Abstract
Deletions of chromosome 8p occur frequently in breast cancers, but analyses of its clinical relevance have been limited to small patient cohorts and provided controversial results. A tissue microarray with 2,197 breast cancers was thus analyzed by fluorescence in-situ hybridization using an 8p21 probe in combination with a centromere 8 reference probe. 8p deletions were found in 50% of carcinomas with no special type, 67% of papillary, 28% of tubular, 37% of lobular cancers and 56% of cancers with medullary features. Deletions were always heterozygous. 8p deletion was significantly linked to advanced tumor stage (P < 0.0001), high-grade (P < 0.0001), high tumor cell proliferation (Ki67 Labeling Index; P < 0.0001), and shortened overall survival (P < 0.0001). For example, 8p deletion was seen in 32% of 290 grade 1, 43% of 438 grade 2, and 65% of 427 grade 3 cancers. In addition, 8p deletions were strongly linked to amplification of MYC (P < 0.0001), HER2 (P < 0.0001), and CCND1 (p = 0.001), but inversely associated with ER receptor expression (p = 0.0001). Remarkably, 46.5% of 8p-deleted cancers harbored amplification of at least one of the analyzed genes as compared to 27.5% amplifications in 8p-non-deleted cancers (P < 0.0001). In conclusion, 8p deletion characterizes a subset of particularly aggressive breast cancers. As 8p deletions are easy to analyze, this feature appears to be highly suited for future DNA based prognostic breast cancer panels. The strong link of 8p deletion with various gene amplifications raises the possibility of a role for regulating genomic stability.
Keywords:
Abbreviations
| ER | = | estrogen receptor |
| FISH | = | fluorescence in situ hybridization |
| HER2 | = | human epidermal growth factor receptor 2 |
| Ki67LI | = | Ki67 Labeling index |
| LOH | = | loss of heterozygosity |
| NGS | = | next generation sequencing |
| NST | = | no special type |
| pN | = | nodal stage |
| PR | = | progesterone receptor |
| pT | = | pathological tumor stage |
| TMA | = | tissue microarray |
Introduction
Breast cancer is the most common carcinoma detected in women, accounting for about one fifth of new cancer cases in females. Surgical removal of the cancer represents the standard of care followed by radiation and adjuvant therapy in patients considered to be at particular risk for systemic disease.Citation1 Estimation of prognosis is of vital importance for tailoring adjuvant therapy in individual breast cancer patients. Conventional pathological parameters, such as histological grade, tumor size and presence of lymph node metastasis are not accurate enough to select subsets of patients who are at sufficiently low risk of progression to be spared extensive adjuvant therapy without compromising the prognosis. Much hope is placed on molecular features analysis of which might eventually help to improve prediction of patient prognosis. Several commercial molecular classifiers, based on multiplexed analyses of the RNAs of 21–70 gene products, have recently been introduced into the market in order to assist in therapy decisions making.Citation2-4 These purely RNA-based tests share the disadvantage that gradual changes of each parameter must be measured, and that these measurements are strongly dependent on tumor cell purity.
As next generation sequencing (NGS) is getting less expensive, it is expected that alternative and potentially better prognostic tests will be increasingly based on DNA analyses including a global assessment of structural rearrangements and gene mutations. NGS tests can analyze biomarkers with yes/no answers such as presence or absence of individual mutations or deletions. In other tumor types, especially in prostate cancer – another important hormone dependent cancer – various chromosomal deletions have been shown to have substantial prognostic relevance.Citation5-9 One of these is deletion of 8p, which is also found in breast cancer. Studies using classical comparative genomic hybridization in 65 – 77 patients,Citation10,11 array-based copy number screening assays in 32 – 71 patients,Citation10,12,13 fluorescence In Situ hybridization (FISH) analysis in 60 – 65 patients,Citation10,14 or loss of heterozygosity (LOH) analysis in 60 – 782 patientsCitation15,16 reported 8p deletions in 18–71 % of breast cancers. 8p deletion typically involves extended areas or even the entire 8p arm, with several putative tumor suppressor genes which are located in this region.Citation17-24 Some breast cancer studies found associations between 8p deletions and advanced tumor stage,Citation25-27 high-grade,Citation28 metastatic growth,Citation25,27,29 familial breast cancer risk,Citation30 and poor prognosis,Citation25,27,29,31 but these associations could not be confirmed by others.Citation14,32-39
To better understand the clinical relevance of 8p deletions in breast cancer, including association to tumor grade, pathological tumor stage, patient survival, hormone receptor status, and amplifications of HER2, MYC and CCND1, we analyzed more than 2,100 breast cancers with clinical follow-up data. FISH was applied for 8p deletion analysis because it is the only method enabling for direct counting of gene copy numbers in histological sections. Our data show that 8p deletion is strongly linked to a subset of particularly aggressive and genetically instable breast cancers.
Results
8p deletion frequency
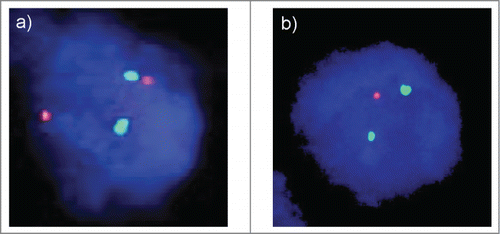
A total of 1,248 (57%) of arrayed cancer samples were analyzable by FISH. A fraction of 50% to 60% analyzable cancers was expected based on previous analyses of the same TMA.Citation40,41 Analysis failure was due to lack of tissue spots in the TMA section (9%), lack of tumor cells in the tissue spot (21%), and lack of FISH signals in the tissue spot (13%) because of insufficient probe penetration. Although the number of FISH interpretable tissue spots can be increased by repeated FISH analysis using more stringent tissue pretreatment conditions, we did not do so in order to avoid a bias that might arise from repeated analyses of some but not all spots. Heterozygous 8p deletions were found in 597 (48%) interpretable breast cancers. Homozygous 8p deletions were not detected.
