Abstract
Preoperative chemoradiotherapy is widely used to improve local control of disease, sphincter preservation and to improve survival in patients with locally advanced rectal cancer. Patients enrolled in the present study underwent preoperative chemoradiotherapy, followed by surgical excision. Response to chemoradiotherapy was evaluated according to Mandard's Tumor Regression Grade (TRG). TRG 3, 4 and 5 were considered as partial or no response while TRG 1 and 2 as complete response. From pretherapeutic biopsies of 84 locally advanced rectal carcinomas available for the analysis, only 42 of them showed 70% cancer cellularity at least. By determining gene expression profiles, responders and non-responders showed significantly different expression levels for 19 genes (P < 0.001). We fitted a logistic model selected with a stepwise procedure optimizing the Akaike Information Criterion (AIC) and then validated by means of leave one out cross validation (LOOCV, accuracy = 95%). Four genes were retained in the achieved model: ZNF160, XRCC3, HFM1 and ASXL2. Real time PCR confirmed that XRCC3 is overexpressed in responders group and HFM1 and ASXL2 showed a positive trend. In vitro test on colon cancer resistant/susceptible to chemoradioterapy cells, finally prove that XRCC3 deregulation is extensively involved in the chemoresistance mechanisms. Protein-protein interactions (PPI) analysis involving the predictive classifier revealed a network of 45 interacting nodes (proteins) with TRAF6 gene playing a keystone role in the network. The present study confirmed the possibility that gene expression profiling combined with integrative computational biology is useful to predict complete responses to preoperative chemoradiotherapy in patients with advanced rectal cancer.
Abbreviations
| RC | = | Rectal cancer |
| pCRT | = | Preoperative chemoradiotherapy |
| CEA | = | carcinoembryonic antigen |
| Gy | = | Gray |
| PPI | = | Protein-protein interaction |
| mRNA | = | mRNA |
| SSB | = | Single-strand breaks |
| DSB | = | Double-strand breaks |
| SNP | = | Single nucleotide polymorphism |
| HT | = | High throughput |
| CRT | = | Chemoradiotherapy |
| RIN | = | RNA integrity number |
| siRNA | = | Small interfering RNA |
Introduction
Preoperative chemoradiotherapy (pCRT) is worldwide accepted as a standard treatment for locally advanced rectal cancer.Citation1-3 After pCRT the complete pathological response is approximately 20%, whereas in 20 to 40% of patients the response is poor or absent.Citation4,5 The prediction of response has the potential to spare unnecessary toxic treatments for non-responders and, in selected cases, to perform a less-radical surgery (e.g. local excision or a wait and see policy).
Several studies have been performed to evaluate potential predictors of response after pCRT for rectal cancer, however findings are still unclear and controversial.Citation6,7 Discrepancies between studies are mainly related to patient selection, sample size, study design, treatments and definitions used for tumor response. Moreover, the only accepted marker to monitor colorectal cancer treatment, progression and relapse is the carcinoembryonic antigen (CEA).Citation8
However, gene signatures using microarray technology may help to predict tumor response after pCRT. Recent studies using microarray technology have shown that gene expression profiles of tumor cells can discriminate responders and non-responders patients after neoadjuvant or adjuvant chemotherapy.Citation9-13
The clinical value of these studies is to identify disease subtypes that represent distinct subphenotypes of rectal cancer in order to better approach opportunities for individualized therapeutics. Despite these advances, few studies have attempted to demonstrate the value in integrating genomic information with the traditional clinical risk factors to provide a more detailed assessment of clinical risk and an improved prediction of response to therapy.
The results we present herein significantly improve the application of gene expression profiling, by biologically dissecting a commonly used clinical predictive classifier in rectal cancer. Using integrative computational biology, we combined multiple data to derive novel interpretations and identifying important players in the prediction of and in the response to treatment.
Results
Patient, tumor and treatment characteristics
A total of 48 patients met all criteria for inclusion in this study. Six samples did not pass our microarray strict quality control standards and had to be excluded. Complete details of the patients, tumor and treatment characteristics are summarized in .
Table 1. Patient, tumor and treatment characteristics of the patients included in the study
Before the CRT, 91% and 88% of patients were clinically staged as T3–4 and lymph nodes positive, respectively; 38 (90%) patients received a total dose of radiotherapy higher than 50 Gy, and 15 out of these cases (36%), drugs other than 5-FU were administered (n = 11, Oxaliplatin; n = 4, Carboplatin). For 33 (79%) patients, 5-FU was administered by continuous venous infusion. The median (range) interval time between the completion of pCRT and surgery was 46 (30–66) days.
With a median follow-up of 81 months, only 6 out of 42 patients had recurrent disease, 9 patients died from disease and 1 patient from unrelated causes. The following TRG distribution was found: TRG 1: n = 8; TRG 2: n = 11; TRG 3: n = 6; TRG 4: n = 10; and TRG 5: n = 7. On the basis of the TRG distribution, 19 (45%) patients were considered responders (TRG 1 to 2), and 23 (55%) were considered non-responders (TRG 3 to 5).
Class comparison and Hierarchical clustering
A total of 45,868 out of 54,675 probe sets with RefSeq annotation were considered. We investigated different expression levels between the two groups of interest (responders and non-responders) by means of the modified F-test statistic with p-values computed by permutations, as described in experimental procedures. Only 19 genes were found to be informative with an adjusted p-value = 0.037 ().
Table 2: List of 19 informative genes (adjusted p-value = 0.037) discriminating responders and non-responders groups
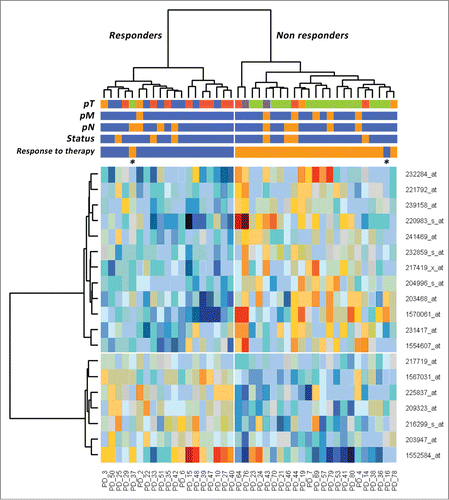
Hierarchical cluster analysis using the 19 informative genes was able to clearly identify the two groups of interest with only two misclassified samples (, “Response to therapy” label). Left branch included 18/19 (94.7%) responders while right branch gathered 22/23 (95.7%) non-responders. Interestingly, non-responders branch correlated with 5/6 (83.3%) cases with pM event and 16/17 (94.1%) cases with a specific pT class. The inspection of clinical data did not suggest any particular explanation about the two misclassified samples; further analyses will be performed to clarify the outliers. The predictive 19 gene classifier from our study were entered into Ingenuity Pathway Analysis Software and, as previously described by Breettingham-Moore,Citation14 TNF signaling pathway was enriched in our network (Fig. S1). Moreover, we tested the 19 genes classifier on patients treated with 5-FU alone (n = 27) and patients treated with other drugs alone (n = 15). Six out of 27 (22%) and 2 out of 15 (13%) outliers resulted in 5-FU alone and other drug association groups, respectively, suggesting similar trend for different treatment protocols.
