Abstract
The adverse effects (diarrhea and neutropenia) of irinotecan (7-ethyl-10-[4-(1-piperidino)-1-piperidino]carbonyloxycamptothecin) are associated with genetic variants of uridine diphosphate glucuronosyltransferase 1A subfamilies (UGT1As). UGT1As are enzymes that metabolize the active form of irinotecan, 7-ethyl-10 hydroxycamptothecin (SN-38), by glucuronidation in the liver. They are widely known as predictive factors of severe adverse effects, such as neutropenia and diarrhea. Some studies have suggested that variants of UGT1As affect SN-38 glucuronidation activities, thus exerting severe adverse effects. We aimed to identify UGT1A isoforms that show SN-38 glucuronidation activity and determine the relationship between UGT1A variants and SN-38 glucuronidation in vitro. We found that UGT1A1 and UGT1A6–UGT1A10 displayed SN-38 glucuronidation activity. Among these, UGT1A1 was the most active. Furthermore, the variants of these isoforms showed decreased SN-38 glucuronidation activity. In our study, we compared the different variants of UGT1As, such as UGT1A1.6, UGT1A1.7, UGT1A1.27, UGT1A1.35, UGT1A7.3, UGT1A8.4, UGT1A10M59I, and UGT1A10T202I, to determine the differences in the reduction of glucuronidation. Our study elucidates the relationship between UGT1A variants and the level of glucuronidation associated with each variant. Therefore, testing can be done before the initiation of irinotecan treatment to predict potential toxicities and adverse effects.
Introduction
Irinotecan (7-ethyl-10-[4-(1-piperidino)-1-piperidino]carbonyloxycamptothecin) is a semisynthetic analog of the cytotoxic alkaloid camptothecin. Irinotecan is widely used for the treatment of colorectal, pancreatic, and lung cancers because of its anticancer activity (de Man et al. Citation2018). It is converted to the active metabolite 7-ethyl-10 hydroxycamptothecin (SN-38) by carboxylesterase (de Man et al. Citation2018). SN-38 is further glucuronidated to the inactive metabolite SN-38 glucuronide (SN-38G) by uridine diphosphate glucuronosyltransferase 1A (UGT1A) subfamily members (Haaz et al. Citation1997; Hanioka et al. Citation2001). In this catalytic reaction, uridine-5′-diphosphoglucuronic acid (UDPGA) acts as a co-substrate for the formation of hydrophilic glucuronides from non-membrane-associated substrates, such as steroids, bile acids, bilirubin, hormones, dietary constituents, and various drugs, environmental xenobiotics, and carcinogens (Tukey and Strassburg Citation2000). Tissue-specific expression varies among UGT1A subfamily members (Tukey and Strassburg Citation2000). UGT1A1, UGT1A3, UGT1A4, UGT1A6, and UGT1A9 are expressed in the human liver, whereas UGT1A5 is expressed in the human thymus, UGT1A7 in the human tonsil and kidney, UGT1A8 in the human bladder, and UGT1A10 is expressed in the human intestine (Izukawa et al. Citation2009; Basit et al. Citation2020; Zhang H et al. Citation2020). Moreover, substances that can be glucuronidated are different for each UGT1A subfamily member (Maruo et al. Citation2010). UGT1A1, UGT1A3, UGT1A6, UGT1A7, UGT1A8, UGT1A9, and UGT1A10 can glucuronidate SN-38 (Hanioka et al. Citation2001; Gagné Citation2002).
The adverse effects of irinotecan include neutropenia, severe diarrhea, and asthenia, depending on the dose (Haaz et al. Citation1997). UGT1A1 variants such as UGT1A1*6 (p.G71R) and UGT1A1*28 (A(TA)7TAA) are considered good predictors of severe adverse effects of irinotecan (de Man et al. Citation2018). Moreover, some studies have reported that UGT1A1*7 (p.Y486D), UGT1A1*27 (p.P229Q), UGT1A1*35 (p.L233R), and UGT1A1*60 (-3279 T > G) are also predictors of severe adverse effects (Iyer et al. Citation1999; Gagné et al. Citation2002; Chen X et al. Citation2017). Other variants of UGT1A, such as UGT1A7*3 (p.N129K;p.R131K;p.W208R), UGT1A7*4 (p.W208R), UGT1A9*2 (p.C3Y), UGT1A9*3 (p.M33T), and UGT1A9*22 (-126_ -118 T9 > T10), may also be predictors of irinotecan-induced toxicity (Gagné Citation2002; Villeneuve Citation2003; Cui Citation2016). However, many UGT1A variants, particularly UGT1A6, UGT1A8, and UGT1A10, have not been investigated sufficiently.
Here, we aimed to identify UGT1A isoforms that show glucuronidation activity toward SN-38, and the relationship between UGT1A variants and SN-38 glucuronidation in vitro. Our study provided basic information for the identification of UGT1A variants that may serve as predictors of adverse effects of irinotecan.
Materials and methods
Chemicals and reagents
UDPGA, p-aminobenzamide dihydrochloride, methanol, and ethanol were obtained from Nacalai Tesque (Kyoto, Japan). SN-38 was obtained from Tokyo Chemical Industry, Co., Ltd. (Tokyo, Japan). SN-38 glucuronide was obtained from Toronto Research Chemicals (Toronto, Ontario, Canada). Acetonitrile Plus was obtained from Kanto Chemical Co., Inc (Tokyo, Japan). Digitonin and sucrose were obtained from Wako Pure Chemicals (Osaka, Japan). Immun-Blot Polyvinylidene Fluoride Membrane, 10× Tris-Glycine buffer, 10× Tris-Glycine-SDS buffer, and 10% Mini-PROTEAN® TGX™ Precast Gels were obtained from Bio-Rad Laboratories (Hercules, CA). Phenylmethylsulphonyl fluoride (PMSF) was obtained from Merck (Darmstadt, Germany). XbaI and HindIII were obtained from Takara Bio Inc. (Kyoto, Japan). Dulbecco’s modified Eagle’s medium (DMEM), low glucose, pyruvate, fetal bovine serum (FBS), and X-tremeGENE 9 DNA transfection reagent were obtained from Sigma-Aldrich (St. Louis, MO). The Q5 Site-Directed Mutagenesis Kit was obtained from New England Biolabs (Ipswich, MA), and wildtype UGT1 plasmid vectors (UGT1A1, UGT1A3, UGT1A4, UGT1A5, UGT1A6, UGT1A7, UGT1A8, UGT1A9, and UGT1A10 cDNA inserted in the pcR3.1 vector, respectively) were used as previously described (Iwai et al. Citation2004; Mori et al. Citation2005; Takahashi et al. Citation2008; Mimura et al. Citation2011). The pCR vector was obtained from Invitrogen (Carlsbad, CA). Mouse anti-human UGT1A antibody and m-IgG κ BP-HRP were obtained from Santa Cruz Biotechnology (Dallas, TX). Mouse anti-human UGT1A antibody was prepared against epitopes in shared exons 2–5 (amino acids 234–533); this antibody recognized all UGT1A subfamily members. The UGT1A positive control (0.05 mg/mL) was obtained from Becton Dickinson and Company (Franklin Lakes, NJ).