Association with breast cancer phenotype
8p deletions were significantly more frequent in cancers of no special type (NST) (50% of 919) as compared to tubular (28% of 29, p = 0.02), mucinous (31% of 35, p = 0.04), and lobular (37% of 126, p = 0.006) cancers. Highest frequencies were found in cancers with medullary features (56% of 39) and papillary cancers (67% of 18), but the difference to cancer of NST did not reach statistical significance. 8p deletion was strongly linked to advanced tumor stage, high histopathological grade (P < 0.0001 each), and presence of lymph node metastases (p = 0.006) in all cancers as well as the subgroup of NST cancers (). In addition, 8p deletion was more frequent in estrogen receptor (ER) negative breast cancers (57%) as compared to ER-positive tumors (45%, P < 0.0001). All results are summarized in .
Table 1. Clinico-pathological association of 8p deletion
Association with amplifications of HER2, CCND1 and MYC
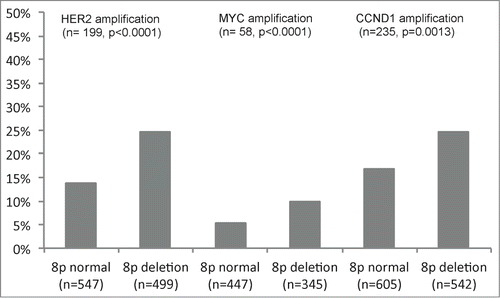
HER2, CCND1 and MYC amplification results were available from a previous study.Citation40 In total, FISH results on both 8p deletions and amplification of HER2, CCND1 and MYC were available in subsets of 1,046 (HER2), 792 (MYC) and 1,147 (CCND1) cancers. There was a positive association between 8p deletion and amplification of HER2, CCND1, and MYC. Amplification was found in 25% (HER2), 26% (CCND1), and 10% (MYC) of 8p-deleted cancers, but only in 14% (HER2, P < 0.0001), 17% (CCND1, p = 0.0013), and 5% (MYC, P < 0.0001) of cancers without 8p deletion (). Overall, 46.5% of 282 8p deleted cancers harbored amplification of at least one of these genes, but only 27.5% of 378 tumors without 8p deletion (P < 0.0001).
Association with cell proliferation
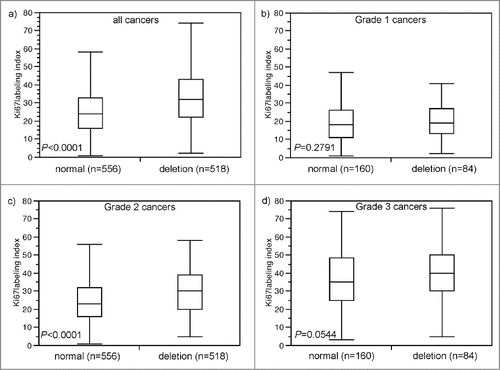
Data on the tumor cell proliferation, as determined by immunohistochemical analysis of the Ki67 antigen, were available from a previous study using the same TMA.Citation40 A total of 556 tumors harboring 8p deletions averaged a Ki67 Labeling Index (Ki67LI) of 32.9 ± 0.6, while the average Ki67LI was 25.8 ± 0.6 in 518 cancers without 8p deletion (P < 0.0001, ). A subset analysis of tumors with different grades revealed, that Ki67LI specifically varied in the intermediate (G2) group (P < 0,0001; ) while differences were only minimal in the subsets of G1 (p = 0.28; ) or G3 cancers (p = 0.05; ).
Prognostic role
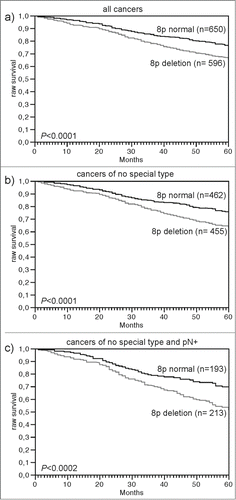
Data on raw survival were available from 1,246 cancers with interpretable 8p FISH results. Presence of 8p deletion was linked to shortened overall survival if all cancers were jointly analyzed (P < 0.0001, ), as well as in the subsets of cancers of NST (P < 0.0001, ), and in the subset of NST cancers with nodal metastases (P < 0.0002), ). Other subsets of cancer such of lobular carcinomas (n = 126; p = 0.28), nodal negative cancers (n = 523, p = 0.05) showed a similar tendency but had too low cases numbers or too few events to reach statistical significance (data not shown).