Figure 1. Hierarchical clustering of 42 patients with rectal carcinomas based on significantly differentially expressed probe sets representing 19 genes (rows) between the subgroup of responders and non-responders (columns) to neoadjuvant chemoradiotherapy. Responders are located on the left branch, Non-responders are clustered on the right branch. Red depicts decreased gene expression; blue indicates increased expression. The two asterisks identify the outliers.
Responders prediction
Considering all the probe sets, we further investigated the capability to predict the patient's outcome. To this aim we fitted a logistic model selected with a stepwise procedure optimizing the AIC and then validated by means of LOOCV. In this way we removed possible redundant information.
Starting from the 19 probe-sets we selected the logistic model maximizing the Akaike Information Criterion. Performance was 95% accuracy by LOOCV. Four genes are representative of the entire set: 1567031_at (ZNF160), 216299_s_at (XRCC3), 241469_at (HFM1) and 231417_at (ASXL2). The target sequence of the 231417_at probe is not defined but it matched 423/424 identities with “putative Polycomb group protein ASXL2” using NCBI BLASTN on all genome assemblies.
These genes were included in the previously identified gene set and were able to correctly predict 40 out of 42 outcomes with one false responder and one false non-responder (LOOCV accuracy = 0.952, specificity = 0.9473, sensitivity = 0.9565, positive predictive value = 0.9565, negative predictive value = 0.9473).
Multivariate analysis
To exclude differences in gene expression between responders and non-responders was due to differences in other characteristics of the two groups (), we performed a multivariate analysis including both the four genes identified in their univariate analysis and the clinicopathological potential confounding factors. We considered a linear model where the four identified genes represent the dependent variables while the confounding factors (sex, tumor distance from anal verge, radiotherapeutic dose delivered, ypTNM) represent the independent variables. Multiplicity corrections have been performed using Holm-Bonferroni method. We found no significant results after multiplicity corrections, thus we can exclude putative associations between the 4 genes and possible confounding factors (data not shown).
Quantitative Real-time PCR analysis
In order to confirm data achieved with microarray analysis, we measured XRCC3, ZNF160, HFM1, and ASXL2 transcript levels, which alone are able to correctly predict 40 out of 42 outcomes, using Real Time quantitative polymerase chain reaction with TaqMan® Assay. XRCC3 gene showed a significant correlation between the array-based and quantitative PCR methods (Pearson = 0.85; r2 = 0.7), with high expression on Affymetrix arrays corresponding to low delta threshold cycle (ΔCt) values from TaqMan® Assay.
Also the expression with TaqMan® Assay of ZNF160, HFM1, and ASXL2 genes are in agreement with microarray results because they show the same expression pattern. Unfortunately, these three genes did not reach a sufficient significance to irrefutably confirm microarray results, probably due to a different resolution of the techniques. The authors anyhow, believes that such genes equally have a pivotal role on the determination of response to treatment, especially if we consider their indirect involvement in a complex protein interaction network, as described below for HFM1 and ASXL2.
XRCC3 knockdown restores sensitivity to 5FU in chemoresistant colon cancer cells
In order to validate the relationship between XRCC3 expression and chemoresistance, we investigated the effect of XRCC3 knockdown on HCT116 and HCT116 p53−/− cells. HCT116 cells are known to be sensitive to 5-FU, whereas HCT116 p53−/− are resistant to the 5-FU chemotherapeutic action.Citation15
We performed a kinetic study of XRCC3 knockdown by siRNA, which revealed a significant decrease of XRCC3 protein levels 48 hours after transfection (Fig. S2), both in HCT116 and HCT116 p53−/− cells.
We then evaluated the effect of XRCC3 knockdown on sensitivity of cells to 5-FU. HCT116 p53−/− (chemoresistant) and HCT116 (chemosensitive) cells were transfected with a control siRNA or with a XRCC3-siRNA, and cells were then treated with 5-FU 36 hours after transfection. XRCC3 knockdown in HCT116 cells had no effect on cell viability with or without administration of 5-FU. On the contrary, in HCT116 p53−/− cells the XRCC3 knockdown in combination with 5-FU treatment caused a relevant decrease of cell viability as compared to the control group (0,81 ± 0,09 vs. 2,05 ± 0,14 absorbance ratio respectively, p = 0.001). As expected, in all the other groups it was observed an increase in cell viability (Ctrl siRNA+5-FU 1.22 ± 0.06 absorbance ratio; XRCC3-siRNA 1.80 ± 0.10 absorbance ratio).
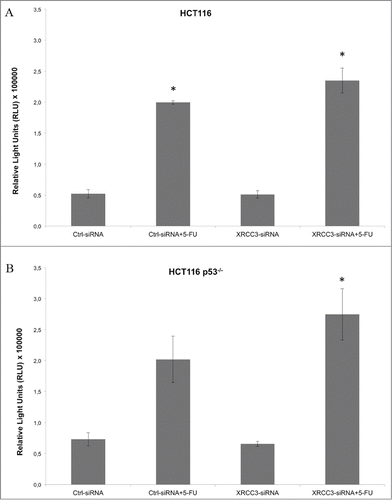
To further characterize the response to 5-FU of the HCT116 or HCT116 p53−/− cells, we performed a caspase 3/7 activation assay which disclosed an increase of caspase activity in XRCC3-siRNA transfected HCT116 p53−/− cells treated with 5-FU. No effect of XRCC3-siRNA on caspase activation was revealed on HCT116 cells ().
