Abstract
Objectives: Antiretroviral regimen switching may be considered for HIV-1-infected, virologically-suppressed patients to enable treatment simplification or improve tolerability, but should be guided by knowledge of pre-existing drug resistance. The current study examined the impact of pre-existing drug resistance mutations on virologic outcomes among virologically-suppressed patients switching to Rilpivirine (RPV)/emtricitabine (FTC)/tenofovir disoproxil fumarate (TDF).
Methods: SPIRIT was a phase 3b study evaluating the safety and efficacy of switching to RPV/FTC/TDF in virologically-suppressed HIV-1-infected patients. Pre-existing drug resistance at baseline was determined by proviral DNA genotyping for 51 RPV/FTC/TDF-treated patients with known mutations by historical RNA genotype and matched controls and compared with clinical outcome at Week 48.
Results: Drug resistance mutations in protease or reverse transcriptase were detected in 62.7% of patients by historical RNA genotype and in 68.6% by proviral DNA genotyping at baseline. Proviral DNA sequencing detected 89% of occurrences of NRTI and NNRTI resistance-associated mutations reported by historical genotype. Mutations potentially affecting RPV activity, including E138A/G/K/Q, Y181C, and H221Y, were detected in isolates from 11 patients by one or both assays. None of the patients with single mutants had virologic failure through Week 48. One patient with pre-existing Y181Y/C and M184I by proviral DNA genotyping experienced virologic failure. Nineteen patients with K103N present by historical genotype were confirmed by proviral DNA sequencing and 18/19 remained virologically-suppressed.
Discussion: Virologic success rates were high among virologically-suppressed patients with pre-existing NRTI and NNRTI resistance-associated mutations who switched to RPV/FTC/TDF in the SPIRIT study. While plasma RNA genotyping remains preferred, proviral DNA genotyping may provide additional value in virologically-suppressed patients for whom historical resistance data are unavailable.
Introduction
With the current availability of numerous safe and effective antiretroviral therapy (ART) options, most HIV-1-infected patients are able to be successfully treated and achieve sustained virologic suppression. Regimen switching may be desirable in the context of virologic suppression in order to benefit from advantages offered by newly available therapies including regimen simplification or improvements in tolerability.Citation1,2 Because maintenance of virologic suppression is essential when considering a regimen switch, the complete resistance profile of the patient should be used to guide next-line regimen selection. However, because routine genotyping of viral RNA present in plasma usually requires a viral load of ≥500 copies/mL, it is not possible to use these resistance assays in patients with very low or undetectable levels of circulating HIV-1 RNA. Clinicians must therefore rely on the patient’s medical records and historical HIV-1 RNA genotyping results to determine if resistance mutations may be archived in the HIV-1 reservoir that could potentially re-emerge under selective drug pressure of a new ART regimen. If resistance data are unavailable or incomplete, clinicians may be left to infer resistance from the patient’s treatment history, potentially increasing the risk of virologic failure if a suboptimal regimen is inadvertently chosen. Alternatively, if uncertainty exists about a patient’s prior drug resistance, clinicians may elect not to switch the patient from their current suppressive regimen, potentially resulting in missed opportunities for increasing ART convenience or tolerability.
Sequencing the integrated proviral HIV-1 DNA present in infected cells of individuals with undetectable plasma HIV-1 RNA levels can provide information about resistance-associated mutations acquired in the past and archived in the HIV-1 reservoir. Circulating peripheral blood mononuclear cells (PBMCs) are a source of cellular proviral DNA that can be used for genotyping to detect previously transmitted or emergent drug resistance mutations in virologically-suppressed patients.Citation3−Citation5 Proviral DNA sequencing may be a valuable tool when used in combination with or in the absence of historical RNA genotype data to guide regimen switching in the setting of virologic suppression.
