Abstract
Previous studies indicate that the role of B lymphoma Mo-MLV insertion region 1 homolog (Bmi-1) is responsible for multiple cancer progression. However, Bmi-1 in controlling gene expression in non-small cell lung cancer (NSCLC) development is not well explored. Here we report that the Bmi-1 level is highly increased in primary NSCLC tissues compared to matched adjacent non-cancerous tissues and required for lung tumor growth in xenograft model. Furthermore, we also demonstrate that Bmi-1 level is lower in matched involved lymph node cancerous tissues than the respective primary NSCLC tissues. We find that Bmi-1 does not affect cell cycle and apoptosis in lung cancer cell lines as it does not affect the expression of p16/p19, Pten, AKT and P-AKT. Mechanistic analyses note that reduction of Bmi-1 expression inversely regulates invasion and metastasis of NSCLC cells in vitro and in vivo, followed by induction of epithelial-mesenchymal transition (EMT). Using genome microarray assays, we find that RNAi-mediated silence of Bmi-1 modulates some important molecular genetics or signaling pathways, potentially associated with NSCLC development. Taken together, our findings disclose for the first time that Bmi-1 level accumulates strongly in early stage and then declines in late stage, which is potentially important for NSCLC cell invasion and metastasis during progression.
Abbreviations
| NSCLC | = | non-small cell lung cancer |
| EMT | = | epithelial-mesenchymal transition |
| Bmi-1 | = | B lymphoma Mo-MLV insertion region 1 homolog |
| IHC | = | immunohistochemistry |
| FBS | = | fetal calf serum |
| qRT-PCR | = | quantitative real-time PCR |
Introduction
Lung cancer is the major cause of malignancy –related death worldwide, with a 5-year survival rate of only 15%.Citation1 The high mortality of lung cancer is mainly due to the difficulty of early diagnosis and the highly metastatic potential.Citation2 NSCLC accounts for 80% of lung cancer cases. About 70% of patients in NSCLC will develop lymph node and distant metastasis. Therefore, understanding the mechanism of metastasis is beneficial for the development of efficient tumor therapies in NSCLC.
The polycomb group (PcG) proteins are epigenetic gene-silencing proteins with important roles in embryonic development, stem cell biology and oncogenesis.Citation3 B lymphoma Mo-MLV insertion region 1 homolog (Bmi-1) is an epigenetic chromatin modifier that acts as a key component of PcG proteins, which plays important roles in mammalian stem cell self-renewal, cellular differentiation and neoplasia.Citation4 It was initially identified as a proto-oncogene that cooperates with c-myc in generation of B- and T-cell lymphomas.Citation5,6 Bmi-1 is overexpressed in NSCLC and other epithelial malignancies, including colorectal cancer, liver cancer, breast cancer and nasopharyngeal cancer.Citation7 Katerina et al. indicated that Bmi-1 was significantly associated with progression of NSCLC from a tissue microarray study.Citation8 Hu et al. showed that the combination of USP22, PTEN, Bmi-1 and p-AKT markers was the independent prognostic indicator of overall survival in non-small-cell lung cancer.Citation9 The findings display that Bmi-1 is a significant prognostic factor of poor overall survival in lung adenocarcinoma patients.Citation10 However, one group reported that Bmi-1 expression wasn't a significant prognostic factor, and was not correlated with any clinical pathological factors, only relative with early pathologic tumor classification in NSCLC.Citation11 Based on these controversial findings, further exploring the role of Bmi-1 in progression of lung cancer is necessary.
In this study, we explored the significance of Bmi-1 in lung cancer tumorigenesis and metastasis. Bmi-1 expression in 31 surgically resected primary NSCLC tissues and matched involved lymph node cancerous tissues was detected. Our results suggest that Bmi-1 is highly increased in NSCLC tissues compared with matched involved lymph node cancerous tissues. Furthermore, we also shed light on the biological impact of Bmi-1 on the invasive and metastatic properties of NSCLC cells. The overexpression of Bmi-1 reduced the invasiveness of H460 cells. In contrast, inhibiting Bmi-1 expression in NSCLC cells markedly enhanced cell invasion and lung metastases in nude mice, involved in EMT. Our results show that repression of Bmi-1 expression reduces proliferation and tumorigenesis in vivo, but does not affect cell cycle, apoptosis, p16 / p19,PTEN and AKT in vitro. Taken together, these results provide evidence that Bmi-1 inhibits aggressive and metastatic properties of NSCLC.
Results
Expression of Bmi-1 in primary NSCLC tissues is higher than in the matched lymph node cancerous tissues
To disclose the role of Bmi-1 in the progression of NSCLC, immunohistochemistry was performed to assay Bmi-1 expression in 31 surgically resected primary NSCLC tissues and the matched lymph node cancerous tissues. Nuclear Bmi1 expression was negative in normal lung tissues according to the criteria (). Bmi-1 expression in primary NSCLC tissue was positively correlated with that of the matched involved lymph node cancerous tissues (P = 0.076, ). Bmi-1 expression was significantly increased in primary NSCLC tissues compared with the matched lymph node cancerous tissues (P < 0.001, , ).These results suggest that Bmi-1 level accumulates strongly in early stage and then declines in late stage, which is reversely correlated with lymph node metastasis in NSCLC.