Discussion
The analysis of more than 1,200 tumors by FISH revealed 8p deletion in 47.5% of breast cancers. Our definition of deletion as “fewer 8p signals than centromere 8 signals in at least 60% of all tumor cells” was based on an earlier study where a 100% concordance was seen between FISH and array CGH data for PTEN in prostate cancer.Citation8 Given that the risk of false deletion calling mainly depends on the thickness, which is identical in our present study in breast cancer and our previous study in prostate cancer, we strongly feel that the same scoring criteria are justified. This is also supported by the concordance of our data with 2 previous FISH studies reporting 8p deletion in 39% of 60Citation14 and 45% of 65Citation10 analyzed breast cancers using a FISH probe either in the same area or in an adjacent area located only 5.5 megabases telomeric to our FISH probe. Our findings are also in the range of studies using alternative methods for deletion analysis. These studies reported 8p loss in 19–60% of breast cancers using loss of heterozygosity (LOH) analysis,Citation15,16,26,27,32 18–42%Citation42 of cancers analyzed by conventional comparative genomic hybridization (CGH),Citation11,25,29,31,34,38,43 and 32–71% of cancers examined by array-based CGH.Citation12,44 It is noteworthy, that all methods analyzing extracted DNA from disintegrated tissues are influenced by ploidy changes and contamination with normal cells, which inevitably impacts the assay sensitivity and varies markedly between individual tumors.Citation45 In contrast, FISH allows for precise gene copy number determination in individual cells, rendering it independent from the purity of cancer tissues or presence of aneusomy.Citation46-48
8p deletions were unevenly distributed between breast cancer subtypes, providing further support for the existence of biological differences between different subtypes of breast cancer. The higher rate of 8p deletions in NST as compared to lobular cancer is not surprising as most genomic alterations are more frequent in NST than in lobular carcinomas such as amplifications of HER2,Citation41,49 MYC,Citation41 MDM1,Citation41 and AIB1,Citation50 or overexpression of p53.Citation51 A higher rate of 8p deletions in NST than in lobular carcinomas has earlier been suggested by data from Guenther et al.Citation52 finding loss of 8p in 28% of NST but only in 5% of lobular carcinomas (p = 0.05) and by Thor et al.Citation53 reporting 8p in 33% of NST but only in 13% of lobular carcinomas (p = 0.12). Given the general link of 8p deletions with unfavorable disease outcome, the high rate of 8p deletions in papillary cancer and cancers with medullary features was unexpected. Both, papillary cancers and those with medullary features were subtypes considered as prognostic favorable.Citation54-57 Although we cannot explain this finding, it could be possible that papillary and medullary cancers share a particular molecular environment that is non-responsive to 8p deletion. Our observation that 8p deletions were particularly rare in tubular carcinomas fits well to the low grade and good prognosis of these cancers, however.Citation58
In an analysis of all cancers as well as in the largest subgroup of carcinomas with NST, 8p deletions were strongly linked to features of unfavorable tumor phenotype, such as advanced tumor stage, high tumor grade, rapid high tumor cell proliferation rate and poor survival. These findings are in line with previous studies reporting associations between 8p loss/LOH with adverse tumor phenotypeCitation25-29 and poor clinical outcome.Citation25,27,29,31 Studies that could find associations of 8p deletions with tumor progression had involved remarkably small cohorts of 40,Citation35 44,Citation36 46,Citation33 52,Citation38 55,Citation39 60,Citation14 61,Citation32 76Citation34 and 105Citation37 patients. The ability to detect a clinical utility of 8p deletion measurement in small cohorts is obviously caused by the high rate of positive cases (almost 50%) and a particularly strong prognostic impact of this feature. It is noteworthy, however, that several other small cohorts failed to find associations with tumor phenotypeCitation14,16,34-36,59 or patient death.Citation34-36 Limitations of our study include that retrospectively collected data are used for estimating the prognostic value of 8p deletion. Such data may be more applicable when obtained from prospectively designed randomized clinical trials,Citation60 and additional clinical endpoints such as cancer specific survival and metastasis free-survival may be included. In addition, the lack of detailed data on adjuvant/neo-adjuvant therapy made it impossible to adjust our findings for treatment. However, our findings may justify prospective studies to further clarify the suitability of 8p deletion as a putative routine prognostic marker in breast cancer. The strong prognostic impact of 8p deletions makes it very likely, that future prognostic breast cancer tests will include 8p deletion measurement. Given that future next generation sequencing methods may allow for 8p deletion measurement with a comparable accuracy than FISH,Citation61 such an analysis may be integrated in a multi-parametrical NGS-based breast cancer tests.
The strong link between heterozygous 8p deletions and features of unfavorable tumor phenotype implies that 8p may contain one or more genes that do contribute to cancer aggressiveness. Array based copy number profiling studies have shown that 8p deletion typically involves extended areas of 8p or even the entire chromosome arm.Citation13 It is, thus, likely, that the vast majority of 8p-aberrant cancers detected in our study will also harbor larger deletions. Several established or putative tumor suppressor genes are located on 8p, including CSMD1 (8p23),Citation17 DLC1 (8p22),Citation18,19 MSR1 (8p22),Citation20 MTUS1 (8p22),Citation21 TNFRSF10C (8p21),Citation22 LZTS1 (8p21),Citation23 and NRG1 (8p12).Citation24 A biologically relevant role for multiple different genes on 8p is indirectly supported by data from recent whole genome studies showing that none of the putative 8p tumor suppressor genes is A) recurrently mutated in breast cancer, and B) inactivated on both alleles, for example by deletion of one and mutation of the other allele.Citation62 These findings raise the possibility that biologically relevant consequences of 8p deletion might involve subtle expression changes of one or (more likely) several 8p genes resulting from mono-allelic loss. In fact, studies in murine models of hepatocellular carcinomas, another cancer type frequently affected by large 8p deletions, have shown that partial inactivation of several 8p genes can cooperatively drive cancer development,Citation63 thus supporting a model of compound haplo-insufficiency as the main underlying mechanism rendering 8p-deleted breast cancers particularly aggressive. The absence of homozygous deletions in 597 8p deleted cancer is additional indirect support for haplo-insufficiency of 8p genes being effective in breast cancer and suggest that one or several in the vicinity of 8p are essential for survival of breast cancer cells.
In an attempt to find evidence for particular biological effects of 8p deletions, we compared 8p deletion data with cell proliferation and parameters of genomic integrity. Strikingly, 8p deletion was strongly linked to amplification of HER2, MYC or CCND1, and the majority (47%) of 8p-deleted cancers harbored at least one of these amplifications. These findings may suggest a potential role of 8p loss for development of genomic instability. In fact, 8p12 harbors the Werner Syndrome RecQ Helicase-like (WRN) gene, which is important for the maintenance of genome stability. WRN it is a key regulator of fragile site stabilityCitation64 and involved in 2 major DNA repair pathways, the base excision repair (BER) and double strand break repair (DSBR) pathway.Citation65 In addition, POLB (8p11) and NAIL2 (8p23) are also involved in DNA repair as a key element in BER, and WRN likely acts cooperatively with POLB via its helicase and exonuclease activity.Citation66 It can be assumed that all these 3 interacting genes will be co-deleted in many cancers with 8p deletions. CGH and NGS data suggest, that at least 27–55% of 8p deleted cancers extend from 8p11–8p23.Citation10,33,34,67 In an earlier study, we had demonstrated that some breast cancers have a particular tendency to develop multiple gene amplification.Citation41 The simultaneous involvement of at least 3 genes with a role for double strand break repair in case of 8p deletion would make those cancers particularly prone to development of such an “amplifier phenotype.”Citation65 Several studies have suggested that cancers with a high degree of genomic instability might respond better to adjuvant radiation therapy or chemotherapy than genetically stable cancers.Citation68,69 It will thus be interesting to see if breast cancers with 8p-loss might respond better to neoadjuvant chemotherapy as compared to those lacking this alteration. The fact that 8p deletions were markedly associated with rapid cell proliferation is potentially caused by deletion-dependent down-regulation of several well-known cell-cycle control genes located at 8p21, 8p22 and 8p23, including for example LZTS1, an inhibitor of the Cdk1/cyclin B1 complex,Citation23 PINX1, which prolongs G0/S1 phase,Citation70 DLC1, an inducer of apoptosis,Citation71 and MTUS1, which delays progression of mitosis by prolonging metaphase.Citation21 Further steps for identification of putative tumor suppressor genes would include comparison between expression and copy numbers of 8p genes followed by functional studies. Such an analysis has been performed in liver cancer, suggesting that alterations of multiple genes might cooperatively drive tumor progression.Citation63
In conclusion, our results identify 8p deletions as a frequent event in breast cancer with marked prognostic impact. Several lines of evidence suggest, that 8p deletions exert their biological effect through combined haplo-insufficiency of multiple genes. Irrespective of what genes are involved, 8p deletions can easily be measured – for example by next generation sequencing – and thus have high potential to be part of future prognostic breast cancer panels.