Rilpivirine (RPV)/emtricitabine (FTC)/tenofovir disoproxil fumarate (TDF) [Complera®/Eviplera®] single-tablet regimen (STR) is a well-tolerated therapeutic option for the treatment of HIV-1 infection. In Europe, RPV/FTC/TDF is indicated for the treatment of HIV-1-infected adults without known resistance mutations to the components of RPV/FTC/TDF and with a viral load ≤100,000 HIV-1 RNA copies/mL.Citation6 In the US RPV/FTC/TDF is approved for use in HIV-1-infected treatment-naïve adults with baseline viral load ≤100,000 copies/mL and in virologically-suppressed adults to replace their current regimen.Citation7 The Switching Boosted PI to Rilpivirine in Combination with Truvada as a Single-Tablet Regimen (SPIRIT) study was a phase 3b trial to evaluate the safety and efficacy of switching to the RPV/FTC/TDF STR compared with remaining on a multi-pill regimen of a ritonavir (RTV)-boosted protease inhibitor (PI) and two nucleos(t)ide reverse transcriptase inhibitors (NRTIs) in virologically-suppressed HIV-1-infected patients. The primary endpoint of non-inferiority for the proportion of patients with HIV-1 RNA <50 copies/mL at Week 24 for RPV/FTC/TDF vs. PI + RTV + 2 NRTIs was met, with response rates of 93.7% vs. 89.9%, respectively [3.8% treatment difference; 95% confidence interval (CI): −1.6% to 9.1%]. Through Week 48, 89.3% of participants in the group who switched to RPV/FTC/TDF at baseline maintained virologic suppression. Rates of resistance development were low with 0.9% (4/469) of patients who switched to RPV/FTC/TDF and 0.6% (1/159) who maintained their PI-based regimen developing genotypic and/or phenotypic resistance to a study drug.Citation8
The aim of the current study was to determine the impact of pre-existing drug resistance mutations on virologic outcomes in a subset of patients enrolled in the SPIRIT study. Drug resistance mutations documented in historical genotypes were compared to those detected in the PBMC-derived proviral HIV-1 DNA at baseline. Finally, PBMC HIV-1 DNA genotypes along with plasma HIV-1 RNA genotypes in virologic failure patients were analyzed to evaluate the evolution of the HIV-1 viral population over time.
Methods
Study Population
HIV-1-infected individuals included in this study were enrolled in the SPIRIT trial, which has been previously described (GS-US-264-0106; ClinicalTrials.gov Identifier NCT01252940).Citation8 Briefly, SPIRIT was a prospective, randomized, open-label, 48-week study to evaluate the safety and efficacy of switching from a RTV-boosted PI-based (PI + RTV + 2NRTIs) regimen to the RPV/FTC/TDF STR among HIV-1- infected patients who had been virologically-suppressed (plasma HIV-1 RNA levels <50 copies/mL) for at least six months prior to screening.
Enrollment criteria included no history of virologic failure and have undergone genotypic testing prior to starting initial ART with no documented resistance to any of the study drugs including the reverse transcriptase (RT) resistance mutations K65R, K101E/P, E138G/K/R/Q, Y181C/I/V, M184V/I, or H221Y by historical genotype.Citation8 E138A was not an exclusionary RPV-associated resistance mutation at the time of patient enrollment in this study. PBMCs were collected at Baseline, Week 24, Week 48, and early study drug discontinuation (ESDD) visits and stored frozen.
Subset Selection
Patients selected for this analysis included RPV/FTC/TDF-treated patients with K103N present in their historical genotypes that had baseline PBMC samples available along with control patients who lacked the K103N mutation by historical genotype and were matched on baseline CD4 cell count. RPV/FTC/TDF-treated patients with other known RT mutations present in their historical genotypes or who experienced virologic failure with resistance development during the study were also analyzed.
Historical Plasma RNA Genotypes
Historical genotypes were required for all patients and consisted of commercially available and custom assays. Results were reported using a variety of formats, resistance algorithms, and languages. All assays generated genotypic information for the protease (PR) and RT genes, although the specific length of the region sequenced was not always reported. Most assays reported the presence of primary drug resistance mutations to NRTIs, non-nucleoside RT inhibitors (NNRTIs), and PIs that were approved at the time the assays were performed using assay-specific resistance algorithms. Some assays also reported polymorphic mutations for PR and RT.
DNA Genotyping and Data Analysis
Proviral HIV-1 DNA genotyping was performed retrospectively on frozen PBMC samples using a prototype proviral DNA sequencing assay (Monogram Biosciences, South San Francisco, CA). Genomic DNA from purified PBMCs was extracted using the MagNA Pure LC instrument and DNA Isolation Kit (Roche). Nested PCR reactions were performed on the extracted nucleic acid to isolate HIV pol sequences. The resulting 3.2 kb amplicons were purified and sequencing-ready libraries were prepared using the Nextera XT DNA Sample Preparation Kit (Illumina). Samples then underwent 2 × 250 bp paired-end sequencing on the Illumina MiSeq. Resulting FASTQ files were processed through a custom-designed automated bioinformatics analysis pipeline. Briefly, reads were quality trimmed, overlapping paired-end reads were joined, and reads were aligned to a reference sequence (NL43 GenBank: KM390026.1) in a codon-aware manner. Alignments were checked to determine minimal quality metrics: coverage >1000× at all positions, >Q30 average phred score at all positions. Aligned reads were then individually inspected for evidence of APOBEC3G/3F-induced G to A hypermutation using a naïve Bayes classification model.Citation9−Citation13 Hypermutated reads were removed and not used for variant detection. Variants were determined in a codon by codon manner and all SNPs and amino acid variants present at >10% were reported.