Construction of variant UGT1A expression plasmids
Although UGT1A1, UGT1A6, UGT1A7, UGT1A8, UGT1A9, and UGT1A10 can conjugate with SN-38, only the UGT1A1 variants are considered to affect the SN-38 glucuronidation activity. We considered the relationship between the common variants of UGT1A1 and their SN-38 glucuronidation activity. We generated mutant plasmid DNAs using plasmids containing wildtype UGT1A1*1, UGT1A6*1, UGT1A7*1, UGT1A8*1, UGT1A9*1, and UGT1A10*1 (Iwai et al. Citation2004; Mori et al. Citation2005; Takahashi et al. Citation2008; Mimura et al. Citation2011). UGT1A1*28 reduces transcriptional activity and protein levels; however, it does not change amino acid composition. Therefore, we did not examine the relationship between these variants and their SN-38 glucuronidation activity. For the construction of mutant plasmids, we used the Q5 Site-Directed Mutagenesis Kit. We generated UGT1A1*6 (p.G71R), UGT1A1*7 (p.Y486D), UGT1A1*27 (p.P229Q), and UGT1A1*35 (p.L233R) using the UGT1A1*1 plasmid DNA, and UGT1A6*3 (p.S7A) using the UGT1A6*1 plasmid DNA. After generating the UGT1A6*3 plasmid DNA, we generated UGT1A6*4 (p.S7A; p.R184S), then developed UGT1A6*2 (p.S7A; p.T181A; p.R184S) plasmid DNA using the UGT1A6*4 plasmid DNA. We created a mutant plasmid DNA, UGT1A7*2 (p.N129K; p.R131K), and UGT1A7*4 (p.W208R) using the UGT1A7*1 plasmid DNA. UGT1A7*3 (p.N129K; p.R131K; p.W208R) was generated using the UGT1A7*2 plasmid DNA and primers used to generate UGT1A7*4. Similarly, we produced UGT1A8*2 (p.A173G), UGT1A8*3 (p.C277Y), and UGT1A8*4 (p.A144V; p.A173G) using UGT1A8*1 plasmid DNA, UGT1A9*2 (p.C3Y) and UGT1A9*3 (p.M33T) with UGT1A9*1 plasmid DNA, and UGT1A10M59I and UGT1A10T202I using UGT1A10*1 plasmid DNA. Mutagenesis primers were designed using NEBaseChanger™ (Supplemental Table 1). The mutant plasmid DNAs were then transfected with competent Escherichia coli cells. The transfected competent cells were spread on Luria Broth (LB) placed in 100-mm diameter Petri dishes containing 100 mg/mL ampicillin. The colonies on the LB medium were mass cultured with Terrific Broth and then purified using the Genopure Plasmid Maxi Kit (F. Hoffman-La Roche, Basel, Switzerland). Next, the products were digested using XbaI and HindIII to check whether each plasmid DNA was cloned exactly. To check whether sequence variants were generated, we used the BigDye Terminator v1.1 Cycle Sequencing Kit (Thermo Fisher Scientific, Waltham, MA) with the appropriate sequence primers (Supplemental Table 2) (Gagné et al. Citation2002).
Expression of UGT1As
At 24 h before transfection, 6 × 105 COS-7 cells in DMEM and 10% FBS were seeded into a 100-mm diameter cell culture dish. For transfection, we prepared 5 mL of DMEM with 150 μL of X-tremeGENE 9 DNA Transfection Reagent and 50 μg of each plasmid DNA. We spread this solution into a cell culture dish. The cells were incubated for 60 h in a CO2 incubator at 37 °C. After incubation, the transfected COS-7 cells were harvested using a cell scraper and stored at −80 °C with 20 μL of phosphate buffered saline (−) until further use. We added 20 μL of digitonin solution (consisting of 5.4 mg of digitonin per mL of 0.15 M sucrose and 1 mM EDTA), 1 μL of 0.1 mM p-aminobenzamide dihydrochloride, and 1 μL of 2 mM PMSF into the transfected COS-7 cells and ruptured the cells thrice using an ultrasound homogenizer (ASTRASON® Ultrasonic Liquid Processor; Misonix Inc., Farmingdale, NY) for 10 s each. We used this cell homogenate for further reaction and western blotting. We determined the amount of each UGT1A protein in the cell homogenate using immunoblotting.
Reaction of SN-38 with UGTs
Reaction mixtures (100 μL) contained 15 μL of cell homogenate, 10 μL of SN-38 (1, 2, 5, 10, and 20 mM concentrations), 10 μL of 20 mM UDPGA, 15 μL of distilled water, and 50 μL of reaction buffer that consisted of 100 mM Kpi (consisting of 100 mM NaH2PO4 and 100 mM Na2HPO4, adjusted pH 7.4) and 10 mM MgCl2. These solutions were then incubated in a 37 °C water bath for 1 and 2 h. After incubation, 200 μL of 100% ethanol was added to terminate the reaction, vortexed for 10 s, and centrifuged for 15 min at 10 000 × g. We measured the content of this supernatant using a high performance liquid chromatography (HPLC) system (LC-20 AD; Shimadzu, Kyoto, Japan). For analyses of SN-38 and SN-38G, the HPLC conditions were as follows: Wakopak Navi C30-5 column (3.0 mm × 150 mm; FUJIFILM Wako Chemicals, Osaka, Japan), flow rate of 0.8 mL/min, detection at 360 nm, and temperature of 40 °C. The mobile phase consisted of two solvents: A (0.1 M ammonium acetate at pH 4.85) and B (acetonitrile). The gradient conditions were as follows: 0–10 min, 5% B; 10–20 min, 5%–20% B; 20–25 min, 20%–5% B; 25–30 min, 5% B.
