Abstract
Recent studies indicate that post-translational protein neddylation is required for the maintenance of cell viability in several lymphoma cell lines, while inhibition of the neddylation pathway with an NEDD8-activating enzyme (NAE) inhibitor MLN4924 induces apoptosis in lymphoma cells. However, the mechanism by which neddylation inhibition induces apoptosis in lymphoma cells has not been fully elucidated. Moreover, it is unknown whether neddylation inhibition triggers non-apoptotic cell-killing responses, such as cell senescence, in lymphoma cells. Here, we report that MLN4924 specifically inhibited protein neddylation, inactivated cullin-RING E3 ligase (CRL), the best-known neddylation substrate, and induced the accumulation of tumor-suppressive CRL substrates in lymphoma cells. Moreover, MLN4924 potently suppressed the growth of lymphoma cells by inducing G2 cell-cycle arrest, followed by apoptosis or senescence in a cell line-dependent manner. MLN4924-induced apoptosis was mediated by intrinsic apoptotic signaling with substantial up-regulation of pro-apoptotic Bik and Noxa as well as down-regulation of anti-apoptotic XIAP, c-IAP1 and c-IAP2, while senescence induction upon neddylation inhibition seemed dependent on the expression of tumor suppressor p21/p27. Together, these findings expand our understanding on how lymphoma cells respond to neddylation inhibition and support the development of neddylation inhibitors (e.g. MLN4924) for the treatment of lymphoma.
Abbreviations
| NAE | = | NEDD8-activating enzyme |
| CRL | = | cullin-RING E3 ligase |
| NHL | = | non-Hodgkin lymphoma |
| GCB-DLBCL | = | germinal-center B cell-like diffuse large B-cell lymphoma |
| SA-β-gal | = | senescence-associated β-galactosidase |
| IAP | = | inhibitor of apoptosis |
Introduction
Neddylation, a type of protein post-translational modification, regulates a variety of biological processes by affecting the subcellular localization, stability, conformation and function of target proteins.Citation1-5 Neddylation, which conjugates NEDD8,a ubiquitin-like molecule, to target substrates, is a 3-step enzymatic cascade involving NEDD8-activating enzyme (NAE, E1), NEDD8-conjugating enzyme E2 (UBC12 or UBE2F) and substrate-specific E3s.Citation1-5 The best characterized substrates of neddylation are cullin family proteins which serve as essential components of cullin-RING E3 ligases (CRLs), the largest multiunit E3 ubiquitin ligase family that regulates the turnover of numerous proteins in a proteasome-dependent manner.Citation6-9 Covalent modification of cullin by NEDD8 is required for the activation of CRL to regulate diverse processes, such as transcription, signal transduction, cell-cycle progression and stress responses, whereas its dysfunction leads to carcinogenesis.Citation7,10-14
Inhibition of protein neddylation, particularly cullin neddylation, has emerged as a promising anticancer strategy since the discovery of the NAE inhibitor MLN4924. MLN4924 inhibits the neddylation pathway, blocks cullin neddylation, and therefore inactivates CRL, leading to accumulation of tumor-suppressive CRL substrates to suppress the growth of cancer cells by triggering cell-cycle defects, apoptosis or senescence.Citation9,15-21 During the process,MLN4924 induces autophagic response as a pro-survival signal, suggesting that blockage of the protective autophagy may serve as a promising strategy to enhance the efficacy of MLN4924.Citation17,22,23 Recently, we found that inhibition of neddylation with MLN4924 also impairs tumor angiogenesis as a new mechanism of tumor growth suppression.Citation24 Due to its significant anticancer efficacy and well-tolerated toxicity in preclinical studies, MLN4924 is currently in several phase I clinical trials for cancer therapy.Citation5,9,25
As a cancer beginning in the lymphatic system, lymphoma consists of Hodgkin lymphoma (HL) and non-Hodgkin lymphoma (NHL), with the latter accounting for about 90% of all lymphomas.Citation26 Recently, the efficacy of neddylation inhibition with MLN4924 on lymphoma cells has been reported.Citation20 Milhollen et al found that MLN4924 induced apoptosis resulting from either inhibition of the NF-κB pathway due to the accumulation of IκBα in activated B cell-like (ABC)-diffuse large B-cell lymphoma (DLBCL) or DNA re-replication due to accumulation of CRL substrate CDT1 in germinal-center B cell-like (GCB)-DLBCL.Citation20 Dengler et al reported that MLN4924 killed mantle cell lymphoma (MCL) cells by stabilizing pro-apoptotic protein NOXA.Citation27 Interestingly, Godbersen et al found that MLN4924 thwarted microenvironment-driven NF-κB activation and induces apoptosis in chronic lymphocytic leukemia B cells.Citation28 These collective findings demonstrate that the neddylation pathway is required for the growth and survival of lymphoma cells. However, the mechanism of MLN4924-induced apoptosis in lymphoma is still not well defined. Moreover, it remains elusive whether inhibition of neddylation with MLN4924 triggers senescence in lymphoma cells. In this study, we report that, in lymphoma cells, NAE inhibitor MLN4924 specifically inhibited the neddylation pathway and triggered intrinsic apoptosis or senescence in a cell line-dependent manner. These findings support the development of neddylation inhibitors (e.g. MLN4924) as a novel agent for the treatment of lymphoma.
