Abstract
Purpose. The RET fusion gene is a novel oncogene observed in a subset of NSCLC in recent years. Nevertheless, the results of epidemiological studies concerning the gene remain unclear. Thus, a meta-analysis was conducted to evaluate the correlation of RET fusion gene with demographic and clinicopathological features of NSCLC. Methods. PubMed, Embase, and Web of Science databases were searched to identify eligible studies. The association of RET fusion gene occurrence with gender, age, smoking status, histology type and tumor stage were analyzed in meta-analysis. Subgroup analysis according to patients' location (Asian and non-Asian) was also conducted. Odds ratio (OR) and 95% confidence interval (95% CI) were calculated to assess the correlation. Results. Nine studies with a total of 6,899 NSCLC patients met the inclusion criteria. A total of 84 patients with RET fusion gene were detected. The RET fusion gene was identified at significantly higher frequencies in female (OR = 0.55, 95%CI = 0.35–0.85) than male patients and in young (<60 ) patients (OR = 0.43, 95%CI = 0.19–0.99) than old patients (≤60 ), particularly in patients from Asian. A significant higher frequency was also identified in non-smokers (OR = 0.28, 95% CI = 0.16–0.49), and in patients with lung adenocarcinomas (OR = 3.59, 95%CI = 1.50–8.56). Additionally, no association between RET fusion gene and the TNM stage of tumor was observed. Conclusion. RET fusion gene occurred predominantly in Asian females with younger age, in non-smokers, and in lung adenocarcinomas patients. This subset of NSCLC patients might be good candidates for personalized diagnostic and therapeutic approaches.
Abbreviations
| NSCLC | = | Non-Small Cell Lung Cancer |
| EGFR | = | Epidermal Growth Factor Receptor |
| GDNF | = | Glial cell line-derived Neurotrophic Factor |
| ARTN | = | Artemin |
| NRTN | = | Neurturin |
| PSPN | = | Persephin |
| PI3K | = | Phosphatidylinositol 3-kinase |
| TKIs | = | Tyrosine Kinase Inhibitors |
| RT-PCR | = | Real-Time Polymerase Chain Reaction |
| OR | = | Odd Ratio |
| CIs | = | Confidence Intervals |
| ADC | = | Adenocarcinoma |
| NADC | = | Non-adenocarcinoma |
| SCLC | = | Small-cell lung cancer |
Introduction
Non-Small Cell Lung Cancer (NSCLC) accounts for approximately 80% to 85% of lung cancers and is a heterogeneous disease with multiple underlying causative genetic mutations.Citation1 With the progress of molecular oncology, targeted gene therapy has become a prospective field in NSCLC treatment. Epidermal growth factor receptor (EGFR) mutation is the most common molecular driver alteration that occurred in 20 to 30 percent of NSCLC patients.Citation2 The treatment strategies blocking EGFR pathway in EGFR-mutant lung cancer, which effectively repress tumor by inhibition of driver oncogenes, establish a remarkable example of molecular targeted therapies.Citation3 Other molecular alterations, such as KRAS, BRAF mutations and ALK translocations have also been defined as “driver mutations” in lung cancer successively.Citation4 Compare with traditional chemotherapy and radiotherapy, patients with certain mutant gene will benefit from targeted therapy owing to specificity and minimal toxic sideeffects.Citation5 However, nearly half of NSCLC patients still don't have explicit gene mutations.Citation6 Thus, there is a continuing need for the exploration of potential driver mutations and to promote more effective diagnostic and individualized therapeutic strategies.