Western blotting
The cell homogenates were subjected to 10% Mini-PROTEAN® TGX™ Precast Gel electrophoresis. Proteins were then transferred onto a polyvinylidene difluoride membrane. The membrane was incubated for 1 h in blocking solution, followed by incubation for 1 h in a solution of mouse anti-human UGT1A antibody (diluted 1:2000) and 1 h in a solution of m-IgG k BP-HRP (diluted 1:10 000). The detection reagent was added, and the membrane and protein peaks were visualized using a Lumino Image Analyzer LAS4000 (Fujifilm, Tokyo, Japan). The amount of each UGT1A protein in the cell homogenate was measured using ImageJ software.
Data analysis
Kinetic parameters were calculated using GraphPad Prism 8 software (GraphPad Software Inc., San Diego, CA), and non-linear regression was performed using the Michaelis–Menten equation. All data are presented as the mean of three independent experiments. The enzyme activities of each UGT1A series and variant UGT1A series were subjected to a one-way analysis of variance (ANOVA). When the results of the one-way ANOVA were statistically significant, they were subjected to Dunnett’s multiple comparison test. First, the wildtype of each UGT1As was established as a standard; second, UGT1A1.1 was established as a standard. The statistical significance was set at p < 0.05.
Results
Expression and identification of UGT1As
Each UGT1A protein was determined using western blotting with an anti-human UGT1A antibody. All UGT1A proteins, including mutant proteins and the UGT1A-positive control, were detected at approximately 55 kDa (). First, the relative expression of wildtype UGT1As was quantified and normalized to the expression of UGT1A1.1, which was defined as 1 unit. The relative expression of UGT1A3.1, UGT1A4.1, UGT1A5.1, UGT1A6.1, UGT1A7.1, UGT1A8.1 UGT1A9.1, and UGT1A10.1 was quantified as 1.1, 0.8, 1.1, 0.6, 0.8, 0.6, 0.6, and 0.5 units, respectively. Next, the relative expression of variants of each UGT1A was quantified and normalized to the expression of wildtype of each UGT1As, which was defined as 1 unit. When the expression level of UGT1A1.1 was 1.0, the expression levels of UGT1A1.6, UGT1A1.7, UGT1A1.27, and UGT1A1.35 were 0.5, 1.0, 1.4, and 0.5 units, respectively. When the expression of UGT1A6.1 was quantified as 1.0, the expression of UGT1A6.2, UGT1A6.3, and UGT1A6.4 was expressed as 3.1, 1.9, and 1.5 units, respectively. When the expression level of UGT1A7.1 was 1.0, the expression levels of UGT1A7.2, UGT1A7.3, and UGT1A7.4 were 1.0, 0.8, and 1.2 units, respectively. When the expression level of UGT1A8.1 was 1.0, the expression levels of UGT1A8.2, UGT1A8.3, and UGT1A8.4 were 0.7, 1.1, and 1.3 units, respectively. When the expression level of UGT1A9.1 was 1.0, the expression levels of UGT1A9.2 and UGT1A9.3 were 0.5 and 1.2 units, respectively. When the expression level of UGT1A10.1 was 1.0, the expression levels of UGT1A10M59I and UGT1A10T202I were 0.5 and 0.7 units, respectively ().
Figure 1. Identification of expressed UGT1A proteins using western blotting, maximum velocity of each wildtype UGT1A protein (A), and Michaelis–Menten kinetics of SN-38 glucuronidation by the expression of each wildtype UGT1A protein (B). (A) The relative expression levels of each of the UGT1A protein were determined using western blotting with mouse anti-human UGT1A antibody and m-IgG k BP-HRP. Approximately 55-kDa protein bands were detected in all expression models and UGT1A-positive proteins. The columns indicate the maximum velocity of each wildtype of UGT1A protein for SN-38 glucuronidation. The reaction mixture contained 15 μL cell homogenate and 2 mM UDPGA; it was incubated at 37 °C for 1 h at pH 7.4. SN-38 concentrations ranged from 0.1 to 2.0 mM. (B) The line indicates the fitting of the data to the Michaelis–Menten equation using non-linear regression. The reaction mixture contained 15 μL cell homogenate and 2 mM UDPGA; it was incubated at 37 °C for 1 h at pH 7.4. SN-38 concentrations ranged from 0.1 to 2.0 mM. PC: positive control; †p < 0.05 compared with wildtype UGT1A1.
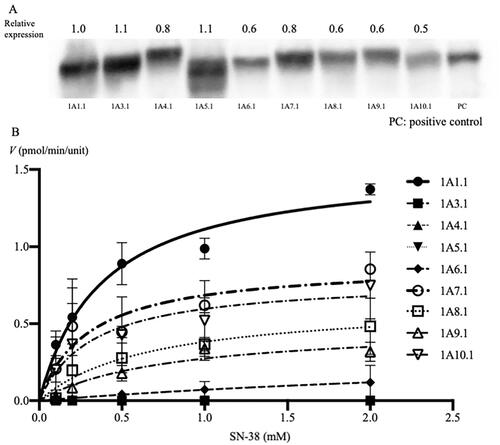
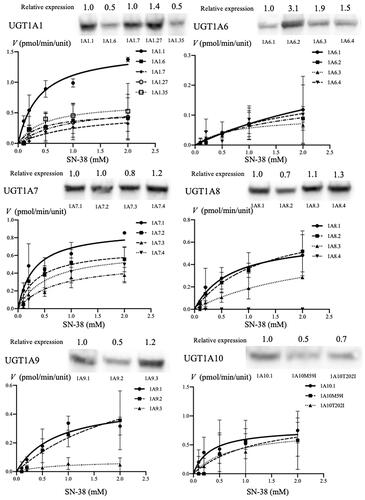
Figure 2. Identification of the expression of each of the UGT1A protein (wildtype and major variant proteins) using western blotting and Michaelis–Menten kinetics of SN-38 glucuronidation, expressed by each wildtype and major variants of UGT1A proteins. The relative expression levels of each of the UGT1A protein were determined using western blotting with mouse anti-human UGT1A antibody and m-IgG k BP-HRP. Approximately 55-kDa protein bands were detected in all wildtype and polymorphic UGT1A proteins. The line indicates the fitting of the data to the Michaelis–Menten equation using non-linear regression. The reaction mixture contained 15 μL cell homogenate and 2 mM UDPGA; it was incubated at 37 °C for 1 h at pH 7.4. SN-38 concentrations ranged from 0.1 to 2.0 mM.