Results
MLN4924 specifically inhibits protein neddylation and inactivates CRL
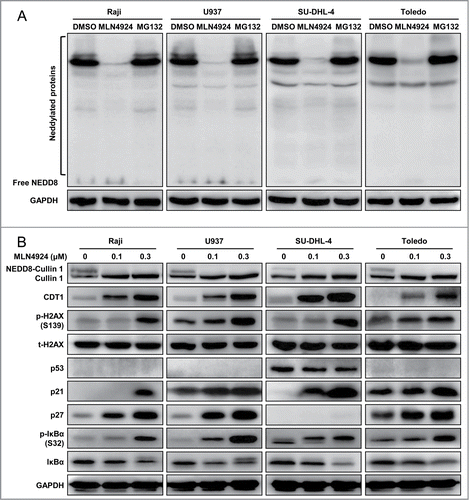
To determine cellular responses of neddylation inhibition, 4 representative human lymphoma cells, including 2 GCB-DLBCL (SU-DHL-4 and Toledo) cell lines, one Burkitt's lymphoma (Raji) cell line and one histiocytic lymphoma (U937) cell line,Citation29 were treated with an NAE inhibitor MLN4924. We first determined the specificity and efficacy of MLN4924 for the inhibition of protein neddylation when compared to MG132, a classical proteasome inhibitor. As shown in , MLN4924, but not MG132, specifically suppressed global protein neddylation. Further, we found that MLN4924 abrogated neddylation of Cullin 1, indicating the inactivation of CRLs (). As a result, MLN4924 induced the accumulation of several CRL substrates, including (a) DNA replication licensing protein CDT1, of which accumulation triggered DNA re-replication stress,Citation9 and DNA damage response, as evidenced by a significant increase of phosphorylated H2AX; (b) cell-cycle inhibitors p21 and p27; and (c) NF-κB inhibitor IκBα, as evidenced by the accumulation of p-IκBα, indicating the inactivation of NF-κB signaling ().Citation20,28 The results indicated that MLN4924 specifically inhibited protein neddylation, inactivated CRLs, and induced accumulation of CRL substrates in lymphoma cells.
Figure 1. MLN4924 specifically inhibits protein neddylation and inactivates CRL E3 ligases. (A) MLN4924 specifically inhibited protein neddylation. Raji, U937, SU-DHL-4 and Toledo cells were seeded into 6-well plates, cultured overnight, treated with 0.3 μM MLN4924, 10 μM proteasome inhibitor MG132 or DMSO for 1 h, and subjected to immunoblotting using antibodies against NEDD8 with GAPDH as a loading control. (B) Effects of MLN4924 on neddylated Cullin 1 and downstream effectors. The four lymphoma cells were treated with MLN4924 at 0.1 and 0.3 μM or DMSO for 48 h, and subjected to immunoblotting using indicated antibodies with GAPDH as a loading control.
Neddylation inhibition by MLN4924 triggers G2 cell-cycle arrest and suppresses the growth of lymphoma cells
After validating the specificity and efficacy of MLN4924 for neddylation inhibition and CRL inactivation, we further evaluated the effect of MLN4924 on cell-cycle profile by propidium iodide (PI) staining and fluorescence-activated cell sorting (FACS) analysis. As shown in , a prominent G2-M cell-cycle arrest was observed in all treated cell lines (Raji, U937, SU-DHL-4 and Toledo) in a dose-dependent manner. To define at which phase (G2 or M) these cells were arrested upon neddylation inactivation, the expression of Wee1, an inhibitor of G2-M phase transition,Citation30 as well as p-H3, a hallmark of M phase cells,Citation31 was detected. As shown in , MLN4924 induced significant accumulation of Wee1 and decrease of p-H3, indicating that MLN4924-treated cells were arrested at the G2 phase.
Figure 2. Neddylation inhibition with MLN4924 triggers G2 cell-cycle arrest and suppresses the growth of lymphoma cells. (A) MLN4924 induced G2-M cell-cycle arrest. Raji, U937, SU-DHL-4 and Toledo cells seeded into 6-well plates were treated with MLN4924 at 0.1 and 0.3 μM or DMSO for 48 h, followed by PI staining and FACS analysis for cell-cycle profile. (B) MLN4924 induced accumulation of Wee1 and decrease of p-H3. The four lymphoma cells were treated with MLN4924 at 0.1 and 0.3 μM or DMSO for 48 h, and subjected to immunoblotting using antibodies against Wee1 and p-H3 (S10) with GAPDH as a loading control. (C) MLN4924 inhibited the proliferation of lymphoma cells. The four cells seeded into 24-well plates were treated with 0.3 μM MLN4924 or DMSO for indicated time points, followed by cell counting (n = 3). (D) MLN4924 impaired cell viability in lymphoma cells. The four cells seeded into 96-well black plates were treated with MLN4924 at 0.1 and 0.3 μM or DMSO for 96 h, followed by cell viability analysis using the ATPlite assay (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001.
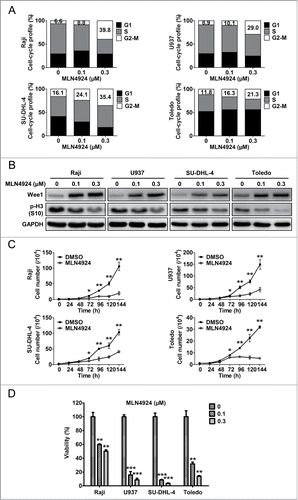
Next, we evaluated the effect of neddylation inhibition on the proliferation of lymphoma cells by cell counting and ATPlite cell viability assay. As shown in , cell counting analysis revealed that MLN4924 significantly inhibited the proliferation of lymphoma cells. Consistently, ATPlite assay revealed that neddylation inactivation with MLN4924 significantly impaired cell viability in a dose-dependent manner (). These findings demonstrated that the inhibition of neddylation with MLN4924 triggered G2 arrest and suppressed the growth of lymphoma cells.
Neddylation inhibition by MLN4924 triggers cell line-dependent induction of apoptosis or senescence in lymphoma cells
To elucidate the underlying mechanisms for the inhibitory effect of neddylation disruption on the proliferation of lymphoma cells, cellular morphological changes upon MLN4924 treatment were first observed by microscopy. As shown in , an increase in cell size with flattened shape, a characteristic of senescence, was observed in Raji and U937 cells, whereas a shrunk morphology in shape, a feature of apoptosis, was noticed in SU-DHL-4 and Toledo cells, suggesting that the different cell fates (senescence or apoptosis) were triggered upon neddylation inhibition. To determine whether MLN4924 indeed induced apoptosis or senescence in a cell-specific manner, the expression of cleaved caspase-3 and cleaved PARP, 2 classical markers of apoptosis, was measured in treated cells. As shown in , both cleaved caspase-3 and cleaved PARP significantly accumulated in apoptotic SU-DHL-4 and Toledo cells, but not in senescent Raji and U937 cells. To further validate the induction of cell senescence in Raji and U937 cells, the expression of senescence-associated β-galactosidase (SA-β-gal), a well-established biochemical marker of senescence, was determined by SA-β-gal staining in MLN4924-treated cells. As shown in , a substantial proportion of MLN4924-treated cells were positively stained. These findings demonstrated that inhibition of neddylation with MLN4924 suppressed the growth of lymphoma cells by inducing apoptosis or senescence in a cell line-dependent manner.