Figure 2. Caspase activation assay on HCT116 and HCT116 p53−/− cells. (A) XRC33 knockdown does not influence caspase activation in HCT116 cells. (B) 5-FU, in combination with XRCC3 knockdown, causes a significant increase of caspase 3/7 activation as compared to control group in HCT116 p53−/− cells. Luminescence is expressed as Relative Light Units (RLU). *: p-value <0.05 compared to control group in t-test with Bonferroni's correction. Error bars represent standard errors of the mean.
p53 Immunohistochemistry
There are many different mechanisms at the basis of chemoresistance. Because XRCC3 in vitro testing was performed on HCT 116 and HCT116 p53−/− cells, the result showed above could be due to XRCC3 deregulation in a p53 mutated background cell line (HCT116 p53−/− cells), rather than to the XRCC3 over/under expression per se. We decided to address this issue characterizing p53 in patient tissues. In 42 preoperative biopsies analyzed, p53 protein expression was not detect in 23 samples (54.7%) whereas it showed different positive degree in 19 samples (45.3%): 4 samples with 11–25%, 4 samples with 26–75% and 11 samples with >75 % of immunostained tumor cells. (Fig. S3). No significant correlation was found between p53 expression and tumor response to therapy.
Network analysis
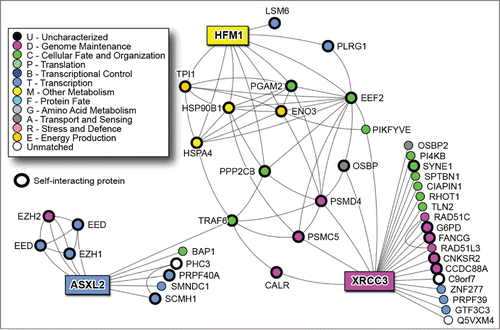
The analysis of the PPI network of the four genes revealed that ZNF160 is a protein with no described interactions while the remaining three are included in a network of 45 nodes (proteins).Citation16 In this network, our most significant protein XRCC3 not only interact with a relevant number of protein per se; but are also related to ASXL2 and HFM1 through indirect interactions. Interestingly, “the heart” of this network seems to be TRAF6 (not relevant by experimental data) that connects ASXL2 to the other two proteins (). The functional annotation and enrichment analysis show a major role of the proteins in the PPI network in DNA repair and recombination, mRNA processing, in sugar catabolic processes and in the organelle lumen organization (Fig. S1).
The microRNA: target analysis using mirDIP shows 472 microRNAs targeting the nodes of the network. Thirty-nine of them are shared by the interactors of the three predictor genes. Twenty-seven have been already described as predictors of response in rectal cancer patients undergoing neoadjuvant therapyCitation17-20 ().
Figure 4. microRNAs targeting the predictor genes PPI network. White squares: microRNAs shared by the 3 genes; pink squares: signature microRNAs described in the literature. The size of the microRNA node corresponds to number of target genes it has. Thick blue lines highlight direct links between predictor genes and corresponding microRNAs.
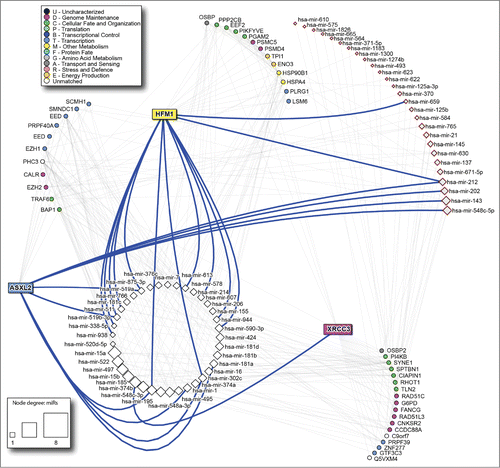
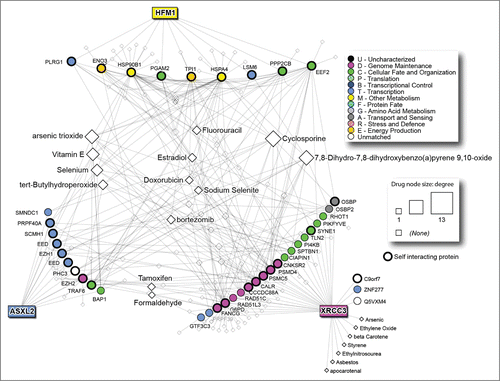
The analysis of drug targets using DrugBank highlighted 130 drugs targeting one or more proteins of the network. The drugs targeting many protein in the network (drug nodes with the highest degree) include cyclosporine, 7,8-Dihydro-7,8-dihydroxybenzo(a)pyrene 9,10-oxide and arsenic trioxide. In this network, fluorouracile affects 6 proteins (PPP2CB, HSPA4, TPI1, PLRG1, PI4KB and FANCG), some of which have a central role. Oxaliplatin targets only one protein, SPTBN1, while carboplatin is not present in the network. Moreover, the central protein TRAF6 is targeted by Estradiol, Folic Acid, Aspirin, Curcumin, Formaldehyde, Hydrogen Peroxide, pirinixic acid and arsenic trioxide ().
Discussion
Currently anti-tumor therapy is predominantly based on the use of chemotherapeutic drugs and leave aside the molecular basis of the disease.
Although these treatments have significantly improved the outcome of many patients, they are ineffective or even toxic for many other types of tumors and in case of metastasis. Recently, new drugs directed against cancer-specific molecular circuits, have been developed and introduced into clinical practice (so-called molecular drugs). However, only selected groups of patients respond to these drugs, and the molecular mechanisms underlying tumor resistance in unresponsive individuals remain to be fully elucidated. In this context, one of the priorities in the field of clinical oncology is the identification of genetic or phenotypic markers able to predict patient responsiveness to treatments.
In an overall perspective of expanding our current capability to tailor personalized therapy, the integrated approach (gene profiling, proteomics, bioinformatics, in vitro and ex-vivo validation) would add an important piece to the puzzle.
Gene expression approach offers the opportunity to evaluate large sets of samples in parallel and has the potential to improve our understanding of tumorigenesis and patients treatment. However, molecular screening alone on different study groups has not achieved sufficient accuracy for the translation into clinical practice. An integrated approach aiming at the interpolation of data collected from protein biomarkers and genetic signatures might offer more reliable predictions. Recent advances in computational science allowed the processing, management and use of large sets of genomic and proteomic information that, properly analyzed, might address us to perform treatment selection and prediction of patient outcome. The molecular profiling of individual patient is a constitutive principle of personalized medicine and is the first step necessary to the clinicians for the selection of the therapeutic regimen.
This study provided a new set of genetic biomarkers associated with the prediction and monitoring of the response to therapy and of tumor chemoradioresistance. Although these tasks are of paramount importance for the development of personalized, mechanism-based anticancer therapies, currently anti-tumor therapy is predominantly based on the use of chemotherapeutic drugs that do not take into account the molecular basis of the disease.