Activities of glucuronidation for SN-38
The HPLC results showed that the peaks of SN-38G were detected when reacted with SN-38 and cell homogenate of UGT1A1.1, UGT1A6.1, UGT1A7.1, UGT1A8.1, UGT1A9.1, and UGT1A10.1. The Vmax/Km values between each wildtype of UGT1A were significantly different (p < 0.001). Among these, the Vmax/Km values of UGT1A7.1 and UGT1A10.1 were statistically the same as those of UGT1A1.1 (p = 0.399 and p = 0.102, respectively). UGT1A6.1, and UGT1A8.1, and UGT1A9.1 showed statistically lower Vmax/Km values than UGT1A1.1 (p < 0.001). The Michaelis–Menten data are shown in and the maximum velocity (Vmax), Michaelis constant (Km), and the Vmax/Km values are listed in .
Table 1. Catalytic activity of each UGT1A.1 isoform.
Variants of UGT and glucuronidation of SN-38
We examined the relationship between the variants of UGT1A1, UGT1A6, UGT1A7, UGT1A8, UGT1A9, and UGT1A10 and SN-38 glucuronidation activity. The Michaelis–Menten data are listed in and Vmax, Km, and Vmax/Km values are provided in .
Table 2. Catalytic activity of each UGT1A variant.
First, we compared the Vmax, Km, and Vmax/Km values, with the values pertaining to UGT1A.1 established as the standard.
For UGT1A1, the Vmax, Km, and Vmax/Km values were significantly different between the wild type and the variants in the one-way ANOVA (p < 0.001, p = 0.028 and p = 0.003, respectively). UGT1A1.6, UGT1A1.7, UGT1A1.27, and UGT1A1.35 showed significantly lower Vmax (33.3%, 34.8%, 41.8%, and 44.2%, respectively, p < 0.001) and Vmax/Km (30.0%; p = 0.008, 11.2%; p = 0.002, 17.4%; p = 0.003, and 35.0%; p = 0.012, respectively) values compared with that of UGT1A1.1.
Among UGT1A6 variants, the Vmax and Km values were significantly different in the one-way ANOVA (p = 0.032 and p = 0.046, respectively). UGT1A6.3 exhibited a significantly lower Vmax (27.0%; p = 0.015) value than the wildtype (UGT1A6.1). Moreover, UGT1A6.2 and UGT1A6.4 had lower Vmax values than UGT1A6.1, but these differences were not statistically significant (45.9%; p = 0.067 and 59.5%; p = 0.170, respectively). In contrast, the Vmax/Km values among UGT1A6 variants were not significantly different (p = 0.090).
Among UGT1A7 variants, the Vmax values were significantly different in the one-way ANOVA (p = 0.017). UGT1A7.3 exhibited significantly lower Vmax (60.7%; p = 0.007) values than the wildtype (UGT1A7.1). In contrast, the Km value among UGT1A7 variants were not statistically different (p = 0.052). The Vmax/Km values among UGT1A7 variants were significantly different (p = 0.037). Among those, the Vmax/Km value of UGT1A7.3 was significantly lower than that of UGT1A7.1 (25.1%; p = 0.020).
Among UGT1A8 variants, the Vmax, Km, and Vmax/Km values were significantly different in the one-way ANOVA (p < 0.001, p = 0.002, and p = 0.024, respectively). The Vmax value of UGT1A8.2 was higher than that of the wildtype (UGT1A8.1, 129.7%), and that of UGT1A8.3 was similar to that of the wildtype (UGT1A8.1, 96.9%). However, UGT1A8.4 lost the SN-38 glucuronidation activity. Among UGT1A8 variants, the Vmax/Km values of UGT1A8.4 was statistically lower (p = 0.020). These results suggest that UGT1A8.4 lost its activity and influenced the glucuronidation of SN-38.
Among UGT1A9 variants, the Vmax and Km values were significantly different in the one-way ANOVA (p = 0.003 and p = 0.031, respectively). UGT1A9.3 had a significantly lower Vmax (16.7%; p = 0.029) value. UGT1A9.2 had a significantly higher Km (316.9%; p = 0.035) value. On the other hand, the Vmax/Km value among UGT1A9 variants was not significantly different (p = 0.081). These results suggest that UGT1A9 variants might not influence the glucuronidation of SN-38.
Among UGT1A10 variants, the Vmax, Km, and Vmax/Km values were not significantly different in the one-way ANOVA (p = 0.450, p = 0.078 and p = 0.102, respectively). UGT1A10M59I had a higher Vmax value than the wildtype (UGT1A10.1, 130.4%, not significant), and UGT1A10T202I had a similar Vmax (98.7%) value as that of the wildtype. However, the Vmax/Km value of UGT1A10M59I and UGT1A10T202I was lower (33.5% and 43.3%, respectively; not significant) than that of the wildtype. These results suggest that major UGT1A10 variants might influence the SN-38 glucuronidation rate to some extent.
Next, we compared the Vmax, Km, and Vmax/Km values when UGT1A1.1 was established as a standard. All values were significantly different in the one-way ANOVA. Compared with those of UGT1A1.1, the Vmax/Km values of UGT1A1.6, UGT1A1.7, UGT1A1.27, UGT1A6.1, UGT1A6.2, UGT1A6.3, UGT1A6.4, UGT1A7.3, UGT1A7.4, UGT1A8.1, UGT1A8.2, UGT1A8.3, UGT1A8.4, UGT1A9.2, UGT1A9.3, UGT1A10M59I, and UGT1A10T202I were significantly lower (30.0%, 11.2%, 17.4%, 2.2%, 2.5%, 3.0%, 2.5%, 18.4%, 29.5%, 23.6%, 16.1%, 6.5%, 15.6%, 8.2%, 2.5%, 20.3%, and 26.3%, respectively). In contrast, the Vmax/Km values of UGT1A7.1 and UGT1A10.1 were not significantly decreased compared to those of UGT1A1.1.