Plasma RNA Genotyping and Viral Load Assays
For post-baseline resistance analyses of viral isolates from patients who experienced virologic failure, PR/RT genotyping (population sequencing) and phenotyping were performed on plasma samples using the PhenoSense GT™ assay (Monogram Biosciences, South San Francisco, CA).Citation14 Viral load (HIV-1 RNA copies/mL) was assessed using COBAS AMPLICOR Monitor Version 1.5 (Roche Diagnostics, Basel, Switzerland).
Results
Baseline Characteristics
A total of 51 PBMC samples collected at baseline from patients enrolled in the SPIRIT study were analyzed by proviral DNA genotyping and compared with historical RNA genotype results. Of the 24 RPV/FTC/TDF-treated patients with K103N reported in their historical genotypes,Citation8 19 had baseline PBMC samples available and were included in this analysis. Twenty-three RPV/FTC/TDF-treated patients who lacked the K103N mutation by historical genotype and were matched on baseline CD4 cell count served as control patients. Five RPV/FTC/TDF-treated patients with other known RPV-associated mutations present in their historical genotypes and four RPV/FTC/TDF-treated patients who experienced virologic failure with resistance development during the study were also analyzed. Baseline characteristics of the patients who had their PBMC provirus analyzed are presented in Table . The 51 virologically-suppressed patients were mostly male (94.1%) and white (76.5%) with a median age of 42. At baseline, individuals had been on treatment for a median of 2.4 years, 98% had HIV-1 RNA <50 copies/mL, and the median CD4 cell count was 537 cells/uL, similar to the overall study population.Citation8 Prior to initiating ART, 51.0% (26/51) of patients had HIV-1 RNA ≤100,000 copies/mL and 58.8% (30/51) had a CD4 cell count of >200 cells/uL. Historical genotype dates for these patients ranged from 5 December 2002 to 21 September 2010 and with a median date of just over 3 years prior to the baseline visit.
Table 1 Baseline characteristics of patients analyzed by proviral DNA genotyping (N = 51).
HIV-1 Subtypes
HIV-1 subtypes were determined by historical RNA and proviral DNA genotype assays. The majority of patients were classified as subtype B by both historical RNA (60.8%) and proviral DNA (96.1%) genotyping. Patients for whom HIV-1 subtype information was not provided by historical RNA genotype (37.3%, 19/51) were found to be mostly subtype B by proviral DNA genotype (18/19 cases). One patient was determined to have subtype D by proviral DNA genotyping. One patient had subtype AG by both proviral DNA and historical RNA genotyping.
Resistance Mutations Detected in Historical RNA Genotypes and Proviral DNA at Baseline
Drug resistance mutations in PR or RT were detected in 62.7% (32/51) of patient samples by historical RNA genotype and in 68.6% (35/51) by proviral DNA genotyping (Table ). NNRTI-associated resistance mutations were detected in 56.9% (29/51) of patient samples by historical RNA genotype and 64.7% (33/51) using proviral DNA genotyping, while NRTI-associated resistance mutations were detected in 9.8% (5/51) of patient samples by historical RNA genotype and 11.8% (6/51) using proviral DNA genotyping. Primary PI-associated resistance mutations were detected in 5.9% (3/51) of patient samples by historical RNA genotype and 7.8% (4/51) by proviral DNA sequencing. Differences between assays in the number of patient samples with resistance mutations detected were not statistically significant (p > 0.05 for all classes of resistance mutations).
Table 2 Protease and reverse transcriptase drug resistance mutations detected by proviral DNA and historical RNA genotypes at baseline
NNRTI- and NRTI-associated drug resistance mutations detected by proviral DNA or historical RNA genotype are listed in Table . At the patient level, concordance of NNRTI- or NRTI-associated mutations by historical RNA and proviral genotyping was observed in 34 of 51 patient samples (66.7%), where 16 patients had no mutations detected by either assay and 18 patients had the same mutations present by both assays. Historical RNA genotyping detected a greater number of mutations for 3 patients, proviral DNA genotyping detected a greater number of mutations for 12 patients, and there were 2 patient samples in which each assay detected mutations not present by the other. At the mutational level, of the 46 occurrences of 14 unique resistance-associated NNRTI- or NRTI-associated substitutions reported by historical RNA genotyping, 41 were also detected by proviral DNA genotyping (89.1% concordance) in the same samples. Mutation occurrences were reported by historical RNA genotype, but were not detected by proviral DNA genotype in five instances, including one unique substitution (K70R). Conversely, proviral DNA genotyping detected an additional 21 mutation occurrences (including nine unique substitutions) that were absent by historical RNA genotyping. Differences between assays in the number of occurrences of each mutation detected were not statistically significant (p > 0.05). More mutations detected by proviral DNA genotyping were reported as mixtures with wild-type (18/62, 29.0%) compared to historical RNA genotyping (8/46, 17.4%).