As shown in the current study, a crucial predictor gene is XRCC3 that codes for a protein involved in homologous recombination repair of DNA double-strand breaks and is required for genomic stability. Ionizing radiation induces both DNA single-strand breaks (SSB) and double-strand breaks (DSB), with the DSBs generally considered the lethal event for cell homeostasis. XRCC3 polymorphisms have been implicated in radiosensitivity mechanisms,Citation21-27 but several studies on rectal cancer patients failed the link between them and sensitivity to radiation treatment. In our study, the expression of XRCC3 supports the importance of its role in the prediction of the response to treatment, suggesting that the mutational analysis limited to very few SNPs in the previous studies has been insufficient to highlight the role of the gene.
The microRNA network reveals a central role of hsa-mir-185, directly targeting XRCC3. As hsa-mir-185 has been correlated with poor survival and metastasis in colorectal cancer,Citation28 the evaluation of the XRCC3 status should be performed not only considering SNPs but also its gene- targeting microRNA expression.
To further investigate the role of XRCC3 gene in the chemoresistance in colon carcinoma, a siRNA-mediated knockdown of this gene was performed in a well-known in vitro model of 5-FU chemoresistance of colon carcinoma, the HCT116 p53−/− cell line.Citation15 The down-regulation of XRCC3 in these cells re-sensitized the chemoresistant cells to 5-FU, suggesting a chemoprotective role of this gene in colon carcinoma settings and supporting the evidence of the up-regulation of this gene in non-responder colon carcinoma patients.
Interestingly another predictor gene, HFM1, is involved in DNA interaction by encoding a putative DNA helicase homolog (S. cerevisiae). Its probe was down regulated in responders group as well as the one related to ASXL2 gene. According to literature, the role of these genes in response to radiochemotherapies remains to be explored.
Approaching to this new kind of study, we must consider that the increasing use of high-throughput (HT) assays shifted research from hypothesis-driven exploration to data-driven hypothesis generation. However, generating substantially more data, HT methods in turn led to shifting from predominantly using statistical tools to depending on computational biology approaches, especially data mining and machine learning algorithms, to aid data analysis and interpretation.Citation29,30 These theoretical paradigm is “on practice translate” in this study through the surprisingly identification of TRAF6 as protein with a pivotal role in XRCC3 network. In fact, basing on experimental data alone we have a partial vision on what really happen in the complex micro-world of cell signaling network. However, thanks to integrated HT approach, if we fall experimental data into a more complex scenario we can see the topics in a new prospective and identify that “hidden players” which better complete our model.
Through this approach, TRAF6 (Tumor necrosis factor (TNF) receptor associated factor 6) has been shown to play a central role in the PPI network of the predictor genes. TRAF6 is a crucial signaling molecule regulating a diverse array of physiological processes, including adaptive and innate immunity, bone metabolism and the development of several tissues including lymph nodes, mammary glands, skin and the central nervous system.Citation16 This protein mediates the signaling not only from the members of the TNF receptor superfamily, but also from the members of the Toll/IL-1 family. It also works as a signal transducer in the NF-kappaB pathway that activates IkappaB kinase (IKK) in response to pro-inflammatory cytokines. Interestingly, TRAF6 is targeted by aspirin, known to reduce risk of rectal cancer Citation31 and by curcumin, a polyphenol known to affect the NF-kappaB pathway in colorectal cancer cells, which is in phase II clinical trial for colorectal cancer prevention.Citation32,33
Afterward, TRAF6, activated by IL-1β or LPS, suppresses TGF-β1/Smad pathways through interaction with TβRIII upon TGF-β1 stimulation. In general, inflammation is tightly regulated and resolved by the induction of anti-inflammatory cytokines.Citation34 Once this regulatory balance is disturbed, non-specific stimulation and activation of inflammatory cells may lead to increased production and release of potently destructive immunological and inflammatory molecules. For instance, improper regulation of IL-1β signaling has been shown to potentiate neoplastic risk and ultimately induce tumor progression.Citation34 In addition, decreased TβRIII expression was closely correlated with tumor progression in various human cancers including breast, lung, prostate, pancreatic, ovarian, and renal cancers,Citation35 supporting the idea that TβRIII-mediated regulation of normal epithelial cells may contribute to prevent tumor progression.
In conclusion, meta-analysis of published gene expression data will be performed to further validate our results and to allow the comparison of data retrieved by different platforms and work groups. Through a coordinated effort, our project could help us in identifying clinically useful biomarkers to predict tumor responsiveness to anti-cancer chemo/radiotherapies and to validate newly identified molecular circuits as potential targets for the development of mechanism-based therapeutic strategies.
Patients and Methods
Patients, samples, and treatment
Between 1998 and 2006, 186 patients with primary adenocarcinoma of the rectum underwent CRT followed by surgery. The pre-treatment evaluation of the patients included a complete clinical history and physical examination, colonoscopy, complete blood cell count, transrectal ultrasound, pelvic computed tomography scan or magnetic resonance imaging, abdominal/chest CT and carcino-embryonic antigen test. The inclusion criteria for CRT were as follows: a) biopsy-proven adenocarcinoma of the mid-low rectum (< 11 cm from the anal verge); b) clinical stage T3–4 and/or node-positive; c) Eastern Cooperative Oncology Group performance status 0–2.
Since most patients received the preoperative CRT elsewhere, only in 84 out of 186 patients who underwent surgery at our institution the pre CRT research biopsies (2–3 mm3) were collected during the initial diagnostic endoscopy, immediately frozen and stored in a liquid nitrogen tank. Biopsies were divided into half, one piece undergoing independent histopathological examination and the other prepared for RNA extraction.
No statistically relevant differences were found between clinical and treatment characteristics of included and excluded patients.
The patients underwent to preoperative external beam radiotherapy using high-energy photons (> 6 MV) with conventional fractionation (≥50 Gy in 28 fractions, 1.8 Gy/day, 5 sessions per week) and 5-fluorouracil (5-FU)-based chemotherapy administered by bolus or continuous venous infusion. A standard total mesorectal excision was performed 4 to 8 weeks after the completion of pCRT.
The study protocol was reviewed and approved by the local ethics committee (protocol number 740 P) and each patient provided written informed consent.
Evaluation of tumor response
The surgical specimens were assessed in a standardized way and reviewed by one pathologist (CM), who was unaware of the patient's outcome. The histopathology findings and definition of radical surgery were reported following the American Joint Committee on Cancer TNM (2002). The tumor response to CRT was defined as the tumor regression grade (TRG) and was scored following the criteria proposed by Mandard et al. Citation36: TRG-1, pathological complete response (pCR), i.e., absence of viable cancer cells in the resected specimen; TRG-2, presence of residual cancer cells; TRG-3, fibrosis outgrowing residual cancer cells; TRG-4, residual cancer cells outgrowing fibrosis; and TRG-5, absence of response. According to the TRG, the patients were classified as responders (TRG 1–2) and non-responders (TRG 3–5).Citation37,38
RNA extraction
After independent histopathology review of sample set, in 52 out of 84 biopsies containing more than 70% tumor, RNA was extracted by phenol/chloroform extraction (TRIzol; Invitrogen) prior to further purification by column chromatography (RNeasy Mini kit; Qiagen). RNA integrity (RIN) was then assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies); 4 samples showed evidence of RNA degradation (RIN<6) and were excluded from the analysis.