Discussion
Glucuronidation activity of each UGT1A
In our study, UGT1A1, UGT1A7, and UGT1A10 showed glucuronidation activity toward SN-38. UGT1A6, UGT1A8, and UGT1A9 also showed SN-38 glucuronidation activity, but their enzyme activity was significantly lower than that of UGT1A. UGT1A3, UGT1A4, and UGT1A5 had no effect on SN-38 glucuronidation. Some studies have reported that UGT1A1 and UGT1A7 play important roles in SN-38 glucuronidation (Ciotti et al. Citation1999; Gagné et al. Citation2002). Although these reports indicate that UGT1A9 also plays an important role in SN-38 glucuronidation (Ciotti et al. Citation1999; Gagné et al. Citation2002), our results revealed that UGT1A9 did not show the same activity toward SN-38 as UGT1A1. In contrast, our study revealed that UGT1A10 also plays an important role in SN-38 glucuronidation. UGT1A10 is expressed mainly in the human intestine (Zhang W et al. Citation2007; Tourancheau et al. Citation2018) and plays an important role in SN-38 glucuronidation, similar to UGT1A1 (Troberg et al. Citation2017). UGT1A1, UGT1A3, UGT1A4, UGT1A6, and UGT1A9 are expressed in the human liver (Izukawa et al. Citation2009), whereas UGT1A1 and UGT1A10 are expressed in the human intestine (Zhang H et al. Citation2020). However, UGT1A7 is not expressed in the human liver or intestine (Tourancheau et al. Citation2018). When the activities of UGT1A1, UGT1A6, UGT1A9, and UGT1A10 decrease, severe adverse effects of irinotecan might occur.
UGT1A variants and their SN-38 glucuronidation activities
We examined the relationship between variants of UGT1As and their SN-38 glucuronidation catalytic activities. The catalytic activities of UGT1A1.6, UGT1A1.7, UGT1A1.27, and UGT1A1.35 were significantly lower than that of UGT1A1.1. Among these variants, UGT1A1.7 had the lowest catalytic activity. These results suggest that UGT1A1.6, UGT1A1.7, UGT1A1.27, and UGT1A1.35 show significantly lower SN-38 glucuronidation activities, causing carriers of these variants to maintain a high serum concentration of SN-38 during chemotherapy, which may cause severe adverse effects. These results are similar to those reported previously (Gagné et al. Citation2002; Jinno et al. Citation2003). UGT1A1*6 results in a high serum SN-38 concentration (Hirose et al. Citation2014) and causes severe irinotecan toxicity (Chen X et al. Citation2017). Ando et al. (Citation2000) and Nakamura et al. (Citation2011) reported that patients with UGT1A1*27 variant experienced severe irinotecan toxicity. However, no clinical studies have examined the relationship between UGT1A1*7 and UGT1A1*35 and irinotecan toxicity. However, there have been certain reports that about 5% of Asian people have UGT1A1*7 and UGT1A1*27 variants (Huang CS et al. Citation2000; Teh et al. Citation2012; Sun et al. Citation2017), and UGT1A1*35 variant has been observed in the European cohort (Gagné et al. Citation2002). These reports suggest that other UGT1A1 variants, except UGT1A1*6, should be taken into consideration when predicting irinotecan toxicities.
Here, the UGT1A6.2, UGT1A6.3, and UGT1A6.4 activities were not significantly different from those of the wildtype. Carlini et al. (Citation2005) reported that UGT1A6 genotypes were not associated with the efficacy or toxicity of irinotecan. Indeed, our study demonstrated that the UGT1A6 variants did not affect the glucuronidation activity toward SN-38.
Among UGT1A7 variants, the catalytic activity of UGT1A7.3 was significantly decreased than that of UGT1A7.1. This results are similar to those reported previously (Ciotti et al. Citation1999; Villeneuve et al. Citation2003). Even though UGT1A7 subfamily members are not expressed in the human liver or intestine (Tourancheau et al. Citation2018), some clinical reports suggest that UGT1A7 polymorphisms, especially UGT1A7*3, are associated with irinotecan toxicities (Cecchin et al. Citation2009; Inoue et al. Citation2013; Valenzuela Jimenez et al. Citation2013; Tziotou et al. Citation2014; Cui et al. Citation2016). Moreover, patients with primary sclerosing cholangitis with UGT1A7 variants (N129K/N131K) presented significantly shorter transplant-free survival than those with the wildtype (Weismuller et al. Citation2020). UGT1A7*3 is expressed more frequently in patients with gastrointestinal cancer and hepatocellular carcinoma than in normal individuals (Vogel et al. Citation2001; Strassburg et al. Citation2002; Zhang Y et al. Citation2017; Hu et al. Citation2021). Chen et al. (Citation2006) reported that the frequencies of UGT1A7*2, UGT1A7*3, and UGT1A7*4 were 16.4%, 24.2%, and 1.8%, respectively, in a Chinese cohort. Checking UGT1A7 variants, especially in patients with colorectal cancer and hepatocellular carcinoma, may be useful for predicting the toxicities of irinotecan.
Among UGT1A8 variants, UGT1A8.4 lost its SN-38 glucuronidation activity. Huang et al. (Citation2002) reported that UGT1A8.3 exhibited reduced catalytic activity compared with UGT1A8.1, whereas the catalytic activity of UGT1A8.2 was similar to that of UGT1A8.1. Our results support these findings. Moreover, our results suggest that the catalytic activity of UGT1A8.4 was lower than that of UGT1A8.3. UGT1A8*4 may affect the risk of irinotecan toxicity. Strassburg et al. (Citation1998) identified UGT1A8 in the colon. The frequencies of UGT1A8*2 and UGT1A8*3 were reported to be 23.8% and 1.2%, respectively, in Caucasians (Thibaudeau et al. Citation2006); thus, investigating for UGT1A8 variants may help predict irinotecan toxicities to some extent.
Among UGT1A9 variants, UGT1A9.3 presented reduced glucuronidation activity toward SN-38. This result agrees with that reported previously (Villeneuve et al. Citation2003). Additionally, this result suggests that the UGT1A9*3 variant may be a predictor of severe toxicities of irinotecan as other UGT1A9 variants (Sandanaraj et al. Citation2008; Valenzuela Jimenez et al. Citation2013; Cui et al. Citation2016). Although our results showed that the glucuronidation activity of UGT1A9 was lower than that of UGT1A1, UGT1A9 contributes to glucuronidation as much as UGT1A1 (Xiao et al. Citation2018). This fact suggests that investigating for UGT1A9 variants is as important as investigating for UGT1A1 variants. UGT1A9*3 may affect the risk of irinotecan toxicity. The frequencies of UGT1A9*2 and UGT1A9*3 have been reported to be 2.5% in Africans and 3.0% in Caucasians (Villeneuve et al. Citation2003; Thibaudeau et al. Citation2006); thus, UGT1A9 variants may affect toxicities of irinotecan.