Table 3 Week 48 virologic outcomes of patients with pre-existing NRTI or NNRTI resistance mutations by baseline proviral DNA or historical RNA genotype
Mutations associated with reduced susceptibility to RPV, including E138A/G/K/Q, Y181C, and H221Y in RT, were detected in 11 patients by one or both assays. There were four patients with mutations at position 138 in RT by historical genotyping (3 E138A and 1 E138Q); all four mutations were confirmed by proviral DNA genotyping. In addition, three more patients had E138 substitutions (1 E138A, 1 E138G, and 1 E138K) detected by proviral DNA genotyping that were not listed in their historical genotypes. All patients with E138 mutations were virologic successes at Week 48 except for the patient with E138Q who discontinued due to a protocol violation (exclusion mutation) at Week 12 with HIV-1 RNA <50 copies/mL and therefore had no data available in the Week 48 window. One patient had H221Y by historical genotype, which was confirmed by proviral DNA sequencing. This patient also discontinued due to a protocol violation (exclusion mutation) at Week 24 while suppressed (HIV-1 RNA <50 copies/mL) and had no data in the Week 48 window. One additional patient was found to have H221H/Y in their proviral DNA that was not present by historical genotype. This patient had missing data in the Week 48 window due to a missed visit but had HIV-1 RNA <50 copies/mL at their last study visit (Week 36). Two patients had Y181C present by proviral DNA genotyping but not by historical RNA genotype; one was a virologic success at Week 48. The other patient had M184I in addition to Y181Y/C by proviral DNA genotyping that was not present in their historical RNA genotype and experienced virologic failure prior to Week 8.
Of the 19 patients with K103N present by historical genotype, all 19 were confirmed to have K103N by proviral DNA sequencing. One additional patient who did not have K103N by historical RNA genotype was found to have K103N by proviral DNA genotype. No patient isolates had the combination of K103N + L100I by either assay. Most patients with pre-existing K103N by proviral DNA or historical RNA genotype were virologic successes at Week 48 (90%, 18/20). One patient had no data available in the Week 48 window but remained suppressed with HIV-1 RNA <50 copies/mL at their last study visit (Week 36). One additional patient who also had V179I detected by both assays experienced virologic failure with emergent resistance at Week 48.
Resistance Mutations Detected by RNA and Proviral DNA Genotyping in Virologic Failure Patients
Through Week 48, there were four RPV/FTC/TDF-treated patients who experienced virologic failure with resistance development detected by plasma RNA genotyping despite having no documented resistance to RPV, FTC, or TDF in their historical RNA genotypes. As described above, one of these patients was found to have the exclusion mutations Y181Y/C and M184I at baseline by proviral DNA sequencing (Figure A). According to their medical history, this patient had been diagnosed with HIV infection 12 years prior to starting treatment and had been on ART for approximately 8 years at the time of study enrollment. However, the historical genotype report for this patient was dated five and a half years prior to the baseline visit, indicating that active viral replication was ongoing at this time, the cause of which is unknown. This patient was discontinued from the SPIRIT study due to protocol violations (taking three NRTIs at study enrollment, more than two previous antiretroviral regimens) and had unconfirmed virologic rebound (single point of HIV-1 RNA of 410 copies/mL) at their early discontinuation visit prior to Week 8. At the virologic failure time point, M184I, but not Y181C, was detectable by plasma RNA genotyping. The PBMC sample from the virologic failure time point failed to amplify in the proviral DNA assay and therefore no results are available.
Figure 1 (A–D) HIV-1 RNA (copies/mL) curves and genotyping results for the four RPV/FTC/TDF-treated patients who experienced virologic failure with emergent resistance. Historical genotype results and timing prior to study enrollment are shown in black boxes. Plasma RNA genotype results from visits on study drug are indicated in white boxes. Proviral DNA genotype results from PBMCs collected during the study are denoted in gray boxes. The baseline visit is indicated as Week 0 with the screening time point occurring within 4 weeks prior. (*) sample was collected after the patient discontinued study drug; (**) result was obtained using an earlier version of the proviral DNA assay that used population sequencing.
