue dTMPNPP, SAMHD1 inhibitor TH6342, or analogue control TH7126, in the enzyme coupled activity assay. Average inhibition values, normalised to solvent controls, from 3 independent experiments each performed in quadruplet shown. Dose-response curves (four-parameter, variable slope) were fitted using Prism 10 (GraphPad Software).
Altogether, the data presented in this short report demonstrate that the N-terminal SAM domain of SAMHD1 is not required for the inhibitory activity of TH6342. This indicates that the binding interface is not located in this region and is possibly within the conserved HD-domain, opening the possibility that this chemical series could target other HD-domain containing SAMHD1 orthologues,[Citation8,Citation9] which could have additional therapeutic implications.[Citation10] It should be noted that whilst we elucidated the mechanism of how TH6342 inhibits full-length SAMHD1, via deterring dimerisation,[Citation5] this remains to be evaluated for inhibition of SAMHD1(113-626), although we would anticipate the mechanism to be the same. Furthermore, these data support the use of SAMHD1(113-626) in structure-based efforts to elucidate the molecular determinants of SAMHD1 inhibition by this series of inhibitor.
3. Materials and methods
3.1. Production of recombinant his-tagged human SAMHD1(113-626)
The human SAMHD1 open reading frame, codon optimised for expression in human cells, was generated synthetically (Life Technologies). The DNA fragment encoding SAMHD1(113-626) was then PCR amplified using forward (5′-TATACATATGGACACCATGAAAGTGATCAACGAC-3′) and reverse (5′-TATAGCGGCCGCCTACATAGGGTCGTCCTTGAAC-3′) primers containing NdeI and NotI restriction sites, respectively. PCR products were then digested and sub-cloned into pET28 in frame with an N-terminal His-tag and the resulting expression construct was verified by sequencing. His-tagged SAMHD1(113-626) was expressed in Escherichia coli BL21 (DE3) T1R pRARE2 following the addition of 0.5 mM IPTG at 18 °C overnight and subsequently purified by chromatography on HisTrap HP and HiLoad 16/60 Superdex 200 columns. The purity of final fractions was assessed by SDS–PAGE. Expression and purification were performed by the Protein Science Facility at Karolinska Institutet/SciLifeLab.
3.2. SDS-PAGE analysis
To assess size and purity of the recombinant SAMHD1 enzymes, 4 µg of protein was combined with sample loading buffer (Thermo Fisher, NP0008) and reducing agent (Thermo Fisher, NP0009) before boiling at 95 °C for five minutes. Using a Tris/Glycine/SDS running buffer (BIO-RAD, #1610732), samples were run on a 4-15% gradient polyacrylamide gel (BIO-RAD, #4561085) at 200 V for 40 min with the Mini-PROTEAN Tetra system. The gel was stained with Coomassie Blue R250 solution (0.1% w/v) for 15 min and destained overnight in a shaking water bath. Imaging was performed with an Odyssey Fc imager (Li-Cor) using the 700 nm channel.
3.3. Enzyme-coupled SAMHD1 activity assay
The enzyme-coupled SAMHD1 activity assay to measure inhibition by small molecules has been described previously in a dedicated methods article.[Citation6] Briefly, 0.35 µM recombinant SAMHD1 is incubated with 25 μM dGTP (GE Healthcare, #27-1870-04) in the reaction buffer (25 mM Tris-acetate pH 8, 40 mM NaCl, 1 mM MgCl2, 0.005% Tween-20, 0.3 mM TCEP) containing 12.5 U/mL E. coli PPase at room temperature for 20 min. The reaction is then quenched by 7.9 mM EDTA (final concentration 3.95 mM), followed by addition of malachite green reagent (final concentration − 0.5 mM malachite green, 2.58 mM ammonium molybdate, 0.036% Tween-20) and incubation at room temperature for a further 20 min. Subsequently, absorption at 630 nm wavelength is measured using a Hidex Sense microplate reader. In each experiment, a standard curve of inorganic phosphate (Sigma Aldrich, # 342483) is included allowing determination of amount of phosphate released. The production of recombinant His-tagged proteins used in the assay, full-length wild-type human SAMHD1 and E. coli PPase, was performed by the Protein Science Facility at Karolinska Institutet/SciLifeLab and has been described previously.[Citation3] The non-hydrolysable dNTP analogue dTMPNPP was purchased from Jena Bioscience (#NU-907-1) as a stock solution in water. SAMHD1 inhibitor TH6342 and the inactive control analogue from this series TH7126 were previously prepared in-house (chemical synthesis described in ref[Citation5]) and stock solutions of 10 mM prepared in DMSO.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Franzolin, E.; Pontarin, G.; Rampazzo, C.; Miazzi, C.; Ferraro, P.; Palumbo, E.; Reichard, P.; Bianchi, V. The Deoxynucleotide Triphosphohydrolase SAMHD1 is a Major Regulator of DNA Precursor Pools in Mammalian Cells. Proc. Natl. Acad. Sci. U S A 2013, 110, 14272–14277. DOI: 10.1073/pnas.1312033110.