RET (located on chromosome 10q11.2) is a proto-oncogene that encodes a receptor tyrosine kinase and acts as a common receptor for members of the glial cell line-derived neurotrophic factor family (GDNF) ligands, including GDNF, artemin (ARTN), neurturin (NRTN), and persephin (PSPN).Citation7 It has a classic receptor tyrosine kinase structure consisting of 3 domains: an extracellular ligand-binding domain, a transmembrane domain and an intracellular tyrosine kinase domain.8 The extracellular binding of receptor and ligand promotes the dimerization of RET and activates intracellular tyrosine kinase domain by phosphorylation. The downstream signal is transducted through multiple pathways like Ras/RAF and phosphatidylinositol 3-kinase (PI3K), and physiologically regulates cell survival, differentiation, proliferation and migration.Citation8-10
Since the beginning of 2012, a series of researches have identified the fusion of RET gene with a couple of other genes such as KIF5B, CCDC6, NCOA4, and TRIM33 in lung cancer.Citation10-13 The mutual exclusivity of this transforming gene to other major driver mutations, like EGFR, KRAS, ALK and BRAF, suggests its potential as a novel driver oncogene.Citation14 Among several types of fusions, the KIF5B-RET is the most common one that formed by a pericentric inversion on chromosome 10. The inversion leads to a coiled coil domain, which induces autophosphorylation of RET tyrosine kinase, thus activating uncontrolled signal transduction and resulting in tumorigenesis.Citation15
To verify whether RET was a feasible target for tumor inhibition, strategies have been used to suppress the activity of RET both in pre-clinical experiments and small clinical trials. In several pre-clinical studies, the growth of RET fusion–positive cells could be inhibited by the multitargeted tyrosine kinase inhibitors (TKIs) with activity against the RET kinase, like sunitinib, sorafenib, vandetanib and alectinib.Citation10,11,16,17 While in 3 independent phase II trials, RET inhibitors including cabozantinib and vandetanib were used to treat a total of 7 RET fusion-positive patients of lung adenocarcinoma.Citation14,18,19 As a result, 4 had confirmed partial responses and 3 had prolonged stable disease, indicating the therapeutic potential of RET inhibitors as targeted drugs tailored for RET fusion-positive patients.
However, RET fusion genes occur in only <2% of NSCLC patients, even when the histological type is limited to lung adenocarcinoma, which makes it difficult for clinicians to screen fusion-positive patients.Citation11,16,20,21 So exploring the specific features of RET-positive patients may help to target people at high risk and narrow the scope of gene test. Heretofore, several studies have reported the distribution of the RET fusion gene in NSCLC patients, however, the results remain uncertain. For example, Tsuta et al. demonstrated that RET rearrangements were significantly more common in younger patients and tended to occur in non-smokers, while Cai et al. revealed that there were no statistically significant difference in age, sex and smoking status.Citation21,22 Therefore, we performed a meta-analysis to quantify the association of RET fusion genes with demographic factors and clinicopathological characteristics of NSCLC, which may facilitate the selection of cases for the check of RET rearrangements and further treatment of RET inhibitors.
Results
Study characteristics
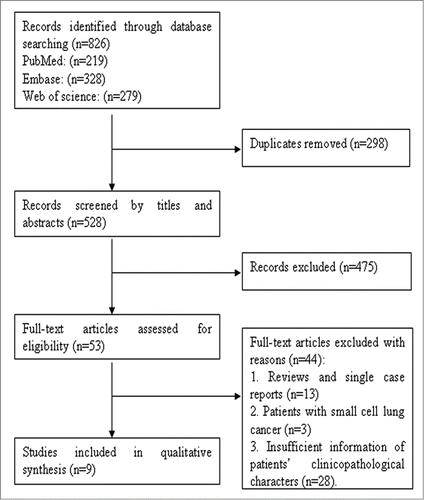
The selection steps were summarized in the flow diagram shown in . A total of 826 relevant articles were retrieved from PubMed, EMBASE, and Web of Science databases and 298 were excluded because of duplication. After screening the titles and abstracts, 475 articles were excluded due to a lack of significant relevance. The remaining 53 articles were further reviewed in detail by full texts. Finally, 9 studies including 6,899 patients were qualified for our meta-analysis.Citation10-12,20-25
Figure 1. Flow diagram of studies selection procedure and specific reasons for exclusion in the meta-analysis.
Main features of 9 eligible studies were summarized in . The median age was reported in 7 studies and the average is 61.4. For patients' location, 7 studies were conducted in Asians, and one study was conducted in patients of USA, while one study included patients from Japan, USA, and Norway. Of the 6,899 patients, 84 had RET fusion genes, giving an overall frequency of 1.2%. Specific information of each subtype of variants was given in 8 studies, including 3 types of fusions: KIF5B-RET fusions (85.6%, 59/69), CCDC6-RET fusions (13.0%, 9/69), and NCOA4-RET fusions (1.4%, 1/69). The detection method was real-time polymerase chain reaction (RT-PCR).