Microarrays preparation
Gene expression analysis was performed using the Affymetrix GeneChip Human Genome U133 Plus 2.0 Array Platform. Preparation of labeled and fragmented RNA targets, hybridization and scanning were carried out according to the manufacturer's protocol (Affymetrix). Briefly, 100 ng of total RNA for each sample was processed using the GeneChip 3′ IVT Express Kit. RNA was reverse transcribed and then converted to double-stranded cDNA prior to biotin labeling during in vitro transcription. Fifteen micrograms of labeled aRNA was then fragmented, and quality control was carried out using the Agilent Bioanalyzer. Fragmented aRNA was then hybridized on GeneChip Human Genome U133 Plus 2.0 Arrays for 16 hours at 45°C. Arrays were then washed and stained using the GeneChip Hybridization, Wash, and Stain Kit on the GeneChip Fluidics Station 450. Chips were then scanned using the Affymetrix GeneChip Scanner 3000. Six out of 48 processed samples did not pass quality controls and were excluded from the analysis; thus, a total of 42 samples were used in the final analysis (19 responders and 23 non-responders).
Class comparison and class prediction analyses
The Affymetrix Human Genome U133 Plus 2.0 Array expressions were preprocessed and normalized using Robust Multi-array Average (RMA) procedure.Citation39 A class-comparison analysis was applied to determine which genes were differentially expressed between responders and non-responders.
To this aim we used the Fss test statistic, which is a modified F test statistic that shrinks both the means and the variance. The Fss test has almost identical power as the Maximum Average Powerful test, but it is computationally less demanding and more powerful than the other modified F-type tests (for more details see Hwang, et al. Citation40).
P-values were computed by means of permutations, hence avoiding any distributional assumption. P-values adjustment for multiple testing was made using the Holm-Bonferroni method to control the family wise error rate. Adjusted p-values < 0.05 were considered significant.
We performed a cluster analysis on the interesting probe-sets to show the discriminant power of the profiles.
To further investigate the predictive capability of genes expression, we selected the logistic model optimizing the Akaike Information Criterion (AIC) considering all the probe-sets.Citation41 LOOCV was then used to estimate the prediction accuracy for the selected model.Citation42,43
Quantitative real-time PCR
The amount of starting RNA was normalized using 18S rRNA as a control transcript. To this end, a QuantumRNA 18S internal standard kit (Ambion) was utilized, followed by quantification of the electrophoretic bands by ImageQuant (MolecularDynamics). Real time PCR was performed on ABI PRISM 7300 (Applied Biosystems Foster City, California, USA) by using specific TaqMan® Gene Expression Assays (Applied Biosystems): XRCC3 (Hs00193725_m1), ASXL2 (Hs00827052_m1), HFM1 (Hs01651101_m1 ), ZNF160 (Hs00369142_m1).
For the amplification, the qPCR core kit was utilized (Applied Biosystem). Real time PCR conditions were set as specified by the manufacturer. All samples were amplified in triplicate and results were analyzed by the 2–DCt method.Citation44
Cell Culture
HCT116 and HCT116 p53−/− colon carcinoma cell lines were a kind gift of Prof. Bert Vogelstein (John Hopkins University, Baltimore, MD USA).
Cells were maintained and cultured in a 37°C incubator at 5% CO2 and grown with McCoy's 5A-Glutamax medium with 10% FBS (Gibco, not Heat Inactivated), 100 U/ml Penicillin and 100 μg/ml Streptomycin.
siRNA mediated knockdown and cell treatments
For siRNA mediated knockdown, HCT116 and HCT116 p53−/− cells were transfected with control siRNA (Negative Control siRNA #1, Life Technologies, final concentration 10 nM) or siRNA against XRCC3 (s14946, Life Technologies, final concentration 10nM), using Lipofectamine™ RNAiMax reagent (Life Technologies) and following manufacturer's protocol optimized for this cell lines.
5-Fluorouracil (5-FU, clinical grade) was administered to cells at the final concentration of 200 µM, 36 hours after transfection.
Cell viability assay and cell death evaluation
Cell viability was evaluated by the Crystal Violet (CV) assay and absorbance was measured with a microplate reader (Tecan Instruments). The cell viability data were calculated and expressed as the ratio between the absorbance read at the end of treatment and the absorbance read 24h after seeding.
To test caspase 3/7 activity it was used the Caspase-Glo© 3/7 Assay (Promega) following manufacturer's protocol. Statistical analysis was performed using IBM SPSS Statistics (version 19). Significant differences between groups were determined by ANOVA with Bonferroni's post-hoc test for multiple comparisons (adjusted p-value <0.05 was considered as significant).
Protein extracts and Immunoblotting
Cells where harvested at determined time points and lysed with a modified RIPA buffer: Tris-HCl pH 8, 50 mM; NaCl 500 mM; IGEPAL 1% v/v; Sodium Deoxycholate 0.5% v/v; EGTA 1 mM; EDTA 1 mM; DTT 1 mM; Protease Inhibitor Cocktail (Sigma-Aldrich) 2% v/v. Quantification of protein lysates was performed using MicroBCA assay (Thermo Scientific).
Protein extracts were separated by SDS-PAGE (NuPAGE, Life Technologies) and blotted on nitrocellulose membranes (iBlot system, Life Technologies). Membranes were then immunodecorated with the following primary antibodies: anti-XRCC3 (mouse monoclonal [10F1/6], Abcam) at a 1:1000 dilution and anti-vinculin (mouse monoclonal [V824], Sigma-Aldrich) at a 1:5000 dilution. The signal detection was performed with a HRP-conjugated secondary anti-mouse antibody (GE Healthcare) and images digitally acquired with G-BOX System (Syngene).
Immunohistochemistry
For each sample, we chosen one slide corresponding to the most representative part of the tumor in order to perform an immunohistochemical evaluation of p53 protein expression. Formalin-fixed, paraffin-embedded sections were deparaffinized and rehydrated and p53 was detected by the mouse monoclonal antibody anti-p53 Ab-2 (clone PAb 1801, Oncogene Research Products) as previously describe in Esposito et al.Citation45 p53 protein expression was graded as: (1) absent or present in ≤10% of tumor cells; (2) present in 11–25%; (3) present in 26 –75%, or (4) present in >75% of tumor cells.