UGT1A10M59I and UGT1A10T202I presented reduced SN-38 glucuronidation activity by 33.5% and 43.3%, respectively. There are no reports on the glucuronidation activity of variants of UGT1A10 toward SN-38. Jinno et al. (Citation2003) reported that the glucuronidation activity of UGT1A10T202I was lower than that of UGT1A10*1 when the glucuronidated substances were 7-Hydroxy-4-(trifluoromethyl)coumarin and 17β-estradiol. UGT1A10 is expressed mainly in the intestine (Basit et al. Citation2020; Zhang H et al. Citation2020), and the influence of UGT1A10 variants may not be ignored.
We compared the catalytic activity when UGT1A1.1 was established as a standard. The catalytic activities of UGT1A7.3, UGT1A7.4, UGT1A10M59I, and UGT1A10T202I were significantly decreased compared to those of UGT1A1.1. We could not find any direct evidence comparing the glucuronidation activity between UGT1A1.1 and other variants of UGT1As. However, UGT1A10 plays important roles in the glucuronidation of SN-38 in the intestine (Ciotti et al. Citation1999; Gagné et al. Citation2002; Oguri et al. Citation2004; Troberg et al. Citation2017; Zhang H et al. Citation2020), and our study revealed that this showed apparent SN-38 glucuronidation activity. Thus, these variants might highly influence the glucuronidation of SN-38.
Our study revealed that UGT1A1 variants, as well as other UGT1A variants, influence the SN-38 glucuronidation rate. Gagné et al. (Citation2002) and Villeneuve et al. (Citation2003) reported that variants of UGT1A7 and UGT1A9 presented reduced SN-38 glucuronidation activity in vitro. Moreover, some clinical reports suggest that variants of UGT1A7 and UGT1A9 are associated with irinotecan toxicity (Hazama et al. Citation2013; Cui et al. Citation2016). To predict the toxicities of irinotecan, we should not only consider UGT1A1 variants, but also other UGT1A variants.
The limitations of our study are as follows. First, we only compared the glucuronidation activity between each UGT1A and UGT1A variant toward SN-38, and did not consider the UGT1A isoform distribution in the human body. If UGT1A10 is expressed more than UGT1A7, the variants of UGT1A10 may be increasingly associated with irinotecan toxicity. Moreover, gastrointestinal toxicity due to SN-38 is caused not only by insufficient SN-38, but also by deconjugation via bacteria (Takasuna et al. Citation1998). Therefore, we should also consider this bacterial effect. Second, we only examined the relationship between UGT1As and their variants and SN-38 glucuronidation, and we did not consider the relationship between UGT2As and glucuronidation activity toward SN-38. Third, we did not examine the effect of multiple variants of UGT1As. Teng et al. (Citation2007) reported that patients with two variants (UGT1A1*6 and UGT1A7*3) were seen in 30% of Gilbert syndrome and 7% of the control cohort. Patients with these variants may have more severe irinotecan toxicity than patients with either variant. To overcome these limitations, further analysis of patients who show severe adverse effects should be performed.
In summary, our results suggest that UGT1A1 variants, UGT1A1*6, UGT1A1*7, UGT1A1*27, and UGT1A1*35 might affect irinotecan toxicity. Moreover, the other UGT1A variants such as UGT1A7 (especially UGT1A7*3), UGT1A8 (especially UGT1A8*4), and UGT1A10 (UGT1A10M59I and UGT1A10T202I) may also affect the SN-38 glucuronidation activity. Our results revealed that many variants of UGT1As may affect toxicities and adverse effects of irinotecan.
Supplemental Material
Download MS Word (18.3 KB)Supplemental Material
Download JPEG Image (62.4 KB)Acknowledgments
The authors express our sincere gratitude to Dr Asami Mori, Dr Masaru Iwai, and Dr Hiroko Takahashi for supplying plasmid vectors. The authors would like to thank Editage (www.editage.com) for English language editing.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are available from the corresponding author, Y. M., upon reasonable request.
References
- Ando Y, Saka H, Ando M, Sawa T, Muro K, Ueoka H, Yokoyama A, Saitoh S, Shimokata K, Hasegawa Y. 2000. Polymorphisms of UDP-glucuronosyltransferase gene and irinotecan toxicity: a pharmacogenetic analysis. Cancer Res. 60(24):6921–6926.
- Basit A, Neradugomma NK, Wolford C, Fan PW, Murray B, Takahashi RH, Khojasteh SC, Smith BJ, Heyward S, Totah RA, et al. 2020. Characterization of differential tissue abundance of major non-CYP enzymes in human. Mol Pharm. 17(11):4114–4124.
- Carlini LE, Meropol NJ, Bever J, Andria ML, Hill T, Gold P, Rogatko A, Wang H, Blanchard RL. 2005. UGT1A7 and UGT1A9 polymorphisms predict response and toxicity in colorectal cancer patients treated with capecitabine/irinotecan. Clin Cancer Res. 11(3):1226–1236.
- Cecchin E, Innocenti F, D'Andrea M, Corona G, De Mattia E, Biason P, Buonadonna A, Toffoli G. 2009. Predictive role of the UGT1A1, UGT1A7, and UGT1A9 genetic variants and their haplotypes on the outcome of metastatic colorectal cancer patients treated with fluorouracil, leucovorin, and irinotecan. J Clin Oncol. 27(15):2457–2465.
- Chen K, Jin M, Zhu Y, Jiang Q, Yu W, Ma X, Yao K. 2006. Genetic polymorphisms of the uridine diphosphate glucuronosyltransferase 1A7 and colorectal cancer risk in relation to cigarette smoking and alcohol drinking in a Chinese population. J Gastroenterol Hepatol. 21(6):1036–1041.
- Chen X, Liu L, Guo Z, Liang W, He J, Huang L, Deng Q, Tang H, Pan H, Guo M, et al. 2017. UGT1A1 polymorphisms with irinotecan-induced toxicities and treatment outcome in Asians with lung cancer: a meta-analysis. Cancer Chemother Pharmacol. 79(6):1109–1117.
- Ciotti M, Basu N, Brangi M, Owens IS. 1999. Glucuronidation of 7-ethyl-10-hydroxycamptothecin (SN-38) by the human UDP-glucuronosyltransferases encoded at the UGT1 locus. Biochem Biophys Res Commun. 260(1):199–202. eng.
- Cui C, Shu C, Cao D, Yang Y, Liu J, Shi S, Shao Z, Wang N, Yang T, Liang H, et al. 2016. UGT1A1*6, UGT1A7*3 and UGT1A9*1b polymorphisms are predictive markers for severe toxicity in patients with metastatic gastrointestinal cancer treated with irinotecan-based regimens. Oncol Lett. 12(5):4231–4237.