Figure 3. Neddylation inhibition with MLN4924 triggers cell line-dependent induction of apoptosis or senescence in lymphoma cells. (A) Changes in cellular morphology upon MLN4924 treatment. Raji, U937, SU-DHL-4 and Toledo cells were treated with 0.1 μM MLN4924 or DMSO for 96 h, followed by morphological observation. Scale bar, 100 μm. (B) MLN4924 significantly induced apoptosis in SU-DHL-4 and Toledo cells, but not Raji and U937 cells. The four lymphoma cells were treated with MLN4924 at 0.1 and 0.3 μM or DMSO for 48 h, and subjected to immunoblotting using antibodies against cleaved caspase-3 and cleaved PARP with GAPDH as a loading control. (C) MLN4924 induced senescence in Raji and U937 cells. Raji and U937 cells, treated with 0.3 μM MLN4924 or DMSO for 96 h, were subjected to senescence-associated β-galactosidase (SA-β-gal) staining assay. Representative pictures were shown (left panel), and positively stained cells were counted and plotted as percentage of total cell numbers (right panel) (n = 3). Scale bar, 100 μm. (D) MLN4924 extended the half-life of p21/p27. Raji and U937 cells were treated with 0.3 μM MLN4924 or DMSO in combination with 50 μg/mL cycloheximide (CHX) for indicated time points, and subjected to immunoblotting using antibodies against p21 and p27 with GAPDH as a loading control. (E) MLN4924 had little effect on the transactivation of p21/p27. Raji and U937 cells were treated with 0.3 μM MLN4924 or DMSO for 6 h, and subjected to real-time PCR for p21 and p27 with GAPDH as a normalizer (n = 3). **P < 0.01, ***P < 0.001.
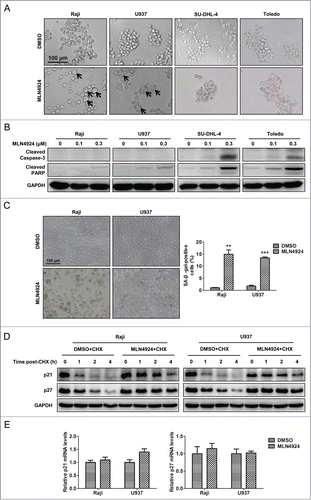
Previous studies have demonstrated a causal role of p21/p27 upregulation in the induction of cell senescence upon neddylation inhibition or CRL inactivation.Citation15,16,32,33 As shown in , p21 and p27, 2 well-known CRL substrates, were accumulated in senescent Raji and U937 cells. To determine the underlying mechanism of p21/p27 regulation by MLN4924, we first applied cycloheximide to block protein translation and determined p21/p27 turnover rate upon MLN4924 treatment. As shown in , MLN4924 significantly delayed p21/p27 turnover and extended the half-life of p21/p27 in both Raji and U937 cells. In contrast, MLN4924 had little effect on the transactivation of p21/p27 (). These findings demonstrated that MLN4924 induced the accumulation of p21/p27 by stabilizing those proteins.
MLN4924-induced apoptosis is mediated through the intrinsic apoptotic signal pathway and associated with a coordinated dysregulation of pro-apoptotic and anti-apoptotic proteins
We next focused on how neddylation inhibition induced apoptosis in SU-DHL-4 and Toledo cells. To determine the kinetics of apoptotic induction upon MLN4924 treatment in these cells, we determined the kinetic activation of cleaved PARP as an apoptotic marker in treated cells. As shown in , the cleaved PARP were obviously detected as early as 24 h post treatment, accumulated over time and reached the peak at 48 h post treatment. We then determined which apoptotic signal pathway, the extrinsic or intrinsic apoptotic signaling, was responsible for MLN4924-triggered apoptosis by measuring the activation of cleaved caspase-8 and cleaved caspase-9, respectively. As shown in , MLN4924 substantially induced the cleavage of caspase-9 but not caspase-8 in both apoptotic SU-DHL-4 and Toledo cells, indicating that the induction of apoptosis upon neddylation inhibition was primarily mediated via the intrinsic apoptotic signal pathway.
Figure 4 (See previous page). MLN4924-induced apoptosis is mediated through the intrinsic apoptotic signaling and associated with a coordinated dysregulation of pro-apoptotic and anti-apoptotic proteins. (A) MLN4924 induced apoptosis at early time post treatment. SU-DHL-4 and Toledo cells were treated with MLN4924 at 0.1 and 0.3 μM or DMSO for indicated time points, and subjected to immunoblotting using the antibody against cleaved PARP with GAPDH as a loading control. (B) MLN4924 induced cell apoptosis mainly via the intrinsic apoptotic signaling pathway. SU-DHL-4 and Toledo cells, treated with MLN4924 at 0.1 and 0.3 μM or DMSO for 48 h, were subjected to immunoblotting using antibodies against caspase-8 and cleaved caspase-9 with GAPDH as a loading control. (C) Effects of MLN4924 on expression of pro-apoptotic and anti-apoptotic proteins. SU-DHL-4 and Toledo cells were treated with MLN4924 at 0.1 and 0.3 μM or DMSO for 48 h, followed by immunoblotting using indicated antibodies against pro-apoptotic (left panel) and anti-apoptotic (right panel) proteins with GAPDH as a loading control. (D) Effects of MLN4924 on transcriptional activation of apoptosis-regulatory proteins. Toledo cells, treated with 0.3 μM MLN4924 or DMSO for 36 h, were subjected to the real-time PCR analysis using human apoptosis PCR array as described in Materials and Methods (n = 3).