Network analysis
We further investigated the molecular pathways involving the predictive classifier using protein-protein interactions (PPIs) and enrichment analysis, as well as the possible common microRNAs and drugs targeting them. We first characterized one part of the classifier by retrieving physical PPIs from I2D database ver. 1.95 Citation46 [http://ophid.utoronto.ca/i2d], creating a PPI network that we visualized and analyzed in NAViGaTOR 2.3 Citation47 [http://ophid.utoronto.ca/navigator]. We then performed a functional annotation and enrichment analysis of all the proteins of the network using DAVID Bioinformatics resources 6.7 Citation48,49 [http://david.abcc.ncifcrf.gov/], a study of the microRNAs targeting the PPI network using mirDIP 1.1 Citation50 [http://ophid.utoronto.ca/mirDIP], and a study of the drugs targeting the same network using DrugBank 3 Citation51 [http://www.drugbank.ca/]. Moreover, to prioritize microRNAs in the network, we collected data from published studies on response to neoadjuvant chemoradiotherapy and microRNA signatures. Citation17-19
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplementary Figures
Download MS Word (1.5 MB)Acknowledgments
Some technical aspects and clinical work was supported by: Alessandro Ambrosi,Citation1 Gaspar C2, Friso ML,Citation3 Lonardi S4, (1University Center for Statistics in the Biomedical Sciences, Università Vita-Salute, San Raffaele Scientific Institute, Milan, Italy; 2Department of Pathology, Josephine Nefkens Institute, Erasmus MC, Rotterdam, The Netherlands; 3Radiotherapy and Nuclear Medicine Unit, Istituto Oncologico Veneto - IRCCS, Padua,Italy; 4First Medical Oncology Unit, Istituto Oncologico Veneto - IRCCS, Padua, Italy). Biological samples were provided by Surgical Clinic, Tumor Tissue Biobank.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Funding
This study was supported in part by grants from the CARIPARO, AIRC Foundation and AIRC Special Program Molecular Clinical Oncology, 5×1000 (No.12214), Fondazione Città della Speranza and MIUR (PON_02782 to ML). Computational analyses were supported in part by Ontario Research Fund (GL2-01-030), Canada Foundation for Innovation (CFI #12301 and CFI #203373), Canada Research chair Program (CRC #203373 and CRC #225404), and IBM to IJ. CP was funded in part by Friuli Exchange Program. This research was funded in part by the Ontario Ministry of Health and Long Term Care. The views expressed do not necessarily reflect those of the OMOHLTC.
Reference
- Sauer R, Becker H, Hohenberger W, Rodel C, Wittekind C, Fietkau R, Martus P, Tschmelitsch J, Hager E, Hess CF, et al. Preoperative versus postoperative chemoradiotherapy for rectal cancer. N Engl J Med 2004; 351:1731-40; PMID:15496622; http://dx.doi.org/10.1056/NEJMoa040694
- Bosset JF, Collette L, Calais G, Mineur L, Maingon P, Radosevic-Jelic L, Daban A, Bardet E, Beny A, Ollier JC. Chemotherapy with preoperative radiotherapy in rectal cancer. N Engl J Med 2006; 355:1114-23; PMID:16971718; http://dx.doi.org/10.1056/NEJMoa060829
- Roh MS, Colangelo LH, O'Connell MJ, Yothers G, Deutsch M, Allegra CJ, Kahlenberg MS, Baez-Diaz L, Ursiny CS, Petrelli NJ, et al. Preoperative multimodality therapy improves disease-free survival in patients with carcinoma of the rectum: NSABP R-03. J Clin Oncol 2009; 27:5124-30; PMID:19770376; http://dx.doi.org/10.1200/JCO.2009.22.0467
- Minsky BD, Cohen AM, Kemeny N, Enker WE, Kelsen DP, Reichman B, Saltz L, Sigurdson ER, Frankel J. Enhancement of radiation-induced downstaging of rectal cancer by fluorouracil and high-dose leucovorin chemotherapy. J Clin Oncol 1992; 10:79-84; PMID:1727928
- Mohiuddin M, Hayne M, Regine WF, Hanna N, Hagihara PF, McGrath P, Marks GM. Prognostic significance of postchemoradiation stage following preoperative chemotherapy and radiation for advanced/recurrent rectal cancers. Int J Radiat Oncol Biol Phys 2000; 48:1075-80; PMID:11072165; http://dx.doi.org/10.1016/S0360-3016(00)00732-X
- Smith FM, Reynolds JV, Miller N, Stephens RB, Kennedy MJ. Pathological and molecular predictors of the response of rectal cancer to neoadjuvant radiochemotherapy. Eur J Surg Oncol 2006; 32:55-64; PMID:16324817; http://dx.doi.org/10.1016/j.ejso.2005.09.010
- Kuremsky JG, Tepper JE, McLeod HL. Biomarkers for response to neoadjuvant chemoradiation for rectal cancer. Int J Radiat Oncol Biol Phys 2009; 74:673-88; PMID:19480968; http://dx.doi.org/10.1016/j.ijrobp.2009.03.003
- Desch CE, Benson AB, 3rd, Somerfield MR, Flynn PJ, Krause C, Loprinzi CL, Minsky BD, Pfister DG, Virgo KS, Petrelli NJ. Colorectal cancer surveillance: 2005 update of an American Society of Clinical Oncology practice guideline. J Clin Oncol 2005; 23:8512-9; PMID:16260687; http://dx.doi.org/10.1200/JCO.2005.04.0063
- Watanabe T, Komuro Y, Kiyomatsu T, Kanazawa T, Kazama Y, Tanaka J, Tanaka T, Yamamoto Y, Shirane M, Muto T, et al. Prediction of sensitivity of rectal cancer cells in response to preoperative radiotherapy by DNA microarray analysis of gene expression profiles. Cancer Res 2006; 66:3370-4; PMID:16585155; http://dx.doi.org/10.1158/0008-5472.CAN-05-3834
- Daemen A, Gevaert O, De Bie T, Debucquoy A, Machiels JP, De Moor B, Haustermans K. Integrating microarray and proteomics data to predict the response on cetuximab in patients with rectal cancer. Pac Symp Biocomput 2008:166-77; PMID:18229684