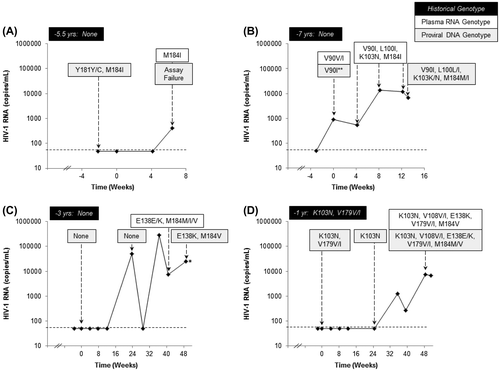
The second patient also had no documented resistance by historical RNA genotype, but entered the study with a detectable HIV-1 RNA of 896 copies/mL at the baseline visit, which continued to increase through Week 12 (Figure B). Plasma RNA genotyping at baseline showed the V90V/I mutation in RT that was confirmed by the proviral DNA assay. Plasma RNA genotyping detected V90I along with emergent L100I, K103N, and M184I at Weeks 4, 8, and 12. Genotyping of PBMCs collected at the ESDD visit just after Week 12 confirmed the presence of these same mutations in the proviral DNA with all but V90I occurring as mixtures with wild-type.
A third patient with no documented resistance at study entry by both historical RNA genotype and proviral DNA genotype at baseline experienced confirmed virologic rebound with emergent E138E/K and M184M/I/V by plasma RNA genotyping at the Week 36 retest (Figure C). The E138K and M184V mutations were also detected by proviral DNA genotyping in the Week 48 PBMC sample, which was collected two months after the patient had discontinued study drug.
The fourth RPV/FTC/TDF-treated virologic failure patient had pre-existing K103N and V179V/I present by both historical RNA and proviral DNA genotyping at baseline (Figure D). Proviral DNA genotyping of PBMCs collected at Week 24 while the patient was still suppressed (HIV-1 RNA <50 copies/mL) on study drug showed the presence of K103N only. This patient subsequently experienced confirmed virologic rebound with K103N, V179V/I, and emergent V108V/I, E138K, and M184V detected in the plasma RNA genotype at Week 48. These same mutations were also detected in the proviral DNA at Week 48 with E138E/K and M184M/V present as mixtures with wild-type.
Discussion
Virologic success rates were high among virologically-suppressed patients with pre-existing NRTI or NNRTI resistance-associated mutations who switched to RPV/FTC/TDF in the SPIRIT study, consistent with the overall study results.Citation8 The lower rate of virologic failure with resistance development observed in the SPIRIT study compared to studies of RPV/FTC/TDF or its components in treatment-naïve patients is likely attributable to the reduced viral burden in virologically-suppressed patients.Citation15,16 Seven patients with pre-existing RPV-associated mutations detected by historical RNA or proviral DNA genotyping (four E138A, one E138G, one E138K, and one Y181C) remained virologically-suppressed through Week 48 after switching to RPV/FTC/TDF. Two additional patients with pre-existing RPV-associated resistance mutations (one E138Q and one H221Y) were discontinued from the study once the protocol-defined resistance mutation was discovered in their historical genotypes, but maintained virologic suppression on RPV/FTC/TDF treatment until discontinuation. Lastly, one patient with H221H/Y by proviral DNA sequencing had missing data in the Week 48 visit window, but remained virologically-suppressed on RPV/FTC/TDF treatment through Week 36. Maintenance of virologic suppression after switching to RPV/FTC/TDF in patients despite pre-existing RPV-associated resistance mutations is notable, particularly since little to no data currently exist regarding clinical outcomes among treatment naïve patients with similar pre-existing mutations from other studies of RPV/FTC/TDF or its components.Citation15,16
Proviral DNA sequencing confirmed the presence of K103N in all patients with K103N reported by historical RNA genotype and also detected one additional occurrence of pre-existing K103N. As previously reported, the presence of pre-existing K103N did not impact virologic outcome for patients switching to RPV/FTC/TDF as most patients were virologic successes.Citation8 These results were confirmed by another published report that documented the successful switch to RPV/FTC/TDF in a small number of virologically-suppressed patients with viral isolates containing K103N.Citation17
Among the four patients treated with RPV/FTC/TDF who experienced virologic failure with resistance development, none had RPV/FTC/TDF resistance-associated mutations present at baseline by historical genotype. Proviral HIV-1 DNA sequencing revealed that one patient had archived Y181Y/C and M184I at baseline with M184I detectable by plasma RNA genotyping at study discontinuation. While the contribution of pre-existing resistance mutations to this patient’s virologic failure is unclear, this patient would have been ineligible for the SPIRIT study if the proviral DNA sequencing results been available at the time of screening. This patient had been diagnosed with HIV infection ~20 years prior to study enrollment and had a complex treatment history with at least one documented period of active viral replication since ART initiation. Since Y181C and M184I are not commonly transmitted drug resistance mutations and were not present on this patient’s historical genotype, it is possible that the patient’s medical history collected at study entry was incomplete and additional information regarding prior treatment would shed some light on this patient’s virologic outcome. The three other patients who experienced virologic failure with treatment-emergent resistance while receiving RPV/FTC/TDF had proviral DNA sequencing results that agreed with their historical RNA genotype results (no RPV/FTC/TDF-associated mutations). All three patients developed RPV and/or FTC-associated resistance mutations while on study treatment that were detected by both plasma RNA and proviral DNA genotyping at the virologic failure time point(s). The resistance mutations that emerged in these patients were consistent with those seen in other studies of RPV/FTC/TDF or its components.Citation15,16