Table 1. The correlation of expression of Bmi-1 protein between primary NSCLC tissues and the matched lymph node cancerous tissues
Table 2. Difference of the expression of Bmi-1 between primary NSCLC tissues and the matched lymph node cancerous tissues
Figure 1. Enhanced expression of Bmi-1 in primary NSCLC tissues compared with the matched lymph node cancerous tissues. (A–C) Bmi-1 shows no or weak staining in the adjacent non-cancerous tissue (A), Strong Bmi-1 staining is detected in the primary NSCLC tissue (B). The matched lymph node cancerous tissues show lower Bmi-1 expression than that of NSCLC tissue (200×) (C). (D)Bmi-1 expression in involved lymph node cancerous tissues, primary NSCLC tissues and the corresponding normal lung tissues.
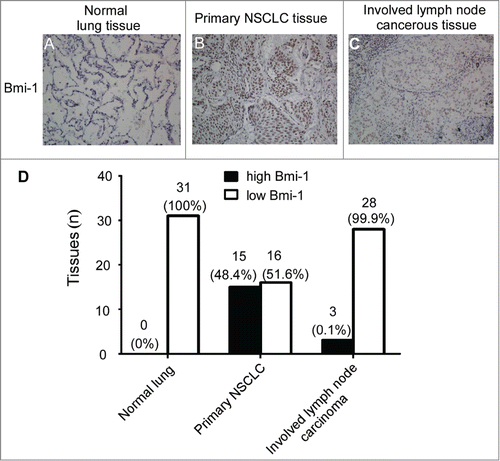
Overexpression of Bmi-1 reduces the invasiveness of H460 and CRL2741 cells in vitro
Based on the above findings, we next explore whether Bmi-1 may play a role in the invasion and metastasis of NSCLC. In this regard, the Bmi-1 expression plasmid was stably transfected into the immortalized lung epithelial cell line CRL2741 and a large-cell lung carcinoma cell line H460. show that restored Bmi-1 expression is confirmed by real-time PCR and Western- blot assays in both cell lines. The matrigel invasion chamber assay was then performed to determine the potential for Bmi-1 on invasion of lung cell. The results showed that the overexpression of Bmi-1 apparently inhibit NSCLC cell invasion compared to the respective control cells ().
Figure 2. The exogenous expression of Bmi-1 reduces the invasiveness in H460 and CRL2741 cells. (A) Quantitative RT-PCR analysis of Bmi-1 gene in CRL2741 and H460 cells expressing control vector pMSCV or pMSCV/Bmi-1.The relative fold increase of transcripts is normalized to the amount of RNA harvested from cells expressing control vector pMSCV. GAPDH served as the internal control. The data are presented as the mean ± SD (n = 3). (B) Western- blot analysis of Bmi-1 protein in CRL2741 and H460 cells expressing control vector pMSCV or pMSCV/Bmi-1. GAPDH is used as a loading control. (C–D) An invasion assay analysis of the invasive properties in CRL2741 and H460 cells expressing control vector pMSCV or pMSCV/Bmi-1. (C) The invasive cells are stained by crystal violet and then photographed by fluorescence inversion microscope system. Original magnification ×200. (D) Invasive cells are plotted as the average number of cells per field of view from 3 different experiments. The data are presented as the mean ± SD (n = 3).
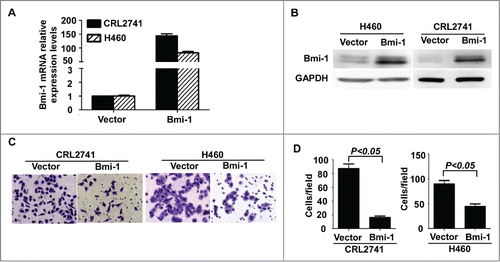
Silencing endogenous Bmi-1 enhances the invasion and metastatic abilities of lung cancer cells
To further investigate the impact of Bmi-1 on invasion and metastasis in NSCLC cells, 2 short hairpin RNA for Bmi-1 were generated to reduce Bmi-1 expression in the A549 cell line as shown in . The Boyden chamber invasion assays revealed that the invasiveness of A549 cells was dramatically increased by the ablation of Bmi-1 () in both shRNAs –mediated silencing of endogenous Bmi-1 protein. Notable, Bmi-1 shRNA #2 was more efficient and therefore chosen for subsequent experiments in vivo (). Moreover, the Bmi-1 shRNA 2# was also used to decrease Bmi-1 expression in H292 and 95D cells and similar results were obtained in both cells as shown in Figure S1. These findings indicate that silencing endogenous Bmi-1 enhances the invasion and metastatic abilities of NSCLC cells.