Material and Methods
Breast cancer tissue microarray (TMA)
A pre-existing TMA was used for the purpose of this study.Citation40 The TMA contained in total 2,197 human breast cancer samples from paraffin-embedded tissue specimens fixed in 4% neutral buffered formalin. The median patient's age was 63 (range 26–101) years. The use of the specimens and data for research purposes was approved by local laws (HmbKHG, §12,1) and the local ethics committee (Ethics commission Hamburg, WF-049/09 and PV3652). According to local laws, informed consent was not required for this study. All work has been carried out in compliance with the Helsinki Declaration. Survival data were either obtained from the cancer registry of Basel or collected from the patients attending physicians. Raw survival data were available from 1,982 patients (713 patients with and 1,508 without event). The mean follow-up time was 63 months (range 1–176 months). Tumor size and nodal status were obtained from the primary pathology reports. No data on adjuvant/neo-adjuvant therapy were available from the arrayed cancers. All slides from the tumors were reviewed by specialized pathologists to define the histologic grade according to Elston and EllisCitation72 and the tumor type according to the WHO classification (WHO 2012). Four μm sections of the TMA blocks were transferred to an adhesive coated slide system (Instrumedics Inc.., Hackensack, New Jersey) for FISH analysis. Molecular data used in this study were available from previously published studies. These included amplification data obtained by FISH for HER2, CCND1, and MYC as well as expression data obtained by immunohistochemistry for estrogen receptor (ER), progesterone receptor (PR) and Ki67.Citation40,41
Fluorescence in situ hybridization
Four micrometer TMA sections were used for FISH. For proteolytic slide pretreatment, a commercial kit was used (paraffin pretreatment reagent kit; Abbott, Wiesbaden, Germany). TMA sections were deparaffinized, air-dried, and dehydrated in 70%, 85%, and 100% ethanol, followed by denaturation for 5 min at 74°C in 70% formamid 2x SSC solution. A FISH probe corresponding to the chromosomal region 8p21 was selected because this region is known to be frequently deleted in breast cancer.Citation10,12-16,43 The FISH probe set consisted of a spectrum-orange labeled 8p21 probe (made from a mixture of BAC RP11–625E02 and BAC RP11–116M17), and a spectrum-green labeled commercial centromere 8 probe (#6J37–08; Abbott, Wiesbaden, Germany) as a reference. Hybridization was performed overnight at 37°C in a humidified chamber. Slides were subsequently washed and counterstained with 0.2µmol/L 4′-6-diamidino-2-phenylindole in antifade solution. Stained slides were manually interpreted with an epifluorescence microscope, and the predominant FISH signal numbers were recorded in each tissue spot. Presence of fewer 8p signals than centromere 8 probe signals in at least 60% tumor nuclei were considered as heterozygous deletion (). Complete absence of 8p signals in the tumor cells, but presence of 8p signals in adjacent normal cells, was considered as homozygous deletion. Tissue spots lacking any detectable 8p signals in all (tumor and normal cells) or lack of any normal cells as an internal control for successful hybridization of the 8p probe were excluded from analysis. These thresholds were based on our previous study analyzing PTEN deletions on a prostate cancer TMA where our approach resulted in a 100% concordance with aCGH data.Citation8,73
Statistics
Statistical calculations were performed with JMP 9 software (SAS Institute Inc.., NC, USA). Fisher's exact test was used to test the statistical significance of associations between 8p deletion status and clinico-pathological variables, hormone receptor status (ER/PR), and HER2, CCND1 and MYC amplification status. Survival analysis was conducted using the Kaplan–Meier method and curves were compared with the log-rank test. Multivariate survival analyses and hazard ratios were calculated using Cox regression analysis. A value of P < 0.05 was considered statistically significant.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors appreciate the excellent technical support of Christina Koop, Sylvia Schnöger and Sasha Eghtessadi.
Funding
This work was supported by the Deutsche Forschungsgemeinschaft, Germany (DFG SI 1347/3–1).