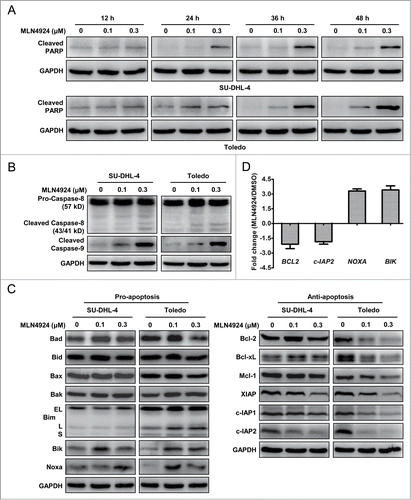
To further investigate the potential mechanisms underlying the activation of intrinsic apoptotic signal pathway, we measured the expression of classical Bcl-2 and IAP (inhibitor of apoptosis) family members, including a panel of pro-apoptotic proteins (Bad, Bid, Bax, Bak, Bim, Bik and Noxa) and anti-apoptotic proteins (Bcl-2, Bcl-xL, Mcl-1, XIAP, c-IAP1 and c-IAP2) in apoptotic SU-DHL-4 and Toledo cells. Among these proteins, pro-apoptotic Bik and Noxa were substantially up-regulated while anti-apoptotic XIAP, c-IAP1 and c-IAP2 were reduced significantly in both cell lines (). To further address how MLN4924 controls the expression of these apoptosis-regulatory proteins, the transcription of 88 apoptosis-regulatory genes was determined by the real-time PCR using human apoptosis PCR array in MLN4924-treated Toledo cells. Consistently, the transcription of Bik and Noxa was significantly activated while the transactivation of c-IAP2 was notably reduced (P < 0.05) ( and data not shown). Together, these results suggested that MLN4924 induced apoptosis by up-regulation of pro-apoptotic Bik and Noxa and downregulation of anti-apoptotic XIAP and c-IAP1/2.
Discussion
Inhibition of protein neddylation with MLN4924 has become an attractive strategy for the treatment of lymphoma, as evidenced by the anti-lymphoma activity in preclinical studies of NAE inhibitor MLN4924.Citation20,27,28 Encouragingly, lymphoma cells, compared to other types of cancer cells, exhibit the highest sensitivity to MLN4924.Citation9 Moreover, normal lymphocytes including T/B cells and peripheral blood mononuclear cells (PBMCs) are resistant to MLN4924 treatment, while lymphoma cells are shown to be very sensitive to this anticancer agent.Citation27,28 Based on these findings, phase I clinical trial of MLN4924 in patients with non-Hodgkin lymphoma has been initiated and the expected pharmacodynamic effects were seen in humans.Citation5,25 These collective findings highlight the therapeutic value of neddylation inhibition with MLN4924 in lymphoma.
Mechanistically, previous and our present studies demonstrated that MLN4924 specifically inhibited the neddylation pathway, blocked the cullin neddylation, inactivated CRLs, induced the accumulation of tumor-suppressive CRL substrates and triggered multiple cell-killing pathways in lymphoma cells.Citation20,27,28 Interestingly, we found that MLN4924 induced G2 cell-cycle arrest, a rare and important type of cell cycle arrest that can be triggered by some cellular stresses.Citation34-37 Our findings seem inconsistent with a previous report showing that treatment of GCB-DLBCL cells with MLN4924 resulted in the accumulation of cells in S-phase.Citation20 A potential explanation for this discrepancy is that the disturbance of cell cycle progression at different phases by MLN4924 is cell line dependent.Citation9,16,17,20,38 Meanwhile, the concentrations of MLN4924 used in the treatments could also determine at which phase (S or G2) the treated cells were arrested, which may also explain the discrepancy.Citation18 In addition to lymphoma cell lines, we and others previously reported that MLN4924 also induced G2 cell-cycle arrest in other types of cancer cells, including liver cancer cells, intrahepatic cholangiocarcinoma cells, prostate cancer cells, pancreatic cancer cells and lung cancer cells.Citation17,38-41 These findings indicate that induction of cell cycle arrest at G2 phase is a general cellular response of cancer cells upon neddylation inhibition with MLN4924. However, it is unclear whether MLN4924-induced G2 arrest is cancer-specific, and future studies are warranted to evaluate the effect of MLN4924 on normal cells.
In this study, we found that neddylation inhibition by MLN4924 induced apoptosis in SU-DHL-4 and Toledo, and the response was mediated mainly through the intrinsic apoptotic pathway. Furthermore, mechanistic study showed that MLN4924 treatment led to significant upregulation of pro-apoptotic proteins Noxa and Bik. Similarly, previous studies reported that the induction of Noxa expression in mantle cell lymphoma (MCL) cells and chronic lymphocytic leukemia (CLL) cells contributed to MLN4924-induced apoptosis,Citation27,28 while the up-regulation of Bik was reported to contribute to apoptotic induction by MLN4924 in combination with cisplatin.Citation42 We also noticed that anti-apoptotic IAP family proteins, including XIAP and c-IAP1/2, were significantly down-regulated upon MLN4924 treatment. So it is likely that the upregulation of Bik and Noxa and down-regulation of IAP proteins induced apoptosis in MLN4924-treated lymphoma cells.
Senescence is an important mechanism to suppress the proliferation of potentially tumorigenic cells, and induction of senescence has become a promising approach for cancer therapy.Citation43,44 In this study, we found that, in addition to apoptosis, MLN4924 triggered cellular senescence as another major mechanism of growth suppression in 2 of 4 tested lymphoma cell lines. Previous studies indicated that senescence is primarily regulated by the p53/p21 and/or p16/pRB pathways.Citation44-46 In this study, the induction of senescence by neddylation inhibition seems to be dependent on the induction of CRL substrate p21/p27 but not p53 or p16/pRB, as (a) p21/p27 was significantly accumulated upon MLN4924 treatment due to the stabilization of these tumor-suppressive proteins (); (b) p53 was not detected in Raji and U937 cells;Citation47,48 and (c) p16 was methylated in Raji and U937 cells leading to inactivation of the p16/pRB pathway.Citation49-51 Together, our findings indicated that the induction of senescence represents a common anticancer mechanism of MLN4924 in lymphoma cells.