Figure 3. Suppression of endogenous Bmi-1enhances cellular invasion in A549. (A–B) Bmi-1 expression is confirmed by quantitative RT-PCR and Western- blot in A549 cells expressing scrambled shRNA or Bmi-1 shRNA. (C) The invasive abilities induced by Bmi-1 are analyzed by using the Matrigel-coated Boyden chamber assay in A549 cells expressing scrambled shRNA or Bmi-1 shRNA (200 ×).
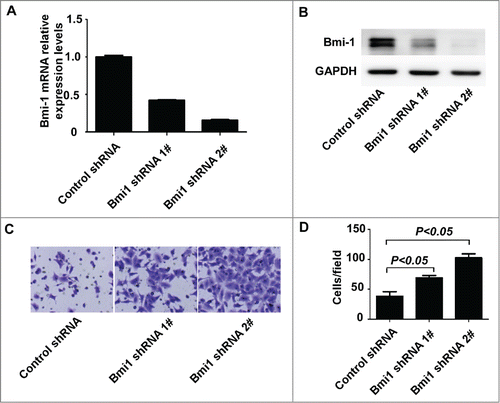
Figure 4. Suppression of endogenous Bmi-1 expression in A549 cells increases the metastasis in vivo. (A) A549-lucscrambledshRNA and A549-lucBmi-1shRNA cells are respectively injected into the caudal vein of BALB/c Nude mice. After injecting for 6 weeks, imaging is executed by a Xenogen IVIS imaging system. Luciferase signals from the tergal surface of the representative mice are shown. The data are presented as the mean ± SD (n = 8). (B) Representative gross images of lung nodules (top) and quantification of lung metastatic nodules are shown (bottom). (C) Representative images of H&E staining for lung micrometastasis (50×).
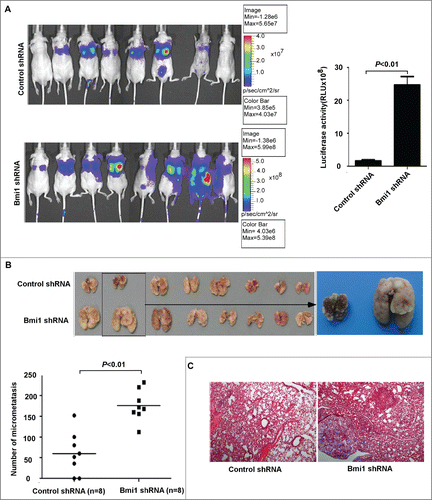
To further explore whether Bmi-1 modulates tumor metastasis in vivo, we transplanted A549-lucshRNA-Bmi-Citation1 and respective A549-lucshRNA-control cells tagged with luciferase into nude balb/c mice by tail vein injection. The results showed that A549-lucshRNA-Bmi-1 cells significantly enhanced lung metastasis compared with A549-lucshRNA-control cells. The bioluminescence imaging noted that A549-lucshRNA-Bmi-1 cells formed obviously more lung metastasis compared with A549-lucshRNA-control cells regardless of whether the animal was imaged from ventral surface () and the dorsal surface ( Fig. S2C) in 6 weeks. Examination of the number of micrometastasis also showed that lung metastasis was markedly enhanced in A549-lucshRNA-Bmi-1 mice compared with control mice ().The macroscopic findings were further confirmed by hematoxylin and eosin (H&E) staining ().These results suggest that reducing Bmi-1 expression has a significant effect on enhancing invasion and metastasis of NSCLC cells.
Identification of downstream genes by Bmi-1. To explore potential downstream targets induced by Bmi-1, we analyzed the genome-wide transcriptome profile of A549shRNA-Bmi-1 and respective A549shRNA-control cells by agilent whole human genome microarrays. The microarray data set has been deposited in the GEO database (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE60480).
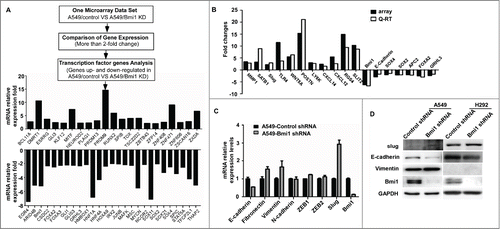
There were 593 up-regulated genes and 505 down-regulated when according to fold-change (×2.0) screening. According to fold-change (×3) screening between expression of Bmi-1 and its respective control, 268 upregulated genes and 247 downregulated genes were observed. Furthermore, silencing endogenous Bmi-1 in A549 cells by shRNAs was associated with upregulating expression of some transcription factors including KLF12,RUNS2, PRDM9, DMRT1, MITF, ZNF471, ZXDA, et al., and reducing the expression of other transcription factors including HOXA9, EGR4, SOX2, SOX4, SOX11, SOX21, FOXA2 and FOXA3, et al. (). Moreover, 18 differentiate genes obtained from microarray analyses were further verified by using qRT-PCR ().