- de Man FM, Goey AKL, van Schaik RHN, Mathijssen RHJ, Bins S. 2018. Individualization of Irinotecan treatment: a review of pharmacokinetics, pharmacodynamics, and pharmacogenetics. Clin Pharmacokinet. 57(10):1229–1254.
- Gagné JF, Montminy V, Belanger P, Journault K, Gaucher G, Guillemette C. 2002. Common human UGT1A polymorphisms and the altered metabolism of irinotecan active metabolite 7-ethyl-10-hydroxycamptothecin (SN-38). Mol Pharmacol. 62(3):608–617.
- Haaz MC, Rivory L, Jantet S, Ratanasavanh D, Robert J. 1997. Glucuronidation of SN-38, the active metabolite of irinotecan, by human hepatic microsomes. Pharmacol Toxicol. 80(2):91–96.
- Hanioka N, Ozawa S, Jinno H, Ando M, Saito Y, Sawada J. 2001. Human liver UDP-glucuronosyltransferase isoforms involved in the glucuronidation of 7-ethyl-10-hydroxycamptothecin. Xenobiotica. 31(10):687–699.
- Hazama S, Mishima H, Tsunedomi R, Okuyama Y, Kato T, Takahashi K, Nozawa H, Ando H, Kobayashi M, Takemoto H, et al. 2013. UGT1A1*6, 1A7*3, and 1A9*22 genotypes predict severe neutropenia in FOLFIRI-treated metastatic colorectal cancer in two prospective studies in Japan. Cancer Sci. 104(12):1662–1669.
- Hirose K, Yamashita K, Takada H, Kaneda N, Fukami K, Maruo E, Kitamura M, Hasegawa J, Maeda Y. 2014. Usefulness of one-point plasma SN-38G/SN-38 concentration ratios as a substitute for UGT1A1 genetic information after irinotecan administration. Int J Clin Oncol. 19(2):397–402.
- Hu DG, Marri S, Mackenzie PI, Hulin JA, McKinnon RA, Meech R. 2021. The expression profiles and deregulation of UDP-glycosyltransferase (UGT) genes in human cancers and their association with clinical outcomes. Cancers (Basel). 13(17):4491.
- Huang CS, Luo GA, Huang ML, Yu SC, Yang SS. 2000. Variations of the bilirubin uridine-diphosphoglucuronosyl transferase 1A1 gene in healthy Taiwanese. Pharmacogenetics. 10(6):539–544.
- Huang YH, Galijatovic A, Nguyen N, Geske D, Beaton D, Green J, Green M, Peters WH, Tukey RH. 2002. Identification and functional characterization of UDP-glucuronosyltransferases UGT1A8*1, UGT1A8*2 and UGT1A8*3. Pharmacogenetics. 12(4):287–297.
- Inoue K, Sonobe M, Kawamura Y, Etoh T, Takagi M, Matsumura T, Kikuyama M, Kimura M, Minami S, Utsuki H, et al. 2013. Polymorphisms of the UDP-glucuronosyl transferase 1A genes are associated with adverse events in cancer patients receiving irinotecan-based chemotherapy. Tohoku J Exp Med. 229(2):107–114.
- Iwai M, Maruo Y, Ito M, Yamamoto K, Sato H, Takeuchi Y. 2004. Six novel UDP-glucuronosyltransferase (UGT1A3) polymorphisms with varying activity. J Hum Genet. 49(3):123–128.
- Iyer L, Hall D, Das S, Mortell MA, Ramírez J, Kim S, Di Rienzo A, Ratain MJ. 1999. Phenotype-genotype correlation of in vitro SN-38 (active metabolite of irinotecan) and bilirubin glucuronidation in human liver tissue with UGT1A1 promoter polymorphism. Clin Pharmacol Ther. 65(5):576–582.
- Izukawa T, Nakajima M, Fujiwara R, Yamanaka H, Fukami T, Takamiya M, Aoki Y, Ikushiro S-i, Sakaki T, Yokoi T. 2009. Quantitative analysis of UDP-glucuronosyltransferase (UGT) 1A and UGT2B expression levels in human livers. Drug Metab Dispos. 37(8):1759–1768.
- Jinno H, Saeki M, Tanaka-Kagawa T, Hanioka N, Saito Y, Ozawa S, Ando M, Shirao K, Minami H, Ohtsu A, et al. 2003. Functional characterization of wild-type and variant (T202I and M59I) human UDP-glucuronosyltransferase 1A10. Drug Metab Dispos. 31(5):528–532.
- Jinno H, Tanaka-Kagawa T, Hanioka N, Saeki M, Ishida S, Nishimura T, Ando M, Saito Y, Ozawa S, Sawada J. 2003. Glucuronidation of 7-ethyl-10-hydroxycamptothecin (SN-38), an active metabolite of irinotecan (CPT-11), by human UGT1A1 variants, G71R, P229Q, and Y486D. Drug Metab Dispos. 31(1):108–113.
- Maruo Y, Takahashi H, Matsui K, Sato H, Takeuchi Y. 2010. Phase II drug metabolism and individualized drug therapy: a focus on functional genetic variation in UDP-glucuronosyltransferases. CPPM. 8(2):146–166.
- Mimura Y, Maruo Y, Ohta Y, Sato H, Takeuchi Y. 2011. Effect of common exon variant (p.P364L) on drug glucuronidation by the human UDP-glucuronosyltransferase 1 family. Basic Clin Pharmacol Toxicol. 109(6):486–493.
- Mori A, Maruo Y, Iwai M, Sato H, Takeuchi Y. 2005. UDP-glucuronosyltransferase 1A4 polymorphisms in a Japanese population and kinetics of clozapine glucuronidation. Drug Metab Dispos. 33(5):672–675.
- Nakamura Y, Soda H, Oka M, Kinoshita A, Fukuda M, Fukuda M, Takatani H, Nagashima S, Soejima Y, Kasai T, et al. 2011. Randomized phase II trial of irinotecan with paclitaxel or gemcitabine for non-small cell lung cancer: association of UGT1A1*6 and UGT1A1*27 with severe neutropenia. J Thorac Oncol. 6(1):121–127.
- Oguri T, Takahashi T, Miyazaki M, Isobe T, Kohno N, Mackenzie PI, Fujiwara Y. 2004. UGT1A10 is responsible for SN-38 glucuronidation and its expression in human lung cancers. Anticancer Res. 24(5A):2893–2896.