Table 1. Main features of studies included in the meta-analysis
Overall analysis of RET fusion genes and demographic features of patients with NSCLC
Gender
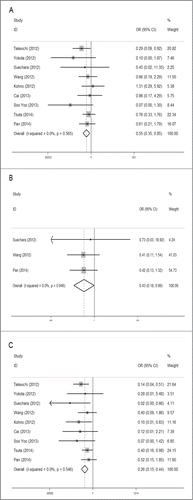
The overall analysis of the association between RET fusion gene and gender was conducted in 9 studies including 6,492 patients, showing that female patients with NSCLC had a higher RET fusion rate (1.7%, 51/3,015) than male patients (0.9%, 31/3,477). The pooled OR was 0.55(95% CI = 0.35–0.85, P = 0.007). For heterogeneity test, chi-squared was 6.74 (P = 0.565), I2 = 0%, showing there was no heterogeneity between studies ().
Age
Three studies with 1,841 patients were analyzed for the association between RET fusion gene and age with the same age-grouping criterion (60 years). Of patients younger than 60 y old, 2.0% (19/957) were RET fusion positive, higher than 1.0% (8/884) of patients 60 y or older. The pooled OR was 0.43 (95% CI = 0.19–0.99, P = 0.046), indicating that younger patients were more likely to carry the RET fusion gene. No obvious heterogeneity was observed (chi-square was 0.11, P = 0.946, and I2 = 0%) ().
Smoking
The overall analysis of the association between RET fusion gene and smoking history showed that non-smokers had a significantly higher rate than smokers with a pooled OR of 0.28 (95% CI = 0.16–0.49, P < 0.001). Among non-smokers, the RET fusion rate was 2.0% (66/3,221), 4 times that of smokers (0.5%, 16/3,268), coming from 9 studies including 6,489 patients. Between-study heterogeneity could be ignored (chi-squared was 6.01, P = 0.538, I2 = 0%) ().
Overall analysis of RET fusion genes and clinicopathological characteristics of patients with NSCLC
Histology type
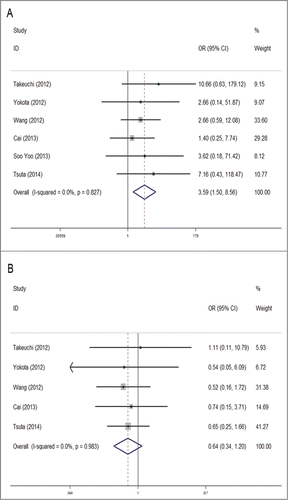
Six studies were analyzed for the association between RET gene fusions and histology type. Three studies were excluded because they specifically selected pulmonary adenocarcinoma (ADC) patients for their original researches. Overall, patients with ADC had a significant higher fusion rate of 1.4% (57/3980), compared with 0.3% (4/1278) in non-adenocarcinoma(NADC) patients. The pooled OR was 3.59 (95% CI = 1.50–8.56, P = 0.004). No obvious heterogeneity was detected (chi-square was 2.16, P = 0.827, I2 = 0%) ().
TNM stage
No statistical significance was observed in correlation between the gene and TNM stage (based on the AJCC classification). The RET fusion rate did not vary much between stage I or II NSCLC and stage III or IV (1.1% Vs 1.6%) with a pooled OR of 0.64 (95% CI = 0.34–1.2, P = 0.164). Between-study heterogeneity could be ignored (chi-squared was 0.39, P = 0.983, I2 = 0%) ().
Subgroup analysis of demographic features
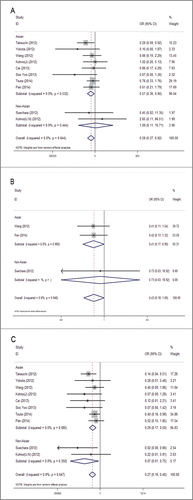
Subgroup analysis was conducted according to patient' location to see whether the demographic characteristics of RET fusion genes varied in NSCLC patients from different regions. Studies were divided into Asian group and non-Asian group. The study of Kohno et al. that included patients from Japan, USA and Norway were also split into Asian and non-Asian groups and were combined with the others. As a result, the occurrence frequency of RET fusion genes were still significantly higher in females than in males in Asian group (1.7% vs. 0.9%) with OR = 0.57, 95%CI = 0.36–0.90, P = 0.017, but not in non-Asian group (OR = 1.09, 95%CI = 0.11–10.71, P = 0.94) (). The same went with the factor of age, that younger patients of Asian group had a higher incidence of gene fusions than elder patients (2.0% vs. 0.8%) with OR = 0.41, 95%CI = 0.17–0.99, P = 0.047, while no statistical difference was detected in non-Asian group (OR = 0.73, 95%CI = 0.03–18.92, P = 0.850) (). The result of subgroup analysis in smoking status accorded with overall analysis, showing that RET fusions were identified at high frequencies in non-smokers than in smokers both in Asian group (2% vs. 0.5%) with OR = 0.29, 95%CI = 0.16–0.50, P < 0.001 and non-Asian group (8.3% vs. 0.6%) with OR = 0.07, 95%CI = 0.01–0.75, P = 0.028 ().