The results of the current study suggest that while certain archived RPV/FTC/TDF-associated resistance mutations may contribute to virologic failure in suppressed patients switching to RPV/FTC/TDF, some patients may be able to maintain virologic suppression in the presence of pre-existing mutations. The only patient with key resistance mutations to both the RPV and FTC components of RPV/FTC/TDF in their proviral DNA (Y181C + M184I) went on to experience virologic failure, indicating that patients with pre-existing resistance to more than one drug within the regimen may have an increased risk of failure upon switching to RPV/FTC/TDF. Conversely, among the small number of patients with a single RPV-associated pre-existing resistance mutation, there were no incidents of virologic failure after switching to RPV/FTC/TDF demonstrating that a sustained virologic response may be possible in a more limited background of drug resistance. It is also possible that the ability to maintain virologic suppression despite archived mutations may vary from patient to patient depending on individual immune system characteristics or other factors. Our results are consistent with findings from another study that showed that some patients with RPV/FTC/TDF-associated resistance mutations by historical genotype were able to maintain virologic suppression after switching to RPV/FTC/TDF, while others experienced virologic failure.Citation18 However, our conclusions are limited by the small number of patients who experienced virologic failure in the SPIRIT study as well as the small number of patients with pre-existing RPV/FTC/TDF-associated resistance mutations enrolled in the trial. More data are needed to determine the effect of pre-existing RPV/FTC/TDF-associated mutations on virologic outcomes among virologically-suppressed patients switching to RPV/FTC/TDF.
Proviral DNA sequencing at the time of regimen switch detected most (89%) of the NRTI- and NNRTI-associated resistance mutations reported in historical RNA genotypes among virologically-suppressed patients in this subgroup analysis of the SPIRIT study. The number of baseline patient samples in which PI or RT resistance-associated mutations were detected was also similar using both methods. However, some resistance mutations were exclusively detected by either proviral DNA or plasma RNA genotyping, and overall, proviral DNA genotyping detected a greater number of total mutations than historical RNA genotypes. The assay concordance of proviral DNA genotyping with plasma RNA genotyping in the current study is generally consistent with that seen in other published studies.Citation19−Citation27 Of note, other studies of proviral DNA genotyping in virologically-suppressed patients have typically detected more mutations by historical plasma RNA genotyping compared to proviral DNA sequencing. The relatively greater number of mutations detected by proviral DNA sequencing in our study may be attributed to differences in drug resistance reporting in the historical genotypes collected. Significant variation between genotyping assays in the level of detail provided in the reports as well as changes in HIV-1 drug resistance algorithms over time may have resulted in the under-reporting of certain mutations, particularly RPV-associated resistance mutations, as RPV was not an approved NNRTI at the time that the historical genotypes were performed.
More data regarding clinical outcomes associated with regimen switching among virologically-suppressed patients with pre-existing drug resistant isolates by historical RNA or proviral DNA genotyping are needed. To date, there have been no large clinical studies prospectively evaluating the predictive value of proviral DNA sequencing prior to switching in order to identify patients appropriate for a regimen switch. Previous studies have retrospectively analyzed patient proviral DNA to evaluate the prevalence of RPV/FTC/TDF-associated resistance mutations in patients treated with regimens other than RPV/FTC/TDF, but these studies lack virologic outcome data in patients that have actually been treated with RPV/FTC/TDF.Citation28,29 While plasma RNA genotyping remains the drug resistance assay of choice, proviral DNA genotyping may provide value as an additional source of information on the total burden of drug resistance present in virologically-suppressed patients, particularly for those who lack a historical RNA genotype and/or have a complex treatment history.