- Helleday, T.; Rudd, S. G. Targeting the DNA Damage Response and Repair in Cancer through Nucleotide Metabolism. Mol. Oncol. 2022, 16, 3792–3810. DOI: 10.1002/1878-0261.13227.
- Herold, N.; Rudd, S. G.; Ljungblad, L.; Sanjiv, K.; Myrberg, I. H.; Paulin, C. B. J.; Heshmati, Y.; Hagenkort, A.; Kutzner, J.; Page, B. D. G.; et al. Targeting SAMHD1 with the Vpx Protein to Improve Cytarabine Therapy for Hematological Malignancies. Nat. Med. 2017, 23, 256–263. DOI: 10.1038/nm.4265.
- Schneider, C.; Oellerich, T.; Baldauf, H.-M.; Schwarz, S.-M.; Thomas, D.; Flick, R.; Bohnenberger, H.; Kaderali, L.; Stegmann, L.; Cremer, A.; et al. SAMHD1 is a Biomarker for Cytarabine Response and a Therapeutic Target in Acute Myeloid Leukemia. Nat. Med. 2017, 23, 250–255. DOI: 10.1038/nm.4255.
- Zhang, S. M.; Paulin, C. B. J.; Shu, H.; Yagüe-Capilla, M.; Michel, M.; Marttila, P.; Ortis, F.; Bwanika, H. C.; Dirks, C.; Venkatram, R. P.; et al. Identification and Evaluation of Small-Molecule Inhibitors against the dNTPase SAMHD1 via a Comprehensive Screening Funnel. iScience 2024, 27, 108907. DOI: 10.1016/j.isci.2024.108907.
- Yagüe-Capilla, M.; Rudd, S. G. A High-Throughput Enzyme-Coupled Activity Assay to Probe Small Molecule Interaction with the dNTPase SAMHD1. J. Vis. Exp. 2021. DOI: 10.3791/62503.
- Morris, E. R.; Caswell, S. J.; Kunzelmann, S.; Arnold, L. H.; Purkiss, A. G.; Kelly, G.; Taylor, I. A. Crystal Structures of SAMHD1 Inhibitor Complexes Reveal the Mechanism of Water-Mediated dNTP Hydrolysis. Nat. Commun. 2020, 11, 3165. DOI: 10.1038/s41467-020-16983-2.
- Maehigashi, T.; Lim, C.; Wade, L. R.; Bowen, N. E.; Knecht, K. M.; Alvarez, N. N.; Kelly, W. G.; Schinazi, R. F.; Kim, D.-H.; Xiong, Y.; et al. Biochemical Functions and Structure of Caenorhabditis elegans ZK177.8 Protein: Aicardi-Goutières Syndrome SAMHD1 dNTPase Ortholog. J. Biol. Chem. 2023, 299, 105148. DOI: 10.1016/j.jbc.2023.105148.
- Antequera-Parrilla, P.; Castillo-Acosta, V. M.; Bosch-Navarrete, C.; Ruiz-Pérez, L. M.; González-Pacanowska, D. A Nuclear Orthologue of the dNTP Triphosphohydrolase SAMHD1 Controls dNTP Homeostasis and Genomic Stability in Trypanosoma brucei. Front. Cell. Infect. Microbiol. 2023, 13, 1241305. DOI: 10.3389/fcimb.2023.1241305.
- Yagüe-Capilla, M.; Castillo-Acosta, V. M.; Bosch-Navarrete, C.; Ruiz-Pérez, L. M.; González-Pacanowska, D. A Mitochondrial Orthologue of the dNTP Triphosphohydrolase SAMHD1 is Essential and Controls Pyrimidine Homeostasis in Trypanosoma brucei. ACS Infect. Dis. 2021, 7, 318–332. DOI: 10.1021/acsinfecdis.0c00551.