References
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin 2011; 61:69-90; PMID:21296855; http://dx.doi.org/10.3322/caac.20107
- Hornberger J, Cosler LE, Lyman GH. Economic analysis of targeting chemotherapy using a 21-gene RT-PCR assay in lymph-node-negative, estrogen-receptor-positive, early-stage breast cancer. Am J Manag Care 2005; 11:313-24; PMID:15898220
- Cobleigh MA, Tabesh B, Bitterman P, Baker J, Cronin M, Liu ML, Borchik R, Mosquera JM, Walker MG, Shak S. Tumor gene expression and prognosis in breast cancer patients with 10 or more positive lymph nodes. Clin Cancer Res 2005; 11:8623-31; PMID:16361546; http://dx.doi.org/10.1158/1078-0432.CCR-05-0735
- van't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 2002; 415:530-6; PMID:11823860; http://dx.doi.org/10.1038/415530a
- Burkhardt L, Fuchs S, Krohn A, Masser S, Mader M, Kluth M, Bachmann F, Huland H, Steuber T, Graefen M, et al. CHD1 is a 5q21 tumor suppressor required for ERG rearrangement in prostate cancer. Cancer Res 2013; 73:2795-805; PMID:23492366; http://dx.doi.org/10.1158/0008-5472.CAN-12-1342
- El Gammal AT, Bruchmann M, Zustin J, Isbarn H, Hellwinkel OJ, Kollermann J, Sauter G, Simon R, Wilczak W, Schwarz J, et al. Chromosome 8p deletions and 8q gains are associated with tumor progression and poor prognosis in prostate cancer. Clin Cancer Res 2010; 16:56-64; PMID:20028754; http://dx.doi.org/10.1158/1078-0432.CCR-09-1423
- Kluth M, Harasimowicz S, Burkhardt L, Grupp K, Krohn A, Prien K, Gjoni J, Haß T, Galal R, Graefen M, et al. Clinical significance of different types of p53 gene alteration in surgically treated prostate cancer. Int J Cancer 2014; 135:1369-80; PMID:24523142; http://dx.doi.org/10.1002/ijc.28784
- Krohn A, Diedler T, Burkhardt L, Mayer PS, De Silva C, Meyer-Kornblum M, Tennstedt P, Huang J, Gerhäuser C, Mader M, et al. Genomic deletion of PTEN is associated with tumor progression and early PSA recurrence in ERG fusion-positive and fusion-negative prostate cancer. Am J Pathol 2012; 181:401-12; PMID:22705054; http://dx.doi.org/10.1016/j.ajpath.2012.04.026
- Krohn A, Seidel A, Burkhardt L, Bachmann F, Mader M, Grupp K, Eichenauer T, Becker A, Adam M, Graefen M, et al. Recurrent deletion of 3p13 targets multiple tumour suppressor genes and defines a distinct subgroup of aggressive ERG fusion-positive prostate cancers. J Pathol 2013; 231:130-41; PMID:23794398; http://dx.doi.org/10.1002/path.4223
- Armes JE, Hammet F, de Silva M, Ciciulla J, Ramus SJ, Soo WK, Mahoney A, Yarovaya N, Henderson MA, Gish K, et al. Candidate tumor-suppressor genes on chromosome arm 8p in early-onset and high-grade breast cancers. Oncogene 2004; 23:5697-702; PMID:15184884; http://dx.doi.org/10.1038/sj.onc.1207740
- Buerger H, Otterbach F, Simon R, Schafer KL, Poremba C, Diallo R, Brinkschmidt C, Dockhorn-Dworniczak B, Boecker W. Different genetic pathways in the evolution of invasive breast cancer are associated with distinct morphological subtypes. J Pathol 1999; 189:521-6; PMID:10629552; http://dx.doi.org/10.1002/(SICI)1096-9896(199912)189:4%3c521::AID-PATH472%3e3.0.CO;2-B
- Han S, Park K, Shin E, Kim HJ, Kim JY, Kim JY, Gwak G. Genomic change of chromosome 8 predicts the response to taxane-based neoadjuvant chemotherapy in node-positive breast cancer. Oncol Rep 2010; 24:121-8; PMID:20514452
- Cooke SL, Pole JC, Chin SF, Ellis IO, Caldas C, Edwards PA. High-resolution array CGH clarifies events occurring on 8p in carcinogenesis. BMC Cancer 2008; 8:288; PMID:18840272; http://dx.doi.org/10.1186/1471-2407-8-288
- Walker LC, McDonald M, Wells JE, Harris GC, Robinson BA, Morris CM. Dual-color fluorescence in situ hybridization reveals an association of chromosome 8q22 but not 8p21 imbalance with high grade invasive breast carcinoma. PLoS One 2013; 8:e70790; PMID:23936250; http://dx.doi.org/10.1371/journal.pone.0070790
- Miller BJ, Wang D, Krahe R, Wright FA. Pooled analysis of loss of heterozygosity in breast cancer: a genome scan provides comparative evidence for multiple tumor suppressors and identifies novel candidate regions. Am J Hum Genet 2003; 73:748-67; PMID:13680524; http://dx.doi.org/10.1086/378522
- Anbazhagan R, Fujii H, Gabrielson E. Allelic loss of chromosomal arm 8p in breast cancer progression. Am J Pathol 1998; 152:815-9; PMID:9502423
- Kamal M, Shaaban AM, Zhang L, Walker C, Gray S, Thakker N, Toomes C, Speirs V, Bell SM. Loss of CSMD1 expression is associated with high tumour grade and poor survival in invasive ductal breast carcinoma. Breast Cancer Res Treat 2010; 121:555-63; PMID:19669408; http://dx.doi.org/10.1007/s10549-009-0500-4
- Yuan BZ, Zhou X, Durkin ME, Zimonjic DB, Gumundsdottir K, Eyfjord JE, Thorgeirsson SS, Popescu NC. DLC-1 gene inhibits human breast cancer cell growth and in vivo tumorigenicity. Oncogene 2003; 22:445-50; PMID:12545165; http://dx.doi.org/10.1038/sj.onc.1206064
- Guan CN, Zhang PW, Lou HQ, Liao XH, Chen BY. DLC-1 expression levels in breast cancer assessed by qRT- PCR are negatively associated with malignancy. Asian Pac J Cancer Prev 2012; 13:1231-3; PMID:22799310; http://dx.doi.org/10.7314/APJCP.2012.13.4.1231