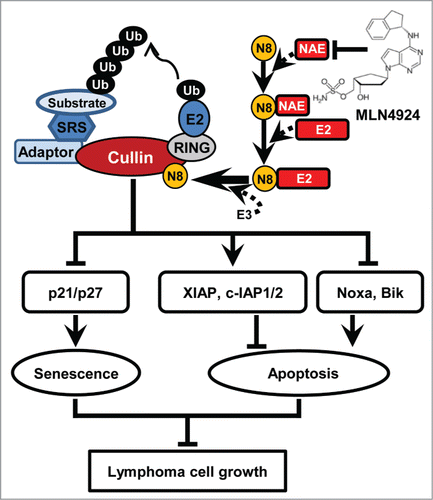
Based on the findings reported in the current and previous studies,Citation20,27,28 we propose a working model regarding the potential role of neddylation pathway in lymphoma tumorigenesis and tumor progression (). To facilitate the development of lymphoma, Neddylation-CRL axis is activated to degrade tumor-suppressive CRL substrates and maintains the malignant phenotypes of lymphoma cells. In contrast, inhibition of neddylation blocks cullin neddylation, inactivates CRL, results in the accumulation of tumor-suppressive CRL substrates, and thus induces apoptosis or senescence to suppress the growth of lymphoma cells.
Materials and Methods
Cell lines and reagents
Human lymphoma cell lines Raji, SU-DHL-4, Toledo and U937 were obtained from the American Type Culture Collection (CCL-86, CRL-2957, CRL-2631, CRL-1593.2), and cultured in RPMI-1640 medium (Thermo Fisher Scientific, SH30809.01B), supplemented with 10% FBS (Biochrom AG, S4115) and 1% penicillin-streptomycin, at 37°C with 5% CO2. MLN4924 was synthesized as previously described.Citation17,52 MG132 (Sigma-Aldrich, M7449) and MLN4924 were dissolved in DMSO and kept in −20°C.
Cell counting and ATPlite cell viability assay
For cell counting, cells were seeded into 24-well plates with 1 × 104 (for Raji, U937 and Toledo) or 2 × 104 (for SU-DHL-4) cells per well, treated with 0.3 μM MLN4924 or DMSO, and counted with Cellometer Auto T4 (Nexcelom Bioscience) at the time points as indicated. For ATPlite assay, cells were seeded at 3,000 cells per well in 96-well black plates, cultured overnight, and treated with MLN4924 or DMSO for 96 h. Cell viability was determined using the ATPlite assay (PerkinElmer, 6016731) according to the manufacturer's instruction.Citation53-55
Cycloheximide-chase assay and immunoblotting
For cycloheximide (CHX)-chase experiments, cells were pretreated with 1 μM MLN4924 for 12 h, washed twice with PBS, and treated with 50 μg/ml CHX (Sigma-Aldrich, C4859) in combination with 0.3 μM MLN4924 or DMSO for indicated time points. Cell lysates were prepared and analyzed by immunoblotting. Antibody against Cullin 1 (sc-11384) was from Santa Cruz Biotechnology. Antibodies against total H2AX (t-H2AX, 3522-1), p21 (2990-1), p-IκBα (Ser32, 2798-1), IκBα (1130-1) were from Epitomics. Antibody against GAPDH (G8795) was from Sigma-Aldrich. Antibody against Noxa (OP180) was from Calbiochem. Antibodies against NEDD8 (2754), CDT1 (8064), p-H2AX (Ser139, 9718), p53 (9282), p27 (3686), Wee1 (4936), p-H3 (Ser10, 3377), cleaved caspase-3 (9661), cleaved PARP (5625), caspase-8 (9746), cleaved caspase-9 (9501), Bad (9239), Bid (2002), Bax (5023), Bak (6947), Bim (2933), Bik (4592), Bcl-2 (2870), Bcl-xL (2764), Mcl-1 (5453), XIAP (2045), c-IAP1 (7065) and c-IAP2 (3130) were from Cell Signaling Technology.
Cell-cycle profile analysis
Cell-cycle profile was evaluated by PI staining and FACS analysis as described previously.Citation17,55 Briefly, cells with the treatment of MLN4924 or DMSO were fixed in 70% ethanol at −20°C overnight, stained with PI (36 μg/mL; Sigma-Aldrich, P4170) containing RNase A (10 μg/mL; Sigma-Aldrich, R6513) at 37°C for 15 min, and analyzed for cell-cycle profile by CyAn ADP (Beckman Coulter). Data were analyzed with ModFit LT software (Verity Software House).
SA-β-gal staining assay
In Raji and U937 cells treated with MLN4924 or DMSO, SA-β-gal staining assay was performed using the SA-β-gal staining kit (Beyotime, C0602). Cells were harvested, fixed and stained according to the manufacturer's instruction.Citation32,56 Positively-stained and total cell numbers were counted under an inverted microscope (Leica Microsystems).Citation15,16
Real-time PCR and gene expression analysis by human apoptosis PCR array
Real-time PCR was performed as described previously.Citation40 Total RNA was isolated, 2.5 μg of total RNA per sample were used in the reverse transcription reaction, and the real-time PCR was performed on the ABI 7500 thermocycler (Applied Biosystems) following the instrument manual. The sequences of primers are as follows (5´ to 3´): human GAPDH: AAAGGGTCATCATCTCTG (forward), GCTGTTGTCATACTTCTC (reverse);Citation57 human p21: GACTCTCAGGGTCGAAAACG (forward), GGATTAGGGCTTCCTCTTGG (reverse);Citation58 human p27: TCCGGCTAACTCTGAGGACAC (forward), TGTTTTGAGTAGAAGAATCGTCGGT (reverse).Citation59
A human apoptosis PCR array (CT Bioscience, PA1) was used to analyze the expression of 88 key genes involved in apoptosis, according to the manufacturer's instruction. Toledo cells were harvested after treatment with 0.3 μM MLN4924 or DMSO for 36 h, and total RNA was extracted and reverse transcribed into cDNA. Gene expression analysis by human apoptosis PCR array was conducted as described.Citation60 The real-time PCR was performed with an initial denaturation at 95°C for 5 min followed by 40 cycles of denaturation at 95°C for 15 s, annealing at 60°C for 15 s and extension at 72°C for 20 s on the LightCycler 480 (Roche). Relative changes in gene expression were calculated using the 2−ΔΔCt method.Citation61
Statistical analysis
All data are represented as the mean ± SD. The statistical significance of difference between groups was assessed using the GraphPad Prism 5 software with the Student′s t test or one-way ANOVA. For all tests, 3 levels of significance (*P < 0.05, **P < 0.01, ***P < 0.001) were used.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by National Basic Research Program of China (973 program, 2012CB910302), National Natural Science Foundation of China (NSFC, grant numbers 81172092, 81372196, 31071204), the Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning and a grant from Fudan University Shanghai Cancer Center in China to Lijun Jia.