Figure 5. The potential target genes identified by global microarray analysis and verified by QRT-PCR. (A) The mRNA expressions of transcription factors are assayed from the agilent whole human genome microarrays. (B) Some potential target genes obtained from microarray analyses are verified by QRT-PCR. (C) Expression of the E-cadherin epithelial markers, the vimentin mesenchymal markers and EMT-inducing transcription factors in Bmi-1 shRNA or scrambled shRNA–expressing A549 cells are detected by quantitative RT-PCR. GAPDH served as the internal control. The data are presented as the mean ± SD (n = 3). (D) Expression of Slug, E-cadherin and vimentin in Bmi-1 shRNA or scrambled shRNA–expressing A549 and H292 cells are detected by Western- blot assay. GAPDH is used as a loading control.
Potential mechanism induced by Bmi-1 in NSCLC progression
Among the genes markedly regulated in A549 cells, we focused on E-cadherin as downregulation of E-cadherin is a key step in Epithelial-mesenchymal transition (EMT).Citation12,13 As shown in , silencing endogenous Bmi-1 in A549 cells decreased expression of E-cadherin epithelial markers and concomitant enhanced expression of vimentin mesenchymal markers. It was also showed that silencing endogenous Bmi-1 in H292 cells decreased expression of E-cadherin. Notable, the real time PCR experiment showed that EMT-inducing transcription factors (EMT-TFs) slug expression was enhanced, whereas the expression of ZEB1 and ZEB2 did not affect (). Furthermore, the protein level of slug was enhanced after reduction of the Bmi-1 expression in A549 and H292 cells (). These results suggest that silencing Bmi-1 expression is potentially responsible for enhancing progression of NSCLC by induction of EMT.
Finally, using the KEGG and Biocarta database, we analyzed some molecular genetics or signaling pathways upon silencing Bmi-1 expression. 98 signal pathways (P < 0.05) after 1098 differentiate genes were involved in as shown in Figure S3, including WNT signaling pathway, TGF-β signaling pathway, Jak-STAT signaling pathway, MAPK signaling pathway and pathways in cancer, et al. However, further work is processing to elucidate whether these signaling pathways are involved in the progression of NSCLC.
Discussion
Efforts to elucidate the factors underlying tumorigenicity, invasion and metastasis of lung cancer are therefore warranted in order to develop novel treatments and cures. Dysregulation of Bmi-1 was often observed in many kinds of cancers.Citation7 In the present study, we demonstrated that Bmi-1 is increased in primary NSCLC tissues compared with adjacent non-cancerous tissues. More importantly, repression of Bmi-1 in NSCLC A549 cells led to inhibition of cell growth and markedly reduces proliferation and tumorigenesis in nude mice (Fig. S4). This result is consistent with the recently report that silencing endogenous Bmi-1 expression in NSCLC cells led to inhibiting cell growth and inhibition of tumorigenesis in vivo.Citation14 It is indicated that Bmi-1 plays an oncogene-role in the early progression of lung cancer. Our result is consistent with that Bmi-1 expression in proliferating neoplastic cells is an early event in lung carcinogenesis.Citation15 It was reported that Bmi-1 did not affect the proliferation of immortalized human mammary epithelial cells (HMECs),Citation16 which is consistent with our result that Bmi-1 doesn't affect the proliferation of immortalized lung epithelial cells CRL2741 cell line (Fig. S4B).
It is well known that Bmi-1 controls P16 / P19 locus, and that the genetic ablation of Bmi1 in murine cell lines leads to a decrease in tumor size, with cell cycle arrest, apoptosis, and resulting upregulation of p16 / p19.Citation17 Here our results showed that repression of Bmi-1 reduces proliferation and tumorigenesis in vivo, but it doesn't affect cell cycle arrest, apoptosis (Fig. S5) and upregulate the expression of p16 / p19 in vitro (Fig. S6A and SB). It was reported that deletion of both p16Ink4a and p19Arf did not affect the growth of Bmi-1−/− mice, suggesting that additional pathways may modulate Bmi-1 downstream, which need to be further elucidated.
Above 90% of mortality from cancer is attributed to metastases, not the primary tumors from which these malignant lesions arise.Citation18,19 In the current study, we showed that the proportion of high Bmi-1 expression in primary NSCLC tissues was higher than the matched involved lymph node cancerous tissues. Furthermore, Bmi-1 inhibited the invasion and metastasis of NSCLC cell in vitro and in vivo. These data suggest that upregulation of Bmi-1 occurred during the process of tumor growth. When tumor cells became more invasive, the Bmi-1 level was subsequently downregulated for inducing appropriate gene expression related to metastasis.