Figure 4. Subgroup meta-analysis of the association between RET fusion genes and demographic features in Asians and Non-Asians: gender (a), age (b), smoking history (c). Studies are divided into Asian and Non-Asian group and plotted in order of publication years. Kohno (J) refers to Japanese patients in the study, and Kohno (U, N) refers to United States and Norway patients in the study.
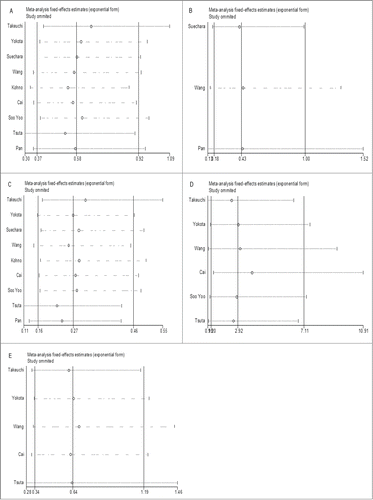
Sensitivity analysis
Sensitivity analysis was conducted by successively omitting each individual study and pooling the remaining studies. The pooled ORs and CIs were not altered significantly by each omitted study, which validated the credibility of outcomes. ()
Figure 5. (See previous page). Sensitive analysis of the effect of individual studies on the pooled OR for association between RET fusion genes and gender (A), age (B), smoking history (C), histology type (D) and TNM stage (E). The vertical axis in the middle indicates the overall OR, and the 2 vertical axes on both sides indicate the 95% CI. Every hollow round indicates the pooled OR when the left study was omitted in a meta-analysis with a random model. The two ends of every broken line represent the respective 95% CI.
Publication bias
We evaluated the publication bias using Begg's funnel plot. The shapes of the funnel plots did not have apparent asymmetry in all meta-analyses, suggesting the absence of significant biases (). This was confirmed by results of Egger's test (), demonstrating that our results were statistically robust.
Figure 6. The funnel plot of studies included in the overall analysis of the association between RET fusion genes and gender (A), age (B), smoking history (C), histology type (D) and TNM stage (E). The funnel plot displays log OR against its standard error (s.e.) for each study included in the overall analysis. The horizontal line indicates the summary estimate of the OR, with the oblique lines indicate the expected 95% CI for a given standard error.
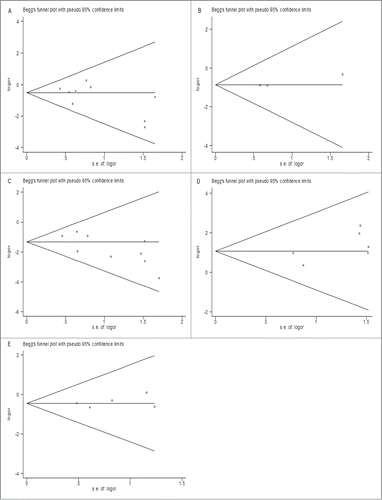
Table 2 Summary of publication bias test
Discussion
In this meta-analysis of 6,899 NSCLC patients, 84 patients were positive in RET fusions, giving an overall frequency of 1.2%. Among 69 RET mutations that classified subtype of variants, KIF5B-RET fusion gene was the major type, accounting for the largest proportion (86%, 59/69). Overall, the frequency and the distribution of mutant types of RET fusion genes in NSCLC patients were similar to other reports in the literature.
To date, several groups have investigated the relationship between the expression of RET fusion genes and demographic or clinicopathological features of NSCLC. Generally, translocations are observed in a higher percentage in young and adenocarcinoma patients.Citation26 However, there are discrepancies among other factors like sex, age, tumor size and stage. Our study, which contained a larger sample size, supported that RET fusion genes were significantly associated with gender, age, smoking status and histology type, but not with TNM stage. Since there might be a potential regional disparity in NSCLC patients with RET fusions like the variance in EGFR mutations, subgroup analysis of demographic features was also conducted among Asians and non-Asians.Citation27 It was not conducted in histology type and TNM stage on account of limited data of non-Asian group.