References
- Xirodimas DP. Novel substrates and functions for the ubiquitin-like molecule NEDD8. Biochem Soc Trans 2008; 36:802-6; PMID:18793140; http://dx.doi.org/10.1042/BST0360802
- Rabut G, Peter M. Function and regulation of protein neddylation. 'Protein modifications: beyond the usual suspects' review series. EMBO Rep 2008; 9:969-76; PMID:18802447; http://dx.doi.org/10.1038/embor.2008.183
- Soucy TA, Dick LR, Smith PG, Milhollen MA, Brownell JE. The NEDD8 Conjugation Pathway and Its Relevance in Cancer Biology and Therapy. Genes Cancer 2010; 1:708-16; PMID:21779466; http://dx.doi.org/10.1177/1947601910382898
- Duncan K, Schafer G, Vava A, Parker MI, Zerbini LF. Targeting neddylation in cancer therapy. Future Oncol 2012; 8:1461-70; PMID:23148618; http://dx.doi.org/10.2217/fon.12.131
- Wang M, Medeiros BC, Erba HP, DeAngelo DJ, Giles FJ, Swords RT. Targeting protein neddylation: a novel therapeutic strategy for the treatment of cancer. Expert Opin Ther Targets 2011; 15:253-64; PMID:21219242; http://dx.doi.org/10.1517/14728222.2011.550877
- Pan ZQ, Kentsis A, Dias DC, Yamoah K, Wu K. Nedd8 on cullin: building an expressway to protein destruction. Oncogene 2004; 23:1985-97; PMID:15021886; http://dx.doi.org/10.1038/sj.onc.1207414
- Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol 2005; 6:9-20; PMID:15688063; http://dx.doi.org/10.1038/nrm1547
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem 2009; 78:399-434; PMID:19489725; http://dx.doi.org/10.1146/annurev.biochem.78.101807.093809
- Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 2009; 458:732-6; PMID:19360080; http://dx.doi.org/10.1038/nature07884
- Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol 2004; 5:739-51; PMID:15340381; http://dx.doi.org/10.1038/nrm1471
- Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell 2008; 134:995-1006; PMID:18805092; http://dx.doi.org/10.1016/j.cell.2008.07.022
- Saha A, Deshaies RJ. Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol Cell 2008; 32:21-31; PMID:18851830; http://dx.doi.org/10.1016/j.molcel.2008.08.021
- Deshaies RJ, Emberley ED, Saha A. Control of cullin-ring ubiquitin ligase activity by nedd8. Subcell Biochem 2010; 54:41-56; PMID:21222272; http://dx.doi.org/10.1007/978-1-4419-6676-6_4
- Jia L, Sun Y. SCF E3 ubiquitin ligases as anticancer targets. Curr Cancer Drug Targets 2011; 11:347-56; PMID:21247385; http://dx.doi.org/10.2174/156800911794519734
- Jia L, Li H, Sun Y. Induction of p21-dependent senescence by an NAE inhibitor, MLN4924, as a mechanism of growth suppression. Neoplasia 2011; 13:561-9; PMID:21677879
- Lin JJ, Milhollen MA, Smith PG, Narayanan U, Dutta A. NEDD8-targeting drug MLN4924 elicits DNA rereplication by stabilizing Cdt1 in S phase, triggering checkpoint activation, apoptosis, and senescence in cancer cells. Cancer Res 2010; 70:10310-20; PMID:21159650; http://dx.doi.org/10.1158/0008-5472.CAN-10-2062
- Luo Z, Yu G, Lee HW, Li L, Wang L, Yang D, Pan Y, Ding C, Qian J, Wu L, et al. The Nedd8-activating enzyme inhibitor MLN4924 induces autophagy and apoptosis to suppress liver cancer cell growth. Cancer Res 2012; 72:3360-71; PMID:22562464; http://dx.doi.org/10.1158/0008-5472.CAN-12-0388
- Mackintosh C, Garcia-Dominguez DJ, Ordonez JL, Ginel-Picardo A, Smith PG, Sacristan MP, de Alava E. WEE1 accumulation and deregulation of S-phase proteins mediate MLN4924 potent inhibitory effect on Ewing sarcoma cells. Oncogene 2013; 32:1441-51; PMID:22641220; http://dx.doi.org/10.1038/onc.2012.153
- Milhollen MA, Narayanan U, Soucy TA, Veiby PO, Smith PG, Amidon B. Inhibition of NEDD8-activating enzyme induces rereplication and apoptosis in human tumor cells consistent with deregulating CDT1 turnover. Cancer Res 2011; 71:3042-51; PMID:21487042; http://dx.doi.org/10.1158/0008-5472.CAN-10-2122
- Milhollen MA, Traore T, Adams-Duffy J, Thomas MP, Berger AJ, Dang L, Dick LR, Garnsey JJ, Koenig E, Langston SP, et al. MLN4924, a NEDD8-activating enzyme inhibitor, is active in diffuse large B-cell lymphoma models: rationale for treatment of NF-{kappa}B-dependent lymphoma. Blood 2010; 116:1515-23; PMID:20525923; http://dx.doi.org/10.1182/blood-2010-03-272567
- Swords RT, Kelly KR, Smith PG, Garnsey JJ, Mahalingam D, Medina E, Oberheu K, Padmanabhan S, O'Dwyer M, Nawrocki ST, et al. Inhibition of NEDD8-activating enzyme: a novel approach for the treatment of acute myeloid leukemia. Blood 2010; 115:3796-800; PMID:20203261; http://dx.doi.org/10.1182/blood-2009-11-254862
- Luo Z, Pan Y, Jeong LS, Liu J, Jia L. Inactivation of the Cullin (CUL)-RING E3 ligase by the NEDD8-activating enzyme inhibitor MLN4924 triggers protective autophagy in cancer cells. Autophagy 2012; 8:1677-9; PMID:22874562; http://dx.doi.org/10.4161/auto.21484