The metastasis of cancer represents the end product of a multistep cell-biological process termed the invasion-metastasis cascade, which involves dissemination of cancer cells to anatomically distant organ sites and their subsequent adaptation to foreign tissue microenvironments.Citation20 After acquiring the ability to undergo EMT, cancers tended to metastasize and build secondary tumors at distant sites.Citation21 It was recently reported that Bmi-1 was involved in the invasion and metastasis of many cancers. Yang et al. demonstrated that the direct regulation of Bmi-1 by twist1, and then these 2 genes were mutually essential to promote EMT and tumor-initiating capability of head and neck cancer cells.Citation22 The findings show that Bmi-1 represses the tumor suppressor Pten and induces EMT in human nasopharyngeal epithelial cells.Citation23 One group reported that the expression of Bmi-1 may play a crucial role in invasion and metastasis in breast cancer.Citation16 In addition, Li et al. provided evidence shows that overexpression of Bmi-1 contributes to the invasion and metastasis of hepatocellular carcinoma.Citation24 However, another group reported that the expression rate of Bmi-1 was lower in breast cancers of axillary lymph node than that of primary breast cancers.Citation25 Therefore, exploring the mechanism of metastasis is critical for the development of efficient tumor therapies in NSCLC.
Our findings indicate that low Bmi-1 expression may contribute to the metastasis of lung cancer. Moreover, the expression of Bmi-1 inhibited invasion and metastasis in lung cancer. Silencing endogenous Bmi-1 in A549 cells and H292 cells led to decreased expression of epithelial markers and concomitant enhanced expression of mesenchymal markers. In addition, the mRNA and protein expression of slug was enhanced. EMT is to be a key mechanism responsible for invasiveness and metastasis of various cancers. Reduction of Bmi-1 expression enhanced the aggressive metastatic phenotype in NSCLC maybe in part attribute to EMT. Meng X et al reported that Bmi-1 depletion inhibits cell migration and metastasis in human A549 cells. Furthermore, Bmi-1 depletion causes Pten upregulation and AKT donwregulation.Citation26 However, our findings indicate that low Bmi-1 expression may contribute to the metastasis of lung cancer. We found that the overexpression of Bmi-1 apparently inhibit NSCLC cell invasion, while silencing endogenous Bmi-1 enhances the invasion and metastatic abilities of NSCLC cells. Moreover, inhibit of Bmi-1 didn't upregulate Pten expression and downregulate AKT expression (Fig. S6C and SD). Therefore, to be fully understood, the mechanisms underlying the role of Bmi-1 level in NSCLC progression is warranty investigated.
Interestingly, some molecular signal pathways changed after inhibiting the expression of Bmi-1 in A549 cells, including WNT signaling pathway, TGF-β signaling pathway, Jak-STAT signaling pathway, MAPK signaling pathway and Hedgehog signaling pathways. Additional work is required to investigate Bmi-1 how to regulate these pathways in the invasion and metastasis of lung cancer.
In summary, our findings disclosed for the first time that Bmi-1 level accumulates strongly in early stage and then declined in late stage, which is potentially important for NSCLC cell invasion and metastasis during progression.
Materials and Methods
Tissue samples
Paraffin-embedded primary NSCLC samples and matched involved lymph node cancerous samples were obtained from 31 Chinese patients (22 men, 9 women, median age: 58 years, range: 37–81 years) diagnosed with lung cancer in 2008-2010 at Cancer Center, Sun Yat-sen University, Guangzhou, China. Patients' consent and approval from the Sun Yat-sen University Cancer Center Institute Research Ethics Committee were obtained before using these clinical materials for research purposes.
Immunohistochemistry in clinical samples
Immunohistochemistry (IHC) was made according to a modified method described previously.Citation16 The 4 μm paraffin-embedded sections of NSCLC tissue were for immunohistochemistry. The mouse monoclonal anti-Bmi-1(1:100, Upstate Biotechnology, Lake Placid, USA) was used. Blinded to the clinical outcome, 2 pathologists scored the results of the staining independently. One score is given according to the percentage of positive cells as: ≤5% of the cells: 1 point; 6%-35% of cells: 2 point; 36%-70% of cells: 3 point; ≥71% of cells: 4 point; another score is given according to the intensity of staining as: negative: 1 point; weak: 2 point; moderate: 3 point; strong: 4 point. A final score is calculated by multiple the 2 scores. If the final score is bigger than 4, the tumor is considered high expression; otherwise, Bmi-1 expression is considered low.