- Sandanaraj E, Jada SR, Shu X, Lim R, Lee SC, Zhou Q, Zhou S, Goh BC, Chowbay B. 2008. Influence of UGT1A9 intronic I399C > T polymorphism on SN-38 glucuronidation in Asian cancer patients. Pharmacogenomics J. 8(3):174–185.
- Strassburg CP, Manns MP, Tukey RH. 1998. Expression of the UDP-glucuronosyltransferase 1A locus in human colon. Identification and characterization of the novel extrahepatic UGT1A8. J Biol Chem. 273(15):8719–8726.
- Strassburg CP, Vogel A, Kneip S, Tukey RH, Manns MP. 2002. Polymorphisms of the human UDP-glucuronosyltransferase (UGT) 1A7 gene in colorectal cancer. Gut. 50(6):851–856.
- Sun L, Li M, Zhang L, Teng X, Chen X, Zhou X, Ma Z, Qi L, Wang P. 2017. Differences in UGT1A1 gene mutations and pathological liver changes between Chinese patients with Gilbert syndrome and Crigler-Najjar syndrome type II. Medicine (Baltimore). 96(45):e8620.
- Takahashi H, Maruo Y, Mori A, Iwai M, Sato H, Takeuchi Y. 2008. Effect of D256N and Y483D on propofol glucuronidation by human uridine 5'-diphosphate glucuronosyltransferase (UGT1A9). Basic Clin Pharmacol Toxicol. 103(2):131–136.
- Takasuna K, Hagiwara T, Hirohashi M, Kato M, Nomura M, Nagai E, Yokoi T, Kamataki T. 1998. Inhibition of intestinal microflora β-glucuronidase modifies the distribution of the active metabolite of the antitumor agent, irinotecan hydrochloride (CPT-11) in rats. Cancer Chemother Pharmacol. 42(4):280–286.
- Teh LK, Hashim H, Zakaria ZA, Salleh MZ. 2012. Polymorphisms of UGT1A1*6, UGT1A1*27 & UGT1A1*28 in three major ethnic groups from Malaysia. Indian J Med Res. 136(2):249–259.
- Teng HC, Huang MJ, Tang KS, Yang SS, Tseng CS, Huang CS. 2007. Combined UGT1A1 and UGT1A7 variant alleles are associated with increased risk of Gilbert's syndrome in Taiwanese adults. Clin Genet. 72(4):321–328.
- Thibaudeau J, Lépine J, Tojcic J, Duguay Y, Pelletier G, Plante M, Brisson J, Têtu B, Jacob S, Perusse L, et al. 2006. Characterization of common UGT1A8, UGT1A9, and UGT2B7 variants with different capacities to inactivate mutagenic 4-hydroxylated metabolites of estradiol and estrone. Cancer Res. 66(1):125–133.
- Tourancheau A, Rouleau M, Guauque-Olarte S, Villeneuve L, Gilbert I, Droit A, Guillemette C. 2018. Quantitative profiling of the UGT transcriptome in human drug-metabolizing tissues. Pharmacogenomics J. 18(2):251–261.
- Troberg J, Jarvinen E, Ge GB, Yang L, Finel M. 2017. UGT1A10 is a high activity and important extrahepatic enzyme: why has its role in intestinal glucuronidation been frequently underestimated? Mol Pharm. 14(9):2875–2883.
- Tukey RH, Strassburg CP. 2000. Human UDP-glucuronosyltransferases: metabolism, expression, and disease. Annu Rev Pharmacol Toxicol. 40:581–616.
- Tziotou M, Kalotychou V, Ntokou A, Tzanetea R, Armenis I, Varsou M, Konstantopoulos K, Tsavaris N, Rombos Y. 2014. Polymorphisms of uridine glucuronosyltransferase gene and irinotecan toxicity: low dose does not protect from toxicity. Ecancermedicalscience. 8:428.
- Valenzuela Jimenez B, Gonzalez Sales M, Escudero Ortiz V, Martinez Navarro E, Perez Ruixo C, Rebollo Liceaga J, Gonzalez Manzano R, Perez Ruixo JJ. 2013. Influence of genetic polymorphisms in UGT1A1, UGT1A7 and UGT1A9 on the pharmacokinetics of irinotecan, SN-38 and SN-38G. Farm Hosp. 37(2):111–127.
- Villeneuve L, Girard H, Fortier LC, Gagne JF, Guillemette C. 2003. Novel functional polymorphisms in the UGT1A7 and UGT1A9 glucuronidating enzymes in Caucasian and African-American subjects and their impact on the metabolism of 7-ethyl-10-hydroxycamptothecin and flavopiridol anticancer drugs. J Pharmacol Exp Ther. 307(1):117–128.
- Vogel A, Kneip S, Barut A, Ehmer U, Tukey RH, Manns MP, Strassburg CP. 2001. Genetic link of hepatocellular carcinoma with polymorphisms of the UDP-glucuronosyltransferase UGT1A7 gene. Gastroenterology. 121(5):1136–1144.
- Weismuller TJ, Zhou T, Kalthoff S, Lenzen H, Manns MP, Strassburg CP. 2020. Genetic variants of UDP-glucuronosyltransferase 1A genes are associated with disease presentation and outcome in primary sclerosing cholangitis. Liver Int. 40(7):1645–1654.
- Xiao L, Zhu L, Li W, Li C, Cao Y, Ge G, Sun X. 2018. New insights into SN-38 glucuronidation: evidence for the important role of UDP glucuronosyltransferase 1A9. Basic Clin Pharmacol Toxicol. 122(4):424–428.
- Zhang H, Wolford C, Basit A, Li AP, Fan PW, Murray BP, Takahashi RH, Khojasteh SC, Smith BJ, Thummel KE, et al. 2020. Regional proteomic quantification of clinically relevant non-cytochrome P450 enzymes along the human small intestine. Drug Metab Dispos. 48(7):528–536.
- Zhang W, Liu W, Innocenti F, Ratain MJ. 2007. Searching for tissue-specific expression pattern-linked nucleotides of UGT1A isoforms. PLoS One. 2(4):e396.
- Zhang Y, Hou J, Feng F, Li D, Jiang Q, Li X, Zhao Q, Li BA. 2017. Genetic polymorphisms in human UDP-glucuronosyltransferases 1A7 and the risk of gastrointestinal carcinomas: a systematic review and network meta-analysis. Oncotarget. 8(39):66371–66381.