An important finding of our study is that female NSCLC patients were more likely to carry the RET fusion genes than males, which has not been declared previously. Reasons for this remain unclear, and it may be associated with different lifestyles, smoking habits and endocrine factors between females and males. However, non-Asian patients did not show this tendency, and no obvious difference of gender was found according to the subgroup analysis. The discrepancy may be due to different environmental and dietary factors in Asian and Non-Asian regions and/or cohort sizes. The meta-analysis also suggested that non-smokers had an increased incidence RET fusion genes (4 times that of smokers), which accorded with most previous studies.Citation12,21 Interestingly, genotype-related sensitivity has also been proven in never smokers of other subgroups of lung cancer like EGFR mutated lung cancer and EML4–ALK-positive lung cancer, suggesting that lung cancer in non-smokers is likely to be an assemblage of molecularly defined subsets.Citation3,28,29 Risk factors for non-smokers in lung cancer have been described in several studies, including second-hand smoking, occupational exposure to carcinogens, hormonal element and cooking or heating fumes. Other factors reported are infections, dietary factors et al.Citation30,31
We also found a statistical difference between RET fusion genes and age, similar to Tsuta's result, demonstrating that RET fusion genes occurred preferentially in younger patients.Citation21 It has been suggested that familial susceptibility and Mendelian inheritance may result in the early onset of lung cancer, which may account for the early onset of the lung cancer in younger patients.Citation32,33 Moreover, the influence of different age-grouping criterion should be taken into consideration. For example, Suechara et al. divided the patients into the younger and older groups by 60 years, while Cai et al. divided by 65 years, thus we failed to combine these data, which affected the sample size and diminish the power of test in meta-analysis.Citation20,22 Subgroup analysis also showed a significant correlation with younger age in Asian group, but not in non-Asian group. The discrepancies could be possibly explained that only one study was available in non-Asian group, so the authenticity of the conclusion remained to be confirmed.
For clinicopathological characteristics, the meta-analysis revealed that RET fusion genes occurred more frequently in ADC patients compared with NADC patients, which was consistent with former reports.Citation11,12 In recent study, Dabir et al. found that small-cell lung cancer(SCLC) cells had a higher RET expression compared with adenocarcinoma lung cells, which deserves further investigation about the emerging role for RET in SCLC.Citation34 Besides, in concordance with previous reports, we did not detect obvious correlation between RET fusions and TNM stage.Citation12,21,22 As to other factors like tumor size or differentiation, meta-analysis was not conducted because we failed to have the access to sufficient original data. The prognostic value of the RET fusion gene in NSCLC was also investigated in some studies. Takeuchi demonstrated that patients carrying fusion genes, including ALK, ROS1 and RET fusions, may have a better prognosis than patients without fusion genes, while the survival analysis of Cai's study showed the opposite result.Citation7,22 Therefore, more studies with precise data are required to illustrate these aspects.
Indeed, the discovery of driver oncogene in NSCLC has dramatically changed today's therapeutic landscape, and the development of molecular profiling technology provides the potential of tailored medical care. The recognition of specific characteristics of RET-positive patients may help to narrow the scope of gene screening and target people for the individualized treatment of RET inhibitors. For example, we should put more emphasis on female adenocarcinoma patients with no smoking history since they are more likely to carry RET fusion genes and probably be sensitive to RET inhibitors.
Overall, the present meta-analysis is the first study to systematically illustrate the correlation of RET fusion genes with demographic and clinicopathological features of NSCLC. We carried out extensive searches to identify studies, and detected no significant heterogeneity or publication bias among eligible studies, indicating that our results were statistically robust and credible. Nevertheless, some limitations of this meta-analysis could not be ignored. On the one hand, our study was univariate analysis, therefore, potential interactions may exist between study variables, like gender and smoking status, for the overwhelming majority of females did not have a smoking history. On the other hand, some other characteristics like tumor size, differentiation and prognosis were not analyzed in our study due to the lack of original data. Further studies are warranted to provide more complete information.
In conclusion, this meta-analysis firstly revealed the significant association between the RET fusion gene and gender, along with age, smoking status and histology type, but not stage. Asian females with younger age, non-smokers and lung adenocarcinomas patients are more likely to carry the RET fusion gene. Our study may have implications for clinicians to select cases with fusion genes and to target patients for better responses to RET inhibitors in NSCLC treatment.