Cell lines, plasmids and stable transduction
Human bronchial epithelial cell line CRL-2741 was grown in keratinocyte serum-free medium (Invitrogen Corp, Carlsbad, CA) supplemented with epidermal growth factor, a gift from Gang Wang professor (Institute of Biochemistry and Cell Biology, SIBS,CAS). Human NSCLC cells A549, H292 and H460 were obtained from the American Type Culture Collection (ATCC, Rockville, MD) and cultured in DMEM containing 10% fetal bovine serum (FBS) (Invitrogen Corp, Carlsbad, CA). A549 cells retrovirally infected with a triple-fusion protein reporter construct encoding herpes simplex virus thymidine kinase 1, green fluorescent protein (GFP) and firefly luciferase (named as A549-luc) and 95D were kindly provided by Guohong Hu professor from the Institute of Health Science (SIBS, CAS) and grown in RPMI 1640 medium (Life Technologies, Carlsbad, CA) containing 10% fetal calf serum (FBS) in a humidified 5% CO2 incubator at 37°C.
The pMSCV-Bmi-1 and Bmi-1 shRNA constructs were generated as described previously.Citation27,28 Retrovirus expressing Bmi-1 was produced and transfected into CRL2741 and H460 cells according to a modified method described previously.Citation29 The plasmid with shBmi-1 was introduced into A549,A549-luc,H460 and 95D cells, and the sequence of short-hairpin RNAs (shRNAs) were as follows:Bmi-1 shRNA 1# GUUCACAAGACCAGACCAC and 2# GACCAGACCACUACUGAAU.Citation28 pMSCV and PRS plasmids were used as controls. Retroviruses were produced by transient transfection according to the manipulate method (Invitrogen Corp, Carlsbad, CA). All retrovirally infected cells were maintained under 1.0 ug/ml Puromycin selection and used as stable cells.
Quantitative real-time PCR and Western-blot analysis
Total RNA was isolated from cell lines using Trizol reagent (Invitrogen, Grand Island, NY) and reversely transcribed using the reverse transcriptase cDNA synthesis kit (Fermentas, St Leon-Rot, Germany) according to the manufacturer's instructions. cDNA was then used as template in the quantitative real-time PCR (qRT-PCR). qRT-PCR was carried out using an ABI PRISM 7500 Sequence Detection System (Applied Biosystems, Foster City, CA).The sequences of primers were listed in Table S1.
Immunoblotting was carried out as previously described.Citation30 The Western-blots were probed with mouse anti-Bmi-1(Upstate Biotechnology, Lake Placid, USA), anti-E-cadherin and anti-vimentin antibodies (BD, Transduction Laboratories, Lexington, UK)as well as with rabbit anti-GAPDH (BD, Transduction Laboratories, Lexington, UK).
In vitro migration assay
This assay measured the ability of cells to invade a matrigel matrix overlying a membrane containing 8-μm pores size polycarbonate filter. Cells were seeded in medium deprived of EGF or added 1% FBS in the top chamber (BD, Transduction Laboratories, Lexington, UK),whereas medium containing EGF or 10% FBS was added to the bottom chamber. After cultivation for 16 h, the chambers were fixed with 1% paraformaldehyde and stained with hematoxylin. Invasive cells were plotted as the average number in 10 random fields of view at 200 × magnification for each filter.
cDNA microarray analysis
The cDNA microarray analysis was carried out as described previously.Citation30 In this study, we used agilent-014850 whole human genome microarrays 4 × 44K G4112F.This Chip targets > 20,000 genes with >40000 probes originated from a broad survey of well known sources such as Ensembl, Goldenpath, RefSeq, Unigene and others. Total RNA (>300 ng) were extracted from A549/control shRNA and A549/Bmi-1shRNA cells. Microarray hybridization, microarray scan, data collection and analysis were performed at Shanghai Biotechnology Corporation.
Experimental tumor and metastasis assay in vivo
The ability to form tumors was analyzed by injecting cells with repressed Bmi-1 into nude mice. Subcutaneous tumors were established as described previously.Citation30 Briefly, 5- to 6-week-old BALB/c nude mice (Slaccas Laboratory Animal, Shanghai, China) were subcutaneously injected with cells (2 × 106/mouse) in PBS solution, and tumor sizes were measured every 5 day in order to calculate tumor volumes. All mice were sacrificed 5 weeks after injection.
The modified transplantation through tail vein injection is referred to in the report of Zheng et al.Citation30 Briefly, Tail vein injections were performed to determine whether that Bmi-1 played a role in metastasis. 5- to 6-week-old BALB/c nude mice (Slaccas Laboratory Animal, Shanghai, China) were injected into tail vein with cells (1 × 106/mouse) in PBS solution. After 6 weeks, bioluminescence was utilized to follow the lung cancer cell-derived tissue metastases. The mice were injected intraperitoneally with 15 mg/ml luciferin prior to the Xenogen IVIS cryogenically cooled imaging system to detect the tumor metastasis.
Statistical analysis
Statistical analyses were performed using SPSS 16.0. Differences among variables were analyzed by 2-tailed Student's t tests. Data were presented as the mean ± SD unless otherwise indicated. Statistical significance was defined as P < 0.05.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Methods, Materials, and Figures
Download Zip (3.4 MB)Funding
Research in the authors' laboratory is supported by the National Natural funding of China (81071747, 81272404), National key program (973) for Basic Research of China (2011CB510106, 2011CB504300), the Program for Professor of Special Appointment (Eastern Scholar to J, Wang) at Shanghai Institutions of Higher Learning, and Program of Shanghai Municipal Health Bureau Subject Chief Scientist (XBR20110052).