- Beuten J, Gelfond JA, Franke JL, Shook S, Johnson-Pais TL, Thompson IM, Leach RJ. Single and multivariate associations of MSR1, ELAC2, and RNASEL with prostate cancer in an ethnic diverse cohort of men. Cancer Epidemiol Biomarkers Prev 2010; 19:588-99; PMID:20086112; http://dx.doi.org/10.1158/1055-9965.EPI-09-0864
- Rodrigues-Ferreira S, Di Tommaso A, Dimitrov A, Cazaubon S, Gruel N, Colasson H, Nicolas A, Chaverot N, Molinié V, Reyal F, et al. 8p22 MTUS1 gene product ATIP3 is a novel anti-mitotic protein underexpressed in invasive breast carcinoma of poor prognosis. PLoS One 2009; 4:e7239; PMID:19794912; http://dx.doi.org/10.1371/journal.pone.0007239
- Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, Quist MJ, Jing X, Lonigro RJ, Brenner JC, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 2012; 487:239-43; PMID:22722839; http://dx.doi.org/10.1038/nature11125
- Lovat F, Ishii H, Schiappacassi M, Fassan M, Barbareschi M, Galligioni E, Gasparini P, Baldassarre G, Croce CM, Vecchione A. LZTS1 downregulation confers paclitaxel resistance and is associated with worse prognosis in breast cancer. Oncotarget 2014; 5:970-7; PMID:24448468
- Chua YL, Ito Y, Pole JC, Newman S, Chin SF, Stein RC, Ellis IO, Caldas C, O'Hare MJ, Murrell A, et al. The NRG1 gene is frequently silenced by methylation in breast cancers and is a strong candidate for the 8p tumour suppressor gene. Oncogene 2009; 28:4041-52; PMID:19802002; http://dx.doi.org/10.1038/onc.2009.259
- Rennstam K, Ahlstedt-Soini M, Baldetorp B, Bendahl PO, Borg A, Karhu R, Tanner M, Tirkkonen M, Isola J. Patterns of chromosomal imbalances defines subgroups of breast cancer with distinct clinical features and prognosis. A study of 305 tumors by comparative genomic hybridization. Cancer Res 2003; 63:8861-8; PMID:14695203
- Seitz S, Werner S, Fischer J, Nothnagel A, Schlag PM, Scherneck S. Refined deletion mapping in sporadic breast cancer at chromosomal region 8p12-p21 and association with clinicopathological parameters. Eur J Cancer 2000; 36:1507-13; PMID:10930798; http://dx.doi.org/10.1016/S0959-8049(00)00135-0
- Sigbjornsdottir BI, Ragnarsson G, Agnarsson BA, Huiping C, Barkardottir RB, Egilsson V, Ingvarsson S. Chromosome 8p alterations in sporadic and BRCA2 999del5 linked breast cancer. J Med Genet 2000; 37:342-7; PMID:10807692; http://dx.doi.org/10.1136/jmg.37.5.342
- Buerger H, Mommers EC, Littmann R, Simon R, Diallo R, Poremba C, Dockhorn-Dworniczak B, van Diest PJ, Boecker W. Ductal invasive G2 and G3 carcinomas of the breast are the end stages of at least two different lines of genetic evolution. J Pathol 2001; 194:165-70; PMID:11400144; http://dx.doi.org/10.1002/path.875
- Weber-Mangal S, Sinn HP, Popp S, Klaes R, Emig R, Bentz M, Mansmann U, Bastert G, Bartram CR, Jauch A. Breast cancer in young women (<or = 35 years): Genomic aberrations detected by comparative genomic hybridization. Int J Cancer 2003; 107:583-92; PMID:14520696; http://dx.doi.org/10.1002/ijc.11460
- Frank B, Bermejo JL, Hemminki K, Sutter C, Wappenschmidt B, Meindl A, Kiechle-Bahat M, Bugert P, Schmutzler RK, Bartram CR, et al. Copy number variant in the candidate tumor suppressor gene MTUS1 and familial breast cancer risk. Carcinogenesis 2007; 28:1442-5; PMID:17301065; http://dx.doi.org/10.1093/carcin/bgm033
- Isola JJ, Kallioniemi OP, Chu LW, Fuqua SA, Hilsenbeck SG, Osborne CK, Waldman FM. Genetic aberrations detected by comparative genomic hybridization predict outcome in node-negative breast cancer. Am J Pathol 1995; 147:905-11; PMID:7573366
- Radford DM, Fair KL, Phillips NJ, Ritter JH, Steinbrueck T, Holt MS, Donis-Keller H. Allelotyping of ductal carcinoma in situ of the breast: deletion of loci on 8p, 13q, 16q, 17p and 17q. Cancer Res 1995; 55:3399-405; PMID:7614479
- Farabegoli F, Hermsen MA, Ceccarelli C, Santini D, Weiss MM, Meijer GA, van Diest PJ. Simultaneous chromosome 1q gain and 16q loss is associated with steroid receptor presence and low proliferation in breast carcinoma. Mod Pathol 2004; 17:449-55; PMID:14976537; http://dx.doi.org/10.1038/modpathol.3800059
- Janssen EA, Baak JP, Guervos MA, van Diest PJ, Jiwa M, Hermsen MA. In lymph node-negative invasive breast carcinomas, specific chromosomal aberrations are strongly associated with high mitotic activity and predict outcome more accurately than grade, tumour diameter, and oestrogen receptor. J Pathol 2003; 201:555-61; PMID:14648658; http://dx.doi.org/10.1002/path.1475
- Hislop RG, Pratt N, Stocks SC, Steel CM, Sales M, Goudie D, Robertson A, Thompson AM. Karyotypic aberrations of chromosomes 16 and 17 are related to survival in patients with breast cancer. Br J Surg 2002; 89:1581-6; PMID:12445070; http://dx.doi.org/10.1046/j.1365-2168.2002.02270.x
- Seute A, Sinn HP, Schlenk RF, Emig R, Wallwiener D, Grischke EM, Hohaus S, Döhner H, Haas R, Bentz M. Clinical relevance of genomic aberrations in homogeneously treated high-risk stage II/III breast cancer patients. Int J Cancer 2001; 93:80-4; PMID:11391625; http://dx.doi.org/10.1002/ijc.1296