- Zhao Y, Xiong X, Jia L, Sun Y. Targeting Cullin-RING ligases by MLN4924 induces autophagy via modulating the HIF1-REDD1-TSC1-mTORC1-DEPTOR axis. Cell Death Dis 2012; 3:e386; PMID:22951983; http://dx.doi.org/10.1038/cddis.2012.125
- Yao WT, Wu JF, Yu GY, Wang R, Wang K, Li LH, Chen P, Jiang YN, Cheng H, Lee HW, et al. Suppression of tumor angiogenesis by targeting the protein neddylation pathway. Cell Death Dis 2014; 5:e1059; PMID:24525735; http://dx.doi.org/10.1038/cddis.2014.21
- Nawrocki ST, Griffin P, Kelly KR, Carew JS. MLN4924: a novel first-in-class inhibitor of NEDD8-activating enzyme for cancer therapy. Expert Opin Investig Drugs 2012; 21:1563-73; PMID:22799561; http://dx.doi.org/10.1517/13543784.2012.707192
- Shankland KR, Armitage JO, Hancock BW. Non-Hodgkin lymphoma. Lancet 2012; 380:848-57; PMID:22835603; http://dx.doi.org/10.1016/S0140-6736(12)60605-9
- Dengler MA, Weilbacher A, Gutekunst M, Staiger AM, Vohringer MC, Horn H, Ott G, Aulitzky WE, van der Kuip H. Discrepant NOXA (PMAIP1) transcript and NOXA protein levels: a potential Achilles' heel in mantle cell lymphoma. Cell Death Dis 2014; 5:e1013; PMID:24457957; http://dx.doi.org/10.1038/cddis.2013.552
- Godbersen JC, Humphries LA, Danilova OV, Kebbekus PE, Brown JR, Eastman A, Danilov AV. The Nedd8-Activating Enzyme Inhibitor MLN4924 Thwarts Microenvironment-Driven NF-kappaB Activation and Induces Apoptosis in Chronic Lymphocytic Leukemia B Cells. Clin Cancer Res 2014; 20:1576-89; PMID:24634471; http://dx.doi.org/10.1158/1078-0432.CCR-13-0987
- Sundstrom C, Nilsson K. Establishment and characterization of a human histiocytic lymphoma cell line (U-937). Int J Cancer 1976; 17:565-77; PMID:178611; http://dx.doi.org/10.1002/ijc.2910170504
- Sarcar B, Kahali S, Prabhu AH, Shumway SD, Xu Y, Demuth T, Chinnaiyan P. Targeting radiation-induced G(2) checkpoint activation with the Wee-1 inhibitor MK-1775 in glioblastoma cell lines. Mol Cancer Ther 2011; 10:2405-14; PMID:21992793; http://dx.doi.org/10.1158/1535-7163.MCT-11-0469
- Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, Hunt DF, Funabiki H, Allis CD. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature 2005; 438:1116-22; PMID:16222246; http://dx.doi.org/10.1038/nature04219
- Yang D, Li L, Liu H, Wu L, Luo Z, Li H, Zheng S, Gao H, Chu Y, Sun Y, et al. Induction of autophagy and senescence by knockdown of ROC1 E3 ubiquitin ligase to suppress the growth of liver cancer cells. Cell Death Differ 2013; 20:235-47; PMID:22935614; http://dx.doi.org/10.1038/cdd.2012.113
- Lin HK, Chen Z, Wang G, Nardella C, Lee SW, Chan CH, Yang WL, Wang J, Egia A, Nakayama KI, et al. Skp2 targeting suppresses tumorigenesis by Arf-p53-independent cellular senescence. Nature 2010; 464:374-9; PMID:20237562; http://dx.doi.org/10.1038/nature08815
- Hoffman RM, Jacobsen SJ. Reversible growth arrest in simian virus 40-transformed human fibroblasts. Proc Natl Acad Sci U S A 1980; 77:7306-10; PMID:6261250; http://dx.doi.org/10.1073/pnas.77.12.7306
- Stern PH, Hoffman RM. Enhanced in vitro selective toxicity of chemotherapeutic agents for human cancer cells based on a metabolic defect. J Natl Cancer Inst 1986; 76:629-39; PMID:3457200
- Sorenson CM, Barry MA, Eastman A. Analysis of events associated with cell cycle arrest at G2 phase and cell death induced by cisplatin. J Natl Cancer Inst 1990; 82:749-55; PMID:1691303; http://dx.doi.org/10.1093/jnci/82.9.749
- Tsao YP, D'Arpa P, Liu LF. The involvement of active DNA synthesis in camptothecin-induced G2 arrest: altered regulation of p34cdc2/cyclin B. Cancer Res 1992; 52:1823-9; PMID:1312900
- Gao Q, Yu GY, Shi JY, Li LH, Zhang WJ, Wang ZC, Yang LX, Duan M, Zhao H, Wang XY, et al. Neddylation pathway is up-regulated in human intrahepatic cholangiocarcinoma and serves as a potential therapeutic target. Oncotarget 2014; 5:7820-32; PMID:25229838
- Wei D, Li H, Yu J, Sebolt JT, Zhao L, Lawrence TS, Smith PG, Morgan MA, Sun Y. Radiosensitization of human pancreatic cancer cells by MLN4924, an investigational NEDD8-activating enzyme inhibitor. Cancer Res 2012; 72:282-93; PMID:22072567; http://dx.doi.org/10.1158/0008-5472.CAN-11-2866
- Li L, Wang M, Yu G, Chen P, Li H, Wei D, Zhu J, Xie L, Jia H, Shi J, et al. Overactivated neddylation pathway as a therapeutic target in lung cancer. J Natl Cancer Inst 2014; 106:dju083; PMID:24853380
- Wang X, Li L, Liang Y, Li C, Zhao H, Ye D, Sun M, Jeong LS, Feng Y, Fu S, et al. Targeting the neddylation pathway to suppress the growth of prostate cancer cells: therapeutic implication for the men's cancer. Biomed Res Int 2014; 2014:974309; PMID:25093192