References
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin 2009; 59(4):225-49; PMID:19474385
- Sone S, Yano S. Molecular pathogenesis and its therapeutic modalities of lung cancer metastasis to bone. Cancer Metast Rev 2007; 26(3-4):685-9; http://dx.doi.org/10.1007/s10555-007-9081-z
- Bracken AP, Helin K. Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat Rev Cancer 2009; 9(11):773-84; PMID:19851313; http://dx.doi.org/10.1038/nrc2736
- Siddique HR, Saleem M. Role of BMI1, a stem cell factor, in cancer recurrence and chemoresistance: preclinical and clinical evidences. Stem Cell 2012; 30(3):372-8; PMID:22252887; http://dx.doi.org/10.1002/stem.1035
- Haupt Y, Alexander WS, Barri G, Klinken SP, Adams JM. Novel zinc finger gene implicated as myc collaborator by retrovirally accelerated lymphomagenesis in E mu-myc transgenic mice. Cell 1991; 65(5):753-63; PMID:1904009; http://dx.doi.org/10.1016/0092-8674(91)90383-A
- van Lohuizen M, Verbeek S, Scheijen B, Wientjens E, van der Gulden H, Berns A. Identification of cooperating oncogenes in E mu-myc transgenic mice by provirus tagging. Cell 1991; 65(5):737-52; PMID:1904008; http://dx.doi.org/10.1016/0092-8674(91)90382-9
- Valk-Lingbeek ME, Bruggeman SW, van Lohuizen M. Stem cells and cancer; the polycomb connection. Cell 2004; 118(4):409-18; PMID:15315754; http://dx.doi.org/10.1016/j.cell.2004.08.005
- Vrzalikova K, Skarda J, Ehrmann J, Murray PG, Fridman E, Kopolovic J, Knizetova P, Hajduch M, Klein J, Kolek V et al. Prognostic value of Bmi-1 oncoprotein expression in NSCLC patients: a tissue microarray study. J Cancer Res Clin Oncol 2008; 134(9):1037-42; PMID:18264721; http://dx.doi.org/10.1007/s00432-008-0361-y
- Hu J, Liu YL, Piao SL, Yang DD, Yang YM, Cai L. Expression patterns of USP22 and potential targets BMI-1, PTEN, p-AKT in non-small-cell lung cancer. Lung Cancer 2012; 77(3):5939; PMID:22717106; http://dx.doi.org/10.1016/j.lungcan.2012.05.112
- Zhang X, Sun J, Wang H, Lou Y, Zhang Y, Sha H, Feng J, Han B. IGF-1R and Bmi-1 expressions in lung adenocarcinoma and their clinicopathologic and prognostic significance. Tumour Biol 2014; 35(1):739-45; PMID:23990442; http://dx.doi.org/10.1007/s13277-013-1100-9
- Kikuchi J, Kinoshita I, Shimizu Y, Kikuchi E, Konishi J, Oizumi S, Kaga K, Matsuno Y, Nishimura M, Dosaka-Akita H. Distinctive expression of the polycomb group proteins Bmi1 polycomb ring finger oncogene and enhancer of zeste homolog 2 in nonsmall cell lung cancers and their clinical and clinicopathologic significance. Cancer 2010; 116(12):3015-24; PMID:20564407; http://dx.doi.org/10.1002/cncr.25128
- Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol 2006; 7(2):131-42; PMID:16493418; http://dx.doi.org/10.1038/nrm1835
- Huber MA, Kraut N, Beug H. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr Opin Cell Biol 2005; 17(5):548-58; PMID:16098727; http://dx.doi.org/10.1016/j.ceb.2005.08.001
- Zheng X, Wang Y, Liu B, Liu C, Liu D, Zhu J, Yang C, Yan J, Liao X, Meng X. Bmi-1-shRNA inhibits the proliferation of lung adenocarcinoma cells by blocking the G1/S phase through decreasing cyclin D1 and increasing p21/p27 levels. NuclAacid Ther 2014; 24(3):210-6; PMID:24552182; http://dx.doi.org/10.1089/nat.2013.0459
- Breuer RH, Snijders PJ, Smit EF, Sutedja TG, Sewalt RG, Otte AP, van Kemenade FJ, Postmus PE, Meijer CJ, Raaphorst FM. Increased expression of the EZH2 polycomb group gene in BMI-1-positive neoplastic cells during bronchial carcinogenesis. Neoplasia 2004; 6(6):736-43; PMID:15720799; http://dx.doi.org/10.1593/neo.04160