- Richard F, Pacyna-Gengelbach M, Schluns K, Fleige B, Winzer KJ, Szymas J, Dietel M, Petersen I, Schwendel A. Patterns of chromosomal imbalances in invasive breast cancer. Int J Cancer 2000; 89:305-10; PMID:10861509; http://dx.doi.org/10.1002/1097-0215(20000520)89:3%3c305::AID-IJC15%3e3.0.CO;2-8
- Loveday RL, Greenman J, Simcox DL, Speirs V, Drew PJ, Monson JR, Kerin MJ. Genetic changes in breast cancer detected by comparative genomic hybridisation. Int J Cancer 2000; 86:494-500; PMID:10797261; http://dx.doi.org/10.1002/(SICI)1097-0215(20000515)86:4%3c494::AID-IJC8%3e3.0.CO;2-O
- Tirkkonen M, Tanner M, Karhu R, Kallioniemi A, Isola J, Kallioniemi OP. Molecular cytogenetics of primary breast cancer by CGH. Genes Chromosomes Cancer 1998; 21:177-84; PMID:9523192; http://dx.doi.org/10.1002/(SICI)1098-2264(199803)21:3%3c177::AID-GCC1%3e3.0.CO;2-X
- Ruiz C, Seibt S, Al Kuraya K, Siraj AK, Mirlacher M, Schraml P, Maurer R, Spichtin H, Torhorst J, Popovska S, et al. Tissue microarrays for comparing molecular features with proliferation activity in breast cancer. Int J Cancer 2006; 118:2190-4; PMID:16331604; http://dx.doi.org/10.1002/ijc.21581
- Al-Kuraya K, Schraml P, Torhorst J, Tapia C, Zaharieva B, Novotny H, Spichtin H, Maurer R, Mirlacher M, Köchli O, et al. Prognostic relevance of gene amplifications and coamplifications in breast cancer. Cancer Res 2004; 64:8534-40; PMID:15574759; http://dx.doi.org/10.1158/0008-5472.CAN-04-1945
- Roylance R, Gorman P, Harris W, Liebmann R, Barnes D, Hanby A, Sheer D. Comparative genomic hybridization of breast tumors stratified by histological grade reveals new insights into the biological progression of breast cancer. Cancer Res 1999; 59:1433-6; PMID:10197608
- Buerger H, Otterbach F, Simon R, Poremba C, Diallo R, Decker T, Riethdorf L, Brinkschmidt C, Dockhorn-Dworniczak B, Boecker W. Comparative genomic hybridization of ductal carcinoma in situ of the breast-evidence of multiple genetic pathways. J Pathol 1999; 187:396-402; PMID:10398097; http://dx.doi.org/10.1002/(SICI)1096-9896(199903)187:4%3c396::AID-PATH286%3e3.0.CO;2-L
- Ruan X, Liu H, Boardman L, Kocher JP. Genome-wide analysis of loss of heterozygosity in breast infiltrating ductal carcinoma distant normal tissue highlights arm specific enrichment and expansion across tumor stages. PLoS One 2014; 9:e95783; PMID:24748104; http://dx.doi.org/10.1371/journal.pone.0095783
- Oros KK, Arcand SL, Bayani J, Squire JA, Mes-Masson AM, Tonin PN, et al. Analysis of genomic abnormalities in tumors: a review of available methods for Illumina two-color SNP genotyping and evaluation of performance. Cancer Genet 2013; 206:103-15; PMID:23623180; http://dx.doi.org/10.1016/j.cancergen.2013.03.001
- Varga Z, Noske A, Ramach C, Padberg B, Moch H. Assessment of HER2 status in breast cancer: overall positivity rate and accuracy by fluorescence in situ hybridization and immunohistochemistry in a single institution over 12 years: a quality control study. BMC Cancer 2013; 13:615; PMID:24377754; http://dx.doi.org/10.1186/1471-2407-13-615
- Leme DE, Souza DH, Mercado G, Pastene E, Dias A, Moretti-Ferreira D. Assessment of clinical scoring systems for the diagnosis of Williams-Beuren syndrome. Genet Mol Res 2013; 12:3407-11; PMID:24065682; http://dx.doi.org/10.4238/2013.September.4.7
- Gamba BF, Vieira GH, Souza DH, Monteiro FF, Lorenzini JJ, Carvalho DR, Morreti-Ferreira D. Smith-Magenis syndrome: clinical evaluation in seven Brazilian patients. Genet Mol Res 2011; 10:2664-70; PMID:22057962; http://dx.doi.org/10.4238/2011.October.31.17
- Rosenthal SI, Depowski PL, Sheehan CE, Ross JS. Comparison of HER-2/neu oncogene amplification detected by fluorescence in situ hybridization in lobular and ductal breast cancer. Appl Immunohistochem Mol Morphol 2002; 10:40-6; PMID:11893034
- Burandt E, Jens G, Holst F, Janicke F, Muller V, Quaas A, Choschzick M, Wilczak W, Terracciano L, Simon R, et al. Prognostic relevance of AIB1 (NCoA3) amplification and overexpression in breast cancer. Breast Cancer Res Treat 2013; 137:745-53; PMID:23322234; http://dx.doi.org/10.1007/s10549-013-2406-4
- Marchetti A, Buttitta F, Pellegrini S, Campani D, Diella F, Cecchetti D, Callahan R, Bistocchi M. p53 mutations and histological type of invasive breast carcinoma. Cancer Res 1993; 53:4665-9; PMID:8402644
- Gunther K, Merkelbach-Bruse S, Amo-Takyi BK, Handt S, Schroder W, Tietze L. Differences in genetic alterations between primary lobular and ductal breast cancers detected by comparative genomic hybridization. J Pathol 2001; 193:40-7; PMID:11169514; http://dx.doi.org/10.1002/1096-9896(2000)9999:9999%3c::AID-PATH745%3e3.0.CO;2-N
- Thor AD, Eng C, Devries S, Paterakos M, Watkin WG, Edgerton S, Moore DH 2nd, Etzell J, Waldman FM. Invasive micropapillary carcinoma of the breast is associated with chromosome 8 abnormalities detected by comparative genomic hybridization. Hum Pathol 2002; 33:628-31; PMID:12152162; http://dx.doi.org/10.1053/hupa.2002.124034