Conflicts of interest
Conflicts of interest: DPP, RK, KA, SKC, MDM, and KLW are employees and shareholders of Gilead Sciences, Inc. JT, YT, OS, SC, and YL are employees of Monogram Biosciences. FP is a consultant and/or on the Speakers’ Bureau for the following: Gilead Sciences, Janssen Pharmaceuticals, Merck, and Bristol Myers Squibb. ClinicalTrials.gov Identifier NCT01252940
Funding
This work was supported by Gilead Sciences, Inc.
Notes
* The SPIRIT study was conducted in accordance with the Declaration of Helsinki. The protocol was reviewed and approved by central of site-specific institutional review boards or ethics committees. All participants gave written informed consent.
References
- Panel on Antiretroviral Guidelines for Adults and Adolescents. Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents. http://aidsinfo.nih.gov/contentfiles/lvguidelines/AdultandAdolescentGL.pdf. Accessed April 8, 2015.
- Huldrych F, Gunthard HF, Aberg JA, et al. Antiretroviral treatment of adult HIV infection: 2014 recommendations of the International Antiviral Society-USA panel. JAMA. 2014;312:410–425.
- Verhofstede C, Noë A, Demecheleer E, et al. Drug-resistant variants that evolve during nonsuppressive therapy persist in HIV-1-infected peripheral blood mononuclear cells after long-term highly active antiretroviral therapy. J Acquir Immune Defic Syndr. 2004;35:473–483.10.1097/00126334-200404150-00005
- Palmisano L, Galluzzo CM, Giuliano M. The importance of testing genotypic resistance in proviral DNA of patients fully responding to highly active antiretroviral therapy. J Acquir Immune Defic Syndr. 2009;51:233–234.10.1097/QAI.0b013e3181a5b247
- Chew CB, Potter SJ, Wang B, et al. Assessment of drug resistance mutations in plasma and peripheral blood mononuclear cells at different plasma viral loads in patients receiving HAART. J Clin Virol. 2005;33:206–216.10.1016/j.jcv.2004.11.006
- Gilead Sciences Ltd. EVIPLERA 200 mg/25 mg/245 mg film coated tablets. Summary of Product Characteristics. Cambridge, UK. Revised December 2013; 2013.
- Gilead Sciences Inc. COMPLERA® (emtricitabine/rilpivirine/tenofovir disoproxil fumarate) tablets. US Prescribing Information. Foster City, CA. Revised June 2014; 2014.
- Palella FJ, Fisher M, Tebas P, et al. Simplification to rilpivirine/emtricitabine/tenofovir disoproxil fumarate from ritonavir-boosted protease inhibitor antiretroviral therapy in a randomized trial of HIV-1 RNA-suppressed participants. AIDS. 2014;28:335–344.10.1097/QAD.0000000000000087
- Armitage AE, Katzourakis A, de Oliveira T, et al. Conserved footprints of APOBEC3G on hypermutated human immunodeficiency virus type 1 and human endogenous retrovirus HERV-K(HML2) sequences. J Virol. 2008;82:8743–8761.10.1128/JVI.00584-08
- Piantadosi A, Humes D, Chohan B, McClelland RS, Overbaugh J. Analysis of the percentage of human immunodeficiency virus type 1 sequences that are hypermutated and markers of disease progression in a longitudinal cohort, including one individual with a partially defective Vif. J Virol. 2009;83:7805–7814.10.1128/JVI.00280-09
- Knoepfel SA, Di Giallonardo F, Däumer M, Thielen A, Metzner kJ. In-depth analysis of G-to-A hypermutation rate in HIV-1 env DNA induced by endogenous APOBEC3 proteins using massively parallel sequencing. J Virol Methods. 2011;171:329–338.10.1016/j.jviromet.2010.11.016
- Kieffer TL, Kwon P, Nettles RE, Han Y, Ray SC, Siliciano RF. G->A hypermutation in protease and reverse transcriptase regions of human immunodeficiency virus type 1 residing in resting CD4+ T cells in vivo. J Virol. 2005;79:1975–1980.10.1128/JVI.79.3.1975-1980.2005
- Armitage AE, Deforche K, Chang CH, et al. APOBEC3G-induced hypermutation of human immunodeficiency virus type-1 is typically a discrete “all or nothing” phenomenon. PLoS Genet. 2012;8:e1002550.10.1371/journal.pgen.1002550
- Petropoulos CJ, Parkin NT, Limoli KL, et al. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob Agents Chemother. 2000;44:920–928.10.1128/AAC.44.4.920-928.2000
- Porter DP, Kulkarni R, Fralich T, Miller MD, White KL. 96-week resistance analyses of the STaR study: Rilpivirine/emtricitabine/tenofovir DF versus efavirenz/emtricitabine/tenofovir DF in antiretroviral-naive, HIV-1-infected subjects. HIV Clin Trials. 2015;16:30–38.10.1179/1528433614Z.0000000009
- Rimsky L, Van Eygen V, Hoogstoel A, et al. 96-week resistance analyses of rilpivirine in treatment-naive, HIV-1-infected adults from the ECHO and THRIVE Phase III trials. Antivir Ther. 2013;18:967–977.