Materials and Methods
Search strategy
PubMed, EMBASE, and Web of Science databases were searched using the keywords “RET fusion” or “RET rearrangement” and “lung cancer” or “pulmonary tumor” or “NSCLC” or “non-small cell lung cancer” with species restricted to human. The last search time was February 1, 2015. Titles and abstracts were screened to identify reports which examined the association of RET fusion genes with demographic and clinicopathological features in NSCLC patients. Reference lists of original articles and review articles were also manually searched for additional eligible studies.
Selection
The inclusion criteria were applied to identify eligible studies: (a) human-based investigations; (b) pathologically confirmed non-small cell lung cancer; (c) the demographic features and clinicopathological characteristics of patients were provided; (d) the availability of data to allow the estimation of the odds ration (OR) with a 95% CI; (e) published in English.
Data extraction
Studies were independently assessed by 2 investigators to extract the following data: surname of the first author, year of publication, study location, sample size, RET mutation status (the numbers of cases with RET mutation, the mutation types), demographic features (age, gender, smoking history), clinicopathological characteristics (histological types, clinical TMN stage) and assay method. Each study was examined fully to eliminate duplicates. Disagreements were resolved by discussion among 3 authors.
Data pooling and statistics
STATA 12.0 (STATA Corp., College Station, TX USA) was used to perform statistical analysis. For each study, the crude ORs and 95% CIs were calculated using χ2 or Fisher's exact tests for fourfold table. To evaluate the overall strength of the association between RET fusions and clinical characteristics, the pooled OR was calculated to estimate the effective value. The chi-square -based Q test and Higgin's I2 statistic were performed to assess the heterogeneity between studies. When strong heterogeneity existed (Q test with P < 0.05 and I2 statistic with values > 50%), the pooled OR was calculated using a random effects model (The DerSimonian and Laird method). Otherwise, a fixed effects model (Mantel-Haenszel method) was more appropriate. Sensitivity analysis was conducted to confirm the credibility of the results in this meta-analysis, by successively omitting each individual study and pooling the remaining studies. We evaluated the publication bias using Begg's funnel plot and tested it with Egger's test. P value <0.05 was considered statistically significant.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
Financial support was provided by project NO. 124119a6200 supported by Shanghai Science and Technology Committee.
References
- D'Addario G, Fruh M, Reck M, Baumann P, Klepetko W, Felip E. Metastatic non-small-cell lung cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2010; 21(Suppl 5):v116-9; PMID:20555059; http://dx.doi.org/10.1093/annonc/mdq189
- Shi Y, Au J, Thongprasert S, Srinivasan S, Tsai CM, Khoa MT, Heeroma K, Itoh Y, Cornelio G, Yang PC. A prospective, molecular epidemiology study of EGFR mutations in asian patients with advanced non–small-cell lung cancer of adenocarcinoma histology (PIONEER). J Thorac Oncol 2014; 9:154-62; PMID:24419411; http://dx.doi.org/10.1097/JTO.0000000000000033
- Lee YJ, Kim JH, Kim SK, Ha SJ, Mok TS, Mitsudomi T, Cho BC. Lung cancer in never smokers: change of a mindset in the molecular era. Lung Cancer 2011; 72:9-15; PMID:21272954; http://dx.doi.org/10.1016/j.lungcan.2010.12.013
- Girard N. Other signalization targets. Targ Oncol 2013; 8(1):3-14; PMID:23307186; http://dx.doi.org/10.1007/s11523-012-0246-5