- Calderaro J, Espie M, Duclos J, Giachetti S, Wehrer D, Sandid W, Cahen-Doidy L, Albiter M, Janin A, de Roquancourt A. Breast intracystic papillary carcinoma: an update. Breast J 2009; 15:639-44; PMID:19735389; http://dx.doi.org/10.1111/j.1524-4741.2009.00823.x
- Solorzano CC, Middleton LP, Hunt KK, Mirza N, Meric F, Kuerer HM, Ross MI, Ames FC, Feig BW, Pollock RE, et al. Treatment and outcome of patients with intracystic papillary carcinoma of the breast. Am J Surg 2002; 184:364-8; PMID:12383904; http://dx.doi.org/10.1016/S0002-9610(02)00941-8
- Wargotz ES, Silverberg SG. Medullary carcinoma of the breast: a clinicopathologic study with appraisal of current diagnostic criteria. Hum Pathol 1988; 19:1340-6; PMID:2846422; http://dx.doi.org/10.1016/S0046-8177(88)80290-9
- Ridolfi RL, Rosen PP, Port A, Kinne D, Mike V. Medullary carcinoma of the breast: a clinicopathologic study with 10 year follow-up. Cancer 1977; 40:1365-85; PMID:907958; http://dx.doi.org/10.1002/1097-0142(197710)40:4%3c1365::AID-CNCR2820400402%3e3.0.CO;2-N
- Min Y, Bae SY, Lee HC, Lee JH, Kim M, Kim J, Lee SK, Kil WH, Kim SW, Lee JE, et al. Tubular carcinoma of the breast: clinicopathologic features and survival outcome compared with ductal carcinoma in situ. J Breast Cancer 2013; 16:404-9; PMID:24454462; http://dx.doi.org/10.4048/jbc.2013.16.4.404
- Yaremko ML, Recant WM, Westbrook CA. Loss of heterozygosity from the short arm of chromosome 8 is an early event in breast cancers. Genes Chromosomes Cancer 1995; 13:186-91; PMID:7669738; http://dx.doi.org/10.1002/gcc.2870130308
- Harris L, Fritsche H, Mennel R, Norton L, Ravdin P, Taube S, Somerfield MR, Hayes DF, Bast RC Jr; American Society of Clinical Oncology. American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J Clin Oncol 2007; 25:5287-312; PMID:17954709; http://dx.doi.org/10.1200/JCO.2007.14.2364
- Hayes JL, Tzika A, Thygesen H, Berri S, Wood HM, Hewitt S, Pendlebury M, Coates A, Willoughby L, Watson CM, et al. Diagnosis of copy number variation by Illumina next generation sequencing is comparable in performance to oligonucleotide array comparative genomic hybridisation. Genomics 2013; 102:174-81; PMID:23598253; http://dx.doi.org/10.1016/j.ygeno.2013.04.006
- Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature 2012; 490:61-70; PMID:23000897; http://dx.doi.org/10.1038/nature11412
- Xue W, Kitzing T, Roessler S, Zuber J, Krasnitz A, Schultz N, Revill K, Weissmueller S, Rappaport AR, Simon J, et al. A cluster of cooperating tumor-suppressor gene candidates in chromosomal deletions. Proc Natl Acad Sci USA 2012; 109:8212-7; PMID:22566646; http://dx.doi.org/10.1073/pnas.1206062109
- Pirzio LM, Pichierri P, Bignami M, Franchitto A. Werner syndrome helicase activity is essential in maintaining fragile site stability. J Cell Biol 2008; 180:305-14; PMID:18209099; http://dx.doi.org/10.1083/jcb.200705126
- Rossi ML, Ghosh AK, Bohr VA. Roles of Werner syndrome protein in protection of genome integrity. DNA Repair (Amst) 2010; 9:331-44; PMID:20075015; http://dx.doi.org/10.1016/j.dnarep.2009.12.011
- Harrigan JA, Wilson DM, 3rd, Prasad R, Opresko PL, Beck G, May A, Wilson SH, Bohr VA. The Werner syndrome protein operates in base excision repair and cooperates with DNA polymerase beta. Nucleic Acids Res 2006; 34:745-54; PMID:16449207; http://dx.doi.org/10.1093/nar/gkj475
- Hermsen MA, Baak JP, Meijer GA, Weiss JM, Walboomers JW, Snijders PJ, van Diest PJ. Genetic analysis of 53 lymph node-negative breast carcinomas by CGH and relation to clinical, pathological, morphometric, and DNA cytometric prognostic factors. J Pathol 1998; 186:356-62; PMID:10209483; http://dx.doi.org/10.1002/(SICI)1096-9896(199812)186:4%3c356::AID-PATH196%3e3.0.CO;2-Z
- Huhn D, Bolck HA, Sartori AA. Targeting DNA double-strand break signalling and repair: recent advances in cancer therapy. Swiss Med Wkly 2013; 143:w13837; PMID:23897299
- Irshad S, Ashworth A, Tutt A. Therapeutic potential of PARP inhibitors for metastatic breast cancer. Expert Rev Anticancer Ther 2011; 11:1243-51; PMID:21916578; http://dx.doi.org/10.1586/era.11.52
- Shi R, Zhou JY, Zhou H, Zhao Z, Liang SH, Zheng WL, Ma WL. The role of PinX1 in growth control of breast cancer cells and its potential molecular mechanism by mRNA and lncRNA expression profiles screening. Biomed Res Int 2014; 2014:978984; PMID:24672800
- Yang C, Wu D, Jia J, Liu D, Li Z, Zhang C, Li M, Xia Y. DLC1 as a regulator of proliferation, invasion, cell cycle, and apoptosis in cutaneous squamous cell carcinoma. Tumour Biol 2013; 34:2633-43; PMID:23625658; http://dx.doi.org/10.1007/s13277-013-0813-0
- Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. C. W. Elston & I. O. Ellis. Histopathology 1991; 19; 403-410. Histopathology 2002; 41:151-2, discussion 2-3; PMID:12405945; http://dx.doi.org/10.1111/j.1365-2559.1991.tb00229.x
- Kallioniemi A. DNA copy number analysis on tissue microarrays. Methods Mol Biol 2010; 664:127-34; PMID:20690059; http://dx.doi.org/10.1007/978-1-60761-806-5_13