- Nawrocki ST, Kelly KR, Smith PG, Espitia CM, Possemato A, Beausoleil SA, Milhollen M, Blakemore S, Thomas M, Berger A, et al. Disrupting Protein NEDDylation with MLN4924 Is a Novel Strategy to Target Cisplatin Resistance in Ovarian Cancer. Clin Cancer Res 2013; 19:3577-90; PMID:23633453; http://dx.doi.org/10.1158/1078-0432.CCR-12-3212
- Schmitt CA, Fridman JS, Yang M, Lee S, Baranov E, Hoffman RM, Lowe SW. A senescence program controlled by p53 and p16INK4a contributes to the outcome of cancer therapy. Cell 2002; 109:335-46; PMID:12015983; http://dx.doi.org/10.1016/S0092-8674(02)00734-1
- Campisi J, d'Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol 2007; 8:729-40; PMID:17667954; http://dx.doi.org/10.1038/nrm2233
- Deng Y, Chan SS, Chang S. Telomere dysfunction and tumour suppression: the senescence connection. Nat Rev Cancer 2008; 8:450-8; PMID:18500246; http://dx.doi.org/10.1038/nrc2393
- Itahana K, Campisi J, Dimri GP. Mechanisms of cellular senescence in human and mouse cells. Biogerontology 2004; 5:1-10; PMID:15138376; http://dx.doi.org/10.1023/B:BGEN.0000017682.96395.10
- Duthu A, Debuire B, Romano J, Ehrhart JC, Fiscella M, May E, Appella E, May P. p53 mutations in Raji cells: characterization and localization relative to other Burkitt's lymphomas. Oncogene 1992; 7:2161-7; PMID:1437144
- Sugimoto K, Toyoshima H, Sakai R, Miyagawa K, Hagiwara K, Ishikawa F, Takaku F, Yazaki Y, Hirai H. Frequent mutations in the p53 gene in human myeloid leukemia cell lines. Blood 1992; 79:2378-83; PMID:1571549
- Chim CS, Wong AS, Kwong YL. Epigenetic inactivation of INK4/CDK/RB cell cycle pathway in acute leukemias. Ann Hematol 2003; 82:738-42; PMID:14513284; http://dx.doi.org/10.1007/s00277-003-0744-8
- Klangby U, Okan I, Magnusson KP, Wendland M, Lind P, Wiman KG. p16/INK4a and p15/INK4b gene methylation and absence of p16/INK4a mRNA and protein expression in Burkitt's lymphoma. Blood 1998; 91:1680-7; PMID:9473234
- Miftakhova R, Sandberg T, Hedblom A, Nevzorova T, Persson JL, Bredberg A. DNA methylation in ATRA-treated leukemia cell lines lacking a PML-RAR chromosome translocation. Anticancer Res 2012; 32:4715-22; PMID:23155234
- Lee HW, Nam SK, Choi WJ, Kim HO, Jeong LS. Stereoselective synthesis of MLN4924, an inhibitor of NEDD8-activating enzyme. J Org Chem 2011; 76:3557-61; PMID:21417215; http://dx.doi.org/10.1021/jo2001897
- Jia L, Soengas MS, Sun Y. ROC1/RBX1 E3 ubiquitin ligase silencing suppresses tumor cell growth via sequential induction of G2-M arrest, apoptosis, and senescence. Cancer Res 2009; 69:4974-82; PMID:19509229; http://dx.doi.org/10.1158/0008-5472.CAN-08-4671
- Jia L, Yang J, Hao X, Zheng M, He H, Xiong X, Xu L, Sun Y. Validation of SAG/RBX2/ROC2 E3 ubiquitin ligase as an anticancer and radiosensitizing target. Clin Cancer Res 2010; 16:814-24; PMID:20103673; http://dx.doi.org/10.1158/1078-0432.CCR-09-1592
- Li L, Liu B, Dong T, Lee HW, Yu J, Zheng Y, Gao H, Zhang Y, Chu Y, Liu G, et al. Neddylation pathway regulates the proliferation and survival of macrophages. Biochem Biophys Res Commun 2013; 432:494-8; PMID:23416079; http://dx.doi.org/10.1016/j.bbrc.2013.02.028
- Itahana K, Campisi J, Dimri GP. Methods to detect biomarkers of cellular senescence: the senescence-associated beta-galactosidase assay. Methods Mol Biol 2007; 371:21-31; PMID:17634571; http://dx.doi.org/10.1007/978-1-59745-361-5_3
- Lubelska K, Milczarek M, Modzelewska K, Krzyszton-Russjan J, Fronczyk K, Wiktorska K. Interactions between drugs and sulforaphane modulate the drug metabolism enzymatic system. Pharmacol Rep 2012; 64:1243-52; PMID:23238480; http://dx.doi.org/10.1016/S1734-1140(12)70920-9
- Cerda SR, Mustafi R, Little H, Cohen G, Khare S, Moore C, Majumder P, Bissonnette M. Protein kinase C delta inhibits Caco-2 cell proliferation by selective changes in cell cycle and cell death regulators. Oncogene 2006; 25:3123-38; PMID:16434969; http://dx.doi.org/10.1038/sj.onc.1209360
- Kim YK, Yu J, Han TS, Park SY, Namkoong B, Kim DH, Hur K, Yoo MW, Lee HJ, Yang HK, et al. Functional links between clustered microRNAs: suppression of cell-cycle inhibitors by microRNA clusters in gastric cancer. Nucleic Acids Res 2009; 37:1672-81; PMID:19153141; http://dx.doi.org/10.1093/nar/gkp002
- Shi W, Li X, Hou X, Peng H, Jiang Q, Shi M, Ji Y, Liu X, Liu J. Differential apoptosis gene expressions of rhabdomyosarcoma cells in response to enterovirus 71 infection. BMC Infect Dis 2012; 12:327; PMID:23191987; http://dx.doi.org/10.1186/1471-2334-12-327
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001; 25:402-8; PMID:11846609; http://dx.doi.org/10.1006/meth.2001.1262