- Guo BH, Feng Y, Zhang R, Xu LH, Li MZ, Kung HF, Song LB, Zeng MS. Bmi-1 promotes invasion and metastasis, and its elevated expression is correlated with an advanced stage of breast cancer. Mol Cancer 2011; 10(1):10; http://dx.doi.org/10.1186/1476-4598-10-10
- Becker M, Korn C, Sienerth AR, Voswinckel R, Luetkenhaus K, Ceteci F, Rapp UR. Polycomb group protein Bmi1 is required for growth of RAF driven non-small-cell lung cancer. PloS One 2009; 4(1):e4230; PMID:19156217; http://dx.doi.org/10.1371/journal.pone.0004230
- Gupta GP, Massague J. Cancer metastasis: building a framework. Cell 2006; 127(4):679-95; PMID:17110329; http://dx.doi.org/10.1016/j.cell.2006.11.001
- Steeg PS. Tumor metastasis: mechanistic insights and clinical challenges. Nat Med 2006; 12(8):895-904; PMID:16892035; http://dx.doi.org/10.1038/nm1469
- Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell 2011; 147(2):275-92; PMID:22000009; http://dx.doi.org/10.1016/j.cell.2011.09.024
- Pantel K, Brakenhoff RH. Dissecting the metastatic cascade. Nat Rev Cancer 2004; 4(6):448-56; PMID:15170447; http://dx.doi.org/10.1038/nrc1370
- Yang MH, Hsu DS, Wang HW, Wang HJ, Lan HY, Yang WH, Huang CH, Kao SY, Tzeng CH, Tai SK et al. Bmi1 is essential in Twist1-induced epithelial-mesenchymal transition. Nat Cell Biol 2010; 12(10):982-92; PMID:20818389; http://dx.doi.org/10.1038/ncb2099
- Song LB, Li J, Liao WT, Feng Y, Yu CP, Hu LJ, Kong QL, Xu LH, Zhang X, Liu WL et al. The polycomb group protein Bmi-1 represses the tumor suppressor PTEN and induces epithelial-mesenchymal transition in human nasopharyngeal epithelial cells. J Clin Invest 2009; 119(12):3626-36; PMID:19884659; http://dx.doi.org/10.1172/JCI39374
- Li X, Yang Z, Song W, Zhou L, Li Q, Tao K, Zhou J, Wang X, Zheng Z, You N et al. Overexpression of Bmi-1 contributes to the invasion and metastasis of hepatocellular carcinoma by increasing the expression of matrix metalloproteinase (MMP)2, MMP-9 and vascular endothelial growth factor via the PTEN/PI3K/Akt pathway. Int J Oncol 2013; 43(3):793-802; PMID:23807724
- Choi YJ, Choi YL, Cho EY, Shin YK, Sung KW, Hwang YK, Lee SJ, Kong G, Lee JE, Kim JS et al. Expression of Bmi-1 protein in tumor tissues is associated with favorable prognosis in breast cancer patients. Breast Cancer Res Treat 2009; 113(1):83-93; PMID:18228133; http://dx.doi.org/10.1007/s10549-008-9909-4
- Meng X, Wang Y, Zheng X, Liu C, Su B, Nie H, Zhao B, Zhao X, Yang H. shRNA-mediated knockdown of Bmi-1 inhibit lung adenocarcinoma cell migration and metastasis. Lung Cancer 2012; 77(1):24-30; PMID:22420951; http://dx.doi.org/10.1016/j.lungcan.2012.02.015
- Song LB, Zeng MS, Liao WT, Zhang L, Mo HY, Liu WL, Shao JY, Wu QL, Li MZ, Xia YF et al. Bmi-1 is a novel molecular marker of nasopharyngeal carcinoma progression and immortalizes primary human nasopharyngeal epithelial cells. Cancer Res 2006; 66(12):6225-32; PMID:16778197; http://dx.doi.org/10.1158/0008-5472.CAN-06-0094
- Guo WJ, Datta S, Band V, Dimri GP. Mel-18, a polycomb group protein, regulates cell proliferation and senescence via transcriptional repression of Bmi-1 and c-Myc oncoproteins. Mol Biol Cell 2007; 18(2):536-46; PMID:17151361; http://dx.doi.org/10.1091/mbc.E06-05-0447
- Dimri GP, Martinez JL, Jacobs JJ, Keblusek P, Itahana K, Van Lohuizen M, Campisi J, Wazer DE, Band V. The Bmi-1 oncogene induces telomerase activity and immortalizes human mammary epithelial cells. Cancer Res 2002; 62(16):4736-45; PMID:12183433
- Zheng J, Wang J, Sun X, Hao M, Ding T, Xiong D, Wang X, Zhu Y, Xiao G, Cheng G et al. HIC1 modulates prostate cancer progression by epigenetic modification. Clin Cancer Res 2013; 19(6):1400-10; PMID:23340301; http://dx.doi.org/10.1158/1078-0432.CCR-12-2888