- Pan Q, Wang Y, Chen J, Xu G, Chen B, Pan J, Huang K. Investigation of the epidermal growth factor receptor mutation rate in non-small cell lung cancer patients and the analysis of associated risk factors using logistic regression. Oncology Letters 2014; 8:813-8; PMID:25013503
- Sequist LV, Heist RS, Shaw AT, Fidias P, Rosovsky R, Temel JS, Lennes IT, Digumarthy S, Waltman BA, Bast E, et al. Implementing multiplexed genotyping of non-small-cell lung cancers into routine clinical practice. Ann Oncol 2011; 22(12):2616-24; PMID:22071650; http://dx.doi.org/10.1093/annonc/mdr489
- Takahashi M, Ritz J, Cooper GM. Activation of a novel human transforming gene, ret, by DNA rearrangement. Cell 1985; 42(2): 581-8; PMID:2992805; http://dx.doi.org/10.1016/0092-8674(85)90115-1
- Phay JE, Shah MH. Targeting RET receptor tyrosine kinase activation in cancer. Clin Cancer Res 2010; 16(24):5936-41; PMID:20930041; http://dx.doi.org/10.1158/1078-0432.CCR-09-0786
- Pützer BM, Drosten M. The RET proto-oncogene: a potential target for molecular cancer therapy. Trends Mol Med 2004; 10(7):351-7; PMID:15242684; http://dx.doi.org/10.1016/j.molmed.2004.06.002
- Kohno T, Ichikawa H, Totoki Y, Yasuda K, Hiramoto M, Nammo T, Sakamoto H, Tsuta K, Furuta K, Shimada Y, et al. KIF5B-RET fusions in lung adenocarcinoma. Nat Med 2012; 18(3):375-7; PMID:22327624; http://dx.doi.org/10.1038/nm.2644
- Takeuchi K, Soda M, Togashi Y, Suzuki R, Sakata S, Hatano S, Asaka R, Hamanaka W, Ninomiya H, Uehara H, et al. RET, ROS1 and ALK fusions in lung cancer. Nat Med 2012; 18(3):378-81; PMID:22327623; http://dx.doi.org/10.1038/nm.2658
- Wang R, Hu H, Pan Y, Li Y, Ye T, Li C, Luo X, Wang L, Li H, Zhang Y, et al. RET fusions define a unique molecular and clinicopathologic subtype of non-small-cell lung cancer. J Clin Oncol 2012; 30(35):4352-9; PMID:23150706; http://dx.doi.org/10.1200/JCO.2012.44.1477
- Drilon A, Wang L, Hasanovic A, Suehara Y, Lipson D, Stephens P, Ross J, Miller V, Ginsberg M, Zakowski MF, et al. Response to cabozantinib in patients with RET fusion-positive lung adenocarcinomas. Cancer Discov 2013; 3(6):630-5; PMID:23533264; http://dx.doi.org/10.1158/2159-8290.CD-13-0035
- Borrello MG, Ardini E, Locati LD, Greco A, Licitra L, Pierotti MA. RET inhibition: implications in cancer therapy. Expert Opin Ther Targets 2013; 17(4):403-19; PMID:23461584; http://dx.doi.org/10.1517/14728222.2013.758715
- Gainor JF, Shaw AT. Novel targets in non-small cell lung cancer:ROS1 and RET fusions. Oncologist 2013; 18:865-75; PMID:23814043; http://dx.doi.org/10.1634/theoncologist.2013-0095
- Lipson D, Capelletti M, Yelensky R, Otto G, Parker A, Jarosz M, Curran JA, Balasubramanian S, Bloom T, Brennan KW, et al. Identication of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med 2012; 18(3):382-4; PMID:22327622; http://dx.doi.org/10.1038/nm.2673
- Kodama T, Tsukaguchi T, Satoh Y, Yoshida M, Watanabe Y, Kondoh O, Sakamoto H. Alectinib shows potent antitumor activity against RET-rearranged non–small cell lung cancer. Mol Cancer Ther 2014; 13(12):1-9; PMID:25349307; http://dx.doi.org/10.1158/1535-7163.MCT-14-0274
- Mukhopadhyay S, Pennell NA, Ali SM, Ross JS, Ma PC, Velcheti V. RET-Rearranged lung adenocarcinomas with lymphangitic spread, psammoma bodies, and clinical responses to cabozantinib. J Thorac Oncol 2014; 9(11):1714-9; PMID:25436805; http://dx.doi.org/10.1097/JTO.0000000000000323
- Gautschi O, Zander T, Keller FA, Strobel K, Hirschmann A, Aebi S, Diebold J. A patient with lung adenocarcinoma and RET fusion treated with vandetanib. J Thorac Oncol 2013; 8(5):e43-4; PMID:23584301; http://dx.doi.org/10.1097/JTO.0b013e31828a4d07