- Rokx C, Verbon A, Rijnders B. Successful switch to rilpivirine/tenofovir/emtricitabine in HIV-1-infected patients with an isolated K103N mutation acquired during prior nonnucleoside reverse transcriptase inhibitor therapy. HIV Med. 2014;15:611–614.
- Amiel C, Schneider V, Guessant S, et al. Initiation of rilpivirine, tenofovir and emtricitabine (RPV/TDF/FTC) regimen in 363 patients with virological vigilance assessment in ‘real life’. J Antimicrob Chemother. 2014;69:3335–3339.10.1093/jac/dku294
- Kabamba-Mukadi B, Duquenne A, Henrivaux P, et al. HIV-1 proviral resistance mutations: usefulness in clinical practice. HIV Med. 2010;11:483–492.
- Ghosn J, Pellegrin I, Goujard C, et al. HIV-1 resistant strains acquired at the time of primary infection massively fuel the cellular reservoir and persist for lengthy periods of time. AIDS. 2006;20:159–170.10.1097/01.aids.0000199820.47703.a0
- Wirden M, Soulie C, Valantin MA, et al. Historical HIV-RNA resistance test results are more informative than proviral DNA genotyping in cases of suppressed or residual viraemia. J Antimicrob Chemother. 2011;66:709–712.10.1093/jac/dkq544
- Delaugerre C, Braun J, Charreau I, et al. Comparison of resistance mutation patterns in historical plasma HIV RNA genotypes with those in current proviral HIV DNA genotypes among extensively treated patients with suppressed replication. HIV Med. 2012;13:517–525.
- Saracino A, Gianotti N, Marangi M, et al. Antiretroviral genotypic resistance in plasma RNA and whole blood DNA in HIV-1 infected patients failing HAART. J Med Virol. 2008;80:1695–1706.10.1002/jmv.v80:10
- Vicenti I, Razzolini F, Saladini F, Romano L, Zazzi M. Use of peripheral blood DNA for genotype antiretroviral resistance testing in drug-naive HIV-infected subjects. Clin Infect Dis. 2007;44:1657–1661.10.1086/518287
- Turriziani O, Bucci M, Stano A, et al. Genotypic resistance of archived and circulating viral strains in the blood of treated HIV-infected individuals. J Acquir Immune Defic Syndr. 2007;44:518–524.10.1097/QAI.0b013e3180315515
- Geretti AM, Conibear T, Hill A, et al. Sensitive testing of plasma HIV-1 RNA and Sanger sequencing of cellular HIV-1 DNA for the detection of drug resistance prior to starting first-line antiretroviral therapy with etravirine or efavirenz. J Antimicrob Chemother. 2014;69:1090–1097.10.1093/jac/dkt474
- Derache A, Shin HS, Balamane M, et al. HIV drug resistance mutations in proviral DNA from a community treatment program. PLOS One. 2015;10:e0117430.10.1371/journal.pone.0117430
- Gallien S, Charreau I, Nere ML, et al. Archived HIV-1 DNA resistance mutations to rilpivirine and etravirine in successfully treated HIV-1-infected individuals pre-exposed to efavirenz or nevirapine. J Antimicrob Chemother. 2015;70:562–565.10.1093/jac/dku395
- Lambert-Niclot S, Allavena C, Grude M, et al. Usefulness of HIV DNA resistance genotypic test in patient candidates for a RPV/TDF/FTC combination switch [Poster 22]. Paper presented at: XXIV International Drug Resistance Workshop; February 21–22, 2015; Seattle, WA.