- Ghadimi BM, Grade M, Difilippantonio MJ, Varma S, Simon R, Montagna C, Fuzesi L, Langer C, Becker H, Liersch T, et al. Effectiveness of gene expression profiling for response prediction of rectal adenocarcinomas to preoperative chemoradiotherapy. J Clin Oncol 2005; 23:1826-38; PMID:15774776; http://dx.doi.org/10.1200/JCO.2005.00.406
- Rimkus C, Friederichs J, Boulesteix AL, Theisen J, Mages J, Becker K, Nekarda H, Rosenberg R, Janssen KP, Siewert JR. Microarray-based prediction of tumor response to neoadjuvant radiochemotherapy of patients with locally advanced rectal cancer. Clin Gastroenterol Hepatol 2008; 6:53-61; PMID:18166477; http://dx.doi.org/10.1016/j.cgh.2007.10.022
- Kim IJ, Lim SB, Kang HC, Chang HJ, Ahn SA, Park HW, Jang SG, Park JH, Kim DY, Jung KH, et al. Microarray gene expression profiling for predicting complete response to preoperative chemoradiotherapy in patients with advanced rectal cancer. Dis Colon Rectum 2007; 50:1342-53; PMID:17665260; http://dx.doi.org/10.1007/s10350-007-277-7
- Brettingham-Moore KH, Duong CP, Greenawalt DM, Heriot AG, Ellul J, Dow CA, Murray WK, Hicks RJ, Tjandra J, Chao M, et al. Pretreatment transcriptional profiling for predicting response to neoadjuvant chemoradiotherapy in rectal adenocarcinoma. Clin Cancer Res 17:3039-47; PMID:21224373; http://dx.doi.org/10.1158/1078-0432.CCR-10-2915
- Bunz F, Hwang PM, Torrance C, Waldman T, Zhang Y, Dillehay L, Williams J, Lengauer C, Kinzler KW, Vogelstein B. Disruption of p53 in human cancer cells alters the responses to therapeutic agents. The Journal of clinical investigation 1999; 104:263-9; PMID:10430607; http://dx.doi.org/10.1172/JCI6863
- Chung JY, Lu M, Yin Q, Lin SC, Wu H. Molecular basis for the unique specificity of TRAF6. Adv Exp Med Biol 2007; 597:122-30; PMID:17633022; http://dx.doi.org/10.1007/978-0-387-70630-6_10
- Della Vittoria Scarpati G, Falcetta F, Carlomagno C, Ubezio P, Marchini S, De Stefano A, Singh VK, D'Incalci M, De Placido S, Pepe S. A Specific miRNA Signature Correlates with Complete Pathological Response to Neoadjuvant Chemoradiotherapy in Locally Advanced Rectal Cancer. Int J Radiat Oncol Biol Phys 2012; 83(4):1113-9; PMID:22172905
- Drebber U, Lay M, Wedemeyer I, Vallbohmer D, Bollschweiler E, Brabender J, Monig SP, Holscher AH, Dienes HP, Odenthal M. Altered levels of the onco-microRNA 21 and the tumor-supressor microRNAs 143 and 145 in advanced rectal cancer indicate successful neoadjuvant chemoradiotherapy. Int J Oncol 2011; 39:409-15; PMID:21567082
- Garajova I, Svoboda M, Slaby O, Kocakova I, Fabian P, Kocak I, Vyzula R. ; Possibilities of resistance prediction to neoadjuvant concomitant chemoradiotherapy in the treatment algorithm of patients with rectal carcinoma. Klin Onkol 2008; 21:330-7; PMID:19382596
- Agostini M, Pucciarelli S, Calore F, Bedin C, Enzo M, Nitti D. miRNAs in colon and rectal cancer: A consensus for their true clinical value. Clin Chim Acta 2010; 411:1181-6; PMID:20452339; http://dx.doi.org/10.1016/j.cca.2010.05.002
- Cecchin E, Agostini M, Pucciarelli S, De Paoli A, Canzonieri V, Sigon R, De Mattia E, Friso ML, Biason P, Visentin M, et al. Tumor response is predicted by patient genetic profile in rectal cancer patients treated with neo-adjuvant chemo-radiotherapy. The pharmacogenomics journal 2011; 11:214-26; PMID:20368715; http://dx.doi.org/10.1038/tpj.2010.25
- Pratesi N, Mangoni M, Mancini I, Paiar F, Simi L, Livi L, Cassani S, Buglione M, Grisanti S, Almici C, et al. Association between single nucleotide polymorphisms in the XRCC1 and RAD51 genes and clinical radiosensitivity in head and neck cancer. Radiotherapy and oncology : journal of the European Society for Therapeutic Radiology and Oncology 2011; 99:356-61; PMID:21704413; http://dx.doi.org/10.1016/j.radonc.2011.05.062
- Mangoni M, Bisanzi S, Carozzi F, Sani C, Biti G, Livi L, Barletta E, Costantini AS, Gorini G. Association between genetic polymorphisms in the XRCC1, XRCC3, XPD, GSTM1, GSTT1, MSH2, MLH1, MSH3, and MGMT genes and radiosensitivity in breast cancer patients. International journal of radiation oncology, biology, physics 2011; 81:52-8; PMID:20708344; http://dx.doi.org/10.1016/j.ijrobp.2010.04.023
- Vral A, Willems P, Claes K, Poppe B, Perletti G, Thierens H. Combined effect of polymorphisms in Rad51 and Xrcc3 on breast cancer risk and chromosomal radiosensitivity. Molecular medicine reports 2011; 4:901-12; PMID:21725594
- Alsbeih G, El-Sebaie M, Al-Harbi N, Al-Hadyan K, Shoukri M, Al-Rajhi N. SNPs in genes implicated in radiation response are associated with radiotoxicity and evoke roles as predictive and prognostic biomarkers. Radiation oncology 2013; 8:125; PMID:23697595; http://dx.doi.org/10.1186/1748-717X-8-125
- Curwen GB, Murphy S, Tawn EJ, Winther, JF, Boice, JD, Jr. A study of DNA damage recognition and repair gene polymorphisms in relation to cancer predisposition and G2 chromosomal radiosensitivity. Environmental and molecular mutagenesis 2011; 52:72-6; PMID:21113933; http://dx.doi.org/10.1002/em.20633
- Farnebo L, Jerhammar F, Ceder R, Grafstrom RC, Vainikka L, Thunell L, Grenman R, Johansson AC, Roberg K. Combining factors on protein and gene level to predict radioresponse in head and neck cancer cell lines. Journal of oral pathology & medicine : official publication of the International Association of Oral Pathologists and the American Academy of Oral Pathology 2011; 40:739-46; http://dx.doi.org/10.1111/j.1600-0714.2011.01036.x