- Suehara Y, Arcila M, Wang L, Hasanovic A, Ang D, Ito T, Kimura Y, Drilon A, Guha U, Rusch V, et al. Identication of KIF5B-RET and GOPC-ROS1 fusions in lung adenocarcinomas through a comprehensive mRNA-based screen for tyrosine kinase fusions. Clin Cancer Res 2012; 18(24):6599-608; PMID:23052255; http://dx.doi.org/10.1158/1078-0432.CCR-12-0838
- Tsuta K, Kohno T, Yoshida A, Shimada Y, Asamura H, Furuta K, Kushima R. RET-rearranged non-small-cell lung carcinoma a clinicopathological and molecular analysis. British J Cancer 2014; 110(6):1571-8; PMID:24504365; http://dx.doi.org/10.1038/bjc.2014.36
- Cai W, Su C, Li X, Fan L, Zheng L, Fei K, Zhou C. KIF5B-RET fusions in Chinese patients with non-small cell lung cancer. Cancer 2013; 119(8):1486-94; PMID:23378251; http://dx.doi.org/10.1002/cncr.27940
- Yokota K, Sasaki H, Okuda K, Shimizu S, Shitara M, Hikosaka Y, Moriyama S, Yano M, Fujii Y. KIF5B/RET fusion gene in surgically-treated adenocarcinoma of the lung. Oncol Rep 2012; 28(4):1187-92; PMID:22797671
- Yoo SS, Jin G, Jung HJ, Hong MJ, Choi JE, Jeon HS, Lee SY, Lim JO, Park JY. RET Fusion genes in korean non-small cell lung cancer. J Korean Med Sci 2013; 28(10):1555-8; PMID:24133367; http://dx.doi.org/10.3346/jkms.2013.28.10.1555
- Pan Y, Zhang Y, Li Y, Hu H, Wang L, Li H, Wang R, Ye T, Luo X, Zhang Y, et al. ALK, ROS1 and RET fusions in 1139 lung adenocarcinomas: A comprehensive study of common and fusion pattern-specificclinicopathologic, histologic and cytologic features. Lung Cancer 2014; 84(2):121-6; PMID:24629636; http://dx.doi.org/10.1016/j.lungcan.2014.02.007
- Emma S, Thomas H, George R, Phillip A, Gregory A, Raphael B, Salgia R. Molecular pathways and therapeutic targets in lung cancer. Oncotarget 2014; 5(6):1392-433; PMID:24722523
- Jeon YK, Sung SW, Chung JH, Park WS, Seo JW, Kim CW, Chung DH. Clinicopathologic features and prognostic implications of epidermal growth factor receptor (EGFR) gene copy number and protein expression in non-small cell lung cancer. Lung Cancer 2006; 54(3):387-98; PMID:17011067; http://dx.doi.org/10.1016/j.lungcan.2006.08.015
- Chang A, Parikh P, Thongprasert S, Tan EH, Perng R, Ganzon D, Yang CH, Tsao CJ, Watkins C, Botwood N, et al. Gefitinib (IRESSA) in patients of Asian origin with refractory advanced non-small cell lung cancer: subset analysis from the ISEL study. J Thorac Oncol 2006; 1:847-55; PMID:17409969; http://dx.doi.org/10.1097/01243894-200610000-00014
- Kwak EL, Bang Y, Camidge DR, Shaw AT, Solomon B, Maki RG, Ou SH, Dezube BJ, Jänne PA, Costa DB, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 2010; 363:1693-703; PMID:20979469; http://dx.doi.org/10.1056/NEJMoa1006448
- Pallis AG, Syrigos KN. Lung cancer in never smokers: disease characteristics and risk factors. Critical Rev Oncol/Hematol 2013; 88:494-503; PMID:23921082; http://dx.doi.org/10.1016/j.critrevonc.2013.06.011
- Couraud S, Zalcman G, Milleron B, Morin F, Souquet PJ. Lung cancer in never smokers-a review. European J Cancer 2012; 48:1299-311; PMID:22464348; http://dx.doi.org/10.1016/j.ejca.2012.03.007
- Coté ML, Kardia SL, Wenzlaff AS, Ruckdeschel JC, Schwartz AG. Risk of lung cancer among white and black relatives of individuals with early-onset lung cancer. JAMA 2005; 293:3036-42; PMID:15972566; http://dx.doi.org/10.1001/jama.293.24.3036
- Kharazmi E, Fallah M, Sundquist K, Hemminki K. Familial risk of early and late onset cancer: nationwide prospective cohort study. BMJ 2012; 345:e8076; PMID:23257063; http://dx.doi.org/10.1136/bmj.e8076
- Dabir S, Babakoohi S, Kluge A, Morrow JJ, Kresak A, Yang M, MacPherson D, Wildey G, Dowlati A. RET Mutation and expression in small-cell lung cancer. J Thorac Oncol 2014; 9(9):1316-23; PMID:25122427; http://dx.doi.org/10.1097/JTO.0000000000000234