- Akcakaya P, Ekelund S, Kolosenko I, Caramuta S, Ozata DM, Xie H, Lindforss U, Olivecrona H, Lui WO. miR-185 and miR-133b deregulation is associated with overall survival and metastasis in colorectal cancer. Int J Oncol 2011; 39:311-8; PMID:21573504
- Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic acids research 2006; 34:D535-9; PMID:16381927; http://dx.doi.org/10.1093/nar/gkj109
- Kerrien S, Alam-Faruque Y, Aranda B, Bancarz I, Bridge A, Derow C, Dimmer E, Feuermann M, Friedrichsen A, Huntley R, et al. IntAct–open source resource for molecular interaction data. Nucleic acids research 2007; 35:D561-5; PMID:17145710; http://dx.doi.org/10.1093/nar/gkl958
- Rothwell PM, Wilson M, Elwin CE, Norrving B, Algra A, Warlow CP, Meade TW. Long-term effect of aspirin on colorectal cancer incidence and mortality: 20-year follow-up of five randomised trials. Lancet 2010; 376:1741-50; PMID:20970847; http://dx.doi.org/10.1016/S0140-6736(10)61543-7
- Carroll RE, Benya RV, Turgeon DK, Vareed S, Neuman M, Rodriguez L, Kakarala M, Carpenter PM, McLaren C, Meyskens FL, Jr., et al. Phase IIa clinical trial of curcumin for the prevention of colorectal neoplasia. Cancer Prev Res (Phila) 2011; 4:354-64; PMID:21372035; http://dx.doi.org/10.1158/1940-6207.CAPR-10-0098
- Sandur SK, Deorukhkar A, Pandey MK, Pabon AM, Shentu S, Guha S, Aggarwal BB, Krishnan S. Curcumin modulates the radiosensitivity of colorectal cancer cells by suppressing constitutive and inducible NF-kappaB activity. Int J Radiat Oncol Biol Phys 2009; 75:534-42; PMID:19735878; http://dx.doi.org/10.1016/j.ijrobp.2009.06.034
- Coussens LM, Werb Z. Inflammation and cancer. Nature 2002; 420:860-7; PMID:12490959; http://dx.doi.org/10.1038/nature01322
- Gatza CE, Oh SY, Blobe GC. Roles for the type III TGF-beta receptor in human cancer. Cell Signal 2010; 22:1163-74; PMID:20153821; http://dx.doi.org/10.1016/j.cellsig.2010.01.016
- Mandard AM, Dalibard F, Mandard JC, Marnay J, Henry-Amar M, Petiot JF, Roussel A, Jacob JH, Segol P, Samama G, et al. Pathologic assessment of tumor regression after preoperative chemoradiotherapy of esophageal carcinoma. Clinicopathologic correlations. Cancer 1994; 73:2680-6; PMID:8194005; http://dx.doi.org/10.1002/1097-0142(19940601)73:11%3c2680::AID-CNCR2820731105%3e3.0.CO;2-C
- Beddy D, Hyland JM, Winter DC, Lim C, White A, Moriarty M, Armstrong J, Fennelly D, Gibbons D, Sheahan K. A simplified tumor regression grade correlates with survival in locally advanced rectal carcinoma treated with neoadjuvant chemoradiotherapy. Ann Surg Oncol 2008; 15:3471-7; PMID:18846402; http://dx.doi.org/10.1245/s10434-008-0149-y
- Ryan R, Gibbons D, Hyland JM, Treanor D, White A, Mulcahy HE, O'Donoghue DP, Moriarty M, Fennelly D, Sheahan K. Pathological response following long-course neoadjuvant chemoradiotherapy for locally advanced rectal cancer. Histopathology 2005; 47:141-6; PMID:16045774; http://dx.doi.org/10.1111/j.1365-2559.2005.02176.x
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 2003; 31:e15; PMID:12582260; http://dx.doi.org/10.1093/nar/gng015
- Hwang JT, Liu P. Optimal tests shrinking both means and variances applicable to microarray data analysis. Stat Appl Genet Mol Biol 2010; 9:Article36; PMID:20887275
- Marchet A, Mocellin S, Belluco C, Ambrosi A, DeMarchi F, Mammano E, Digito M, Leon A, D'Arrigo A, Lise M, et al. Gene expression profile of primary gastric cancer: towards the prediction of lymph node status. Ann Surg Oncol 2007; 14:1058-64; PMID:17106627; http://dx.doi.org/10.1245/s10434-006-9090-0
- Ntzani EE, Ioannidis JP. Predictive ability of DNA microarrays for cancer outcomes and correlates: an empirical assessment. Lancet 2003; 362:1439-44; PMID:14602436; http://dx.doi.org/10.1016/S0140-6736(03)14686-7
- Simon R, Radmacher MD, Dobbin K, McShane LM. Pitfalls in the use of DNA microarray data for diagnostic and prognostic classification. J Natl Cancer Inst 2003; 95:14-8; PMID:12509396; http://dx.doi.org/10.1093/jnci/95.1.14
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001; 25:402-8; PMID:11846609; http://dx.doi.org/10.1006/meth.2001.1262
- Esposito G, Pucciarelli S, Alaggio R, Giacomelli L, Marchiori E, Iaderosa GA, Friso ML, Toppan P, Chieco-Bianchi L, Lise M. P27kip1 expression is associated with tumor response to preoperative chemoradiotherapy in rectal cancer. Annals of surgical oncology 2001; 8:311-8; PMID:11352304; http://dx.doi.org/10.1007/s10434-001-0311-2
- Brown KR, Jurisica I. Online predicted human interaction database. Bioinformatics 2005; 21:2076-82; PMID:15657099; http://dx.doi.org/10.1093/bioinformatics/bti273
- Brown KR, Otasek D, Ali M, McGuffin MJ, Xie W, Devani B, Toch IL, Jurisica I. NAViGaTOR: Network Analysis, Visualization and Graphing Toronto. Bioinformatics 2009; 25:3327-9; PMID:19837718; http://dx.doi.org/10.1093/bioinformatics/btp595
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4:44-57; PMID:19131956; http://dx.doi.org/10.1038/nprot.2008.211
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009; 37:1-13; PMID:19033363; http://dx.doi.org/10.1093/nar/gkn923
- Shirdel EA, Xie W, Mak TW, Jurisica I. NAViGaTing the micronome–using multiple microRNA prediction databases to identify signalling pathway-associated microRNAs. PLoS One 2011; 6:e17429; PMID:21364759; http://dx.doi.org/10.1371/journal.pone.0017429
- Knox C, Law V, Jewison T, Liu P, Ly S, Frolkis A, Pon A, Banco K, Mak C, Neveu V, et al. DrugBank 3.0: a comprehensive resource for ‘omics’ research on drugs. Nucleic Acids Res 2011; 39:D1035-41; PMID:21059682; http://dx.doi.org/10.1093/nar/gkq1126