Abstract
Integrins function in collective migration both as major receptors for extracellular matrix and by crosstalk to adherens junctions. Despite extensive research, important questions remain about how integrin signaling mechanisms are integrated into collective migration programs. Tetraspanins form cell surface complexes with a subset of integrins and thus are good candidates for regulating the balance of integrin functional inputs into cell-matrix and cell-cell interactions. For example, tetraspanin CD151 directly associates with α3β1 integrin in carcinoma cells and promotes rapid α3β1-dependent single cell motility, but CD151 also promotes organized adherens junctions and restrains collective carcinoma cell migration on 2D substrates. However, the individual roles of CD151s integrin partners in CD151s pro-junction activity in carcinoma cells were not well understood. Here we find that CD151 promotes organized carcinoma cell junctions via α3β1 integrin, by a mechanism that requires the a3b1 ligand, laminin-332. Loss of CD151 promotes collective 3D invasion and growth in vitro and in vivo, and the enhanced invasion of CD151-silenced cells is α3 integrin dependent, suggesting that CD151 can regulate the balance between α3β1s pro-junction and pro-migratory activities in collective invasion. An analysis of human cancer cases revealed that changes in CD151 expression can be linked to either better or worse clinical outcomes depending on context, including potentially divergent roles for CD151 in different subsets of breast cancer cases. Thus, the role of the CD151-α3β1 complex in carcinoma progression is context dependent, and may depend on the mode of tumor cell invasion.
Abbreviations
| LM-332 | = | laminin-332 |
| LMα3 | = | laminin-α3 subunit |
| Ecad-GFP | = | E-cadherin-GFP fusion protein |
| EMT | = | epithelial-mesenchymal transition |
| BLI | = | bioluminescence imaging |
| TCGA | = | The Cancer Genome Atlas |
| DAPI | = | 4′,6-diamidino-2-phenylindole. |
Introduction
During development, collective cell migration is a recurring theme encompassing whole tissue rearrangements such as mesendodermal tissue movements; Citation1 migration of groups of cells, such as Drosophila border cell migration.Citation2 and zebrafish lateral line morphogenesis; Citation3 and collaborative invasion, such as angiogenic sprouting.Citation4 and neural crest cell streaming.Citation5 Compelling evidence that tumor cells can co-opt collective migration mechanisms for invasion comes from a combination of histological examination of tumor specimens, intravital microscopy, and in vitro experiments,Citation6-9 (reviewed in refs.Citation10,11 ).
Several factors govern the extent and mode of collective invasion, including the strength of cadherin-based cell-cell junctions.Citation2,12,13 and the ratio of actomyosin contractility to the strength of cell-cell adhesion and substrate rigidity.Citation14 Integrin receptors for extracellular matrix can make both positive and negative contributions to collective invasion not only by participating in matrix-driven motility but also by regulating the strength of cell-cell contacts via crosstalk mechanisms.Citation13,15-17 However, it remains to be determined whether the mechanisms controlling the role of integrin function in collective migration are the same as or different from the mechanisms operating in single cell migration.
Tetraspanin proteins are excellent candidates for regulating the balance between pro- and anti-migratory functions of integrins in collective migration. Tetraspanins are 4 pass transmembrane proteins that interact with each other to assemble multi-molecular cell surface complexes that can include integrins, Ig superfamily proteins, growth factor receptors, 7 transmembrane domain receptors, claudins, cell surface proteases, and receptor tyrosine kinases.Citation18-21 Among tetraspanins, CD151 has received particular attention because of numerous reports linking it to tumor formation and progression.Citation18,19,22-24 In particular, recent studies utilizing pre-clinical in vivo models have advanced CD151 as a potential promoter of breast cancer,Citation25-29 skin squamous cell carcinoma,Citation30 prostate cancer progression,Citation31 and pancreatic tumor cell metastasis.Citation32 CD151s pro-tumor functions may result from CD151s physical association with, and functional modulation of, the laminin-binding integrins α3β1 and α6β4. Loss of CD151 impaired (i) α3β1 integrin-dependent ErbB2 dimerization and activation,Citation33 (ii) α6β4 integrin-dependent growth in 3D Matrigel,Citation26 (iii) PKC or EGF-triggered phosphorylation of the β4 integrin cytoplasmic tail,Citation26,30 (iv) α3β1 and α6β4-dependent scattering and proliferation in 3D Matrigel in response to TGF-β,Citation27 or endothelial cell-conditioned medium,Citation28 and (v) α6 integrin-dependent invasion through Matrigel toward EGF.Citation29 CD151 may also promote metastasis by integrin-independent mechanisms involving modulation of PKC signaling.Citation34
However, CD151 may not promote tumor progression and metastasis in every context. Silencing CD151 promoted the growth of ovarian tumor xenografts,Citation35 and loss of CD151 expression was associated with shorter disease-free survival in endometrial cancer.Citation36 In addition, the loss of CD151-α3β1 integrin co-distribution is a feature of non-cohesively invading oral squamous cell carcinomas.Citation37 CD151 expression was also reduced in a cohort of colorectal carcinomas, and growth of colon carcinoma cells under hypoxic conditions caused CD151 downregulation with reduced cell-cell and cell-matrix adhesion.Citation38 Re-expression of CD151 in colon carcinoma liver metastases suggested a scenario where transient downregulation of CD151 in the primary tumor allows for detachment and dissemination.Citation38 In certain contexts, CD151 can also function as a negative regulator of Ras activity.Citation39
We previously found that, although CD151 is required for rapid, α3β1 integrin-dependent single cell motility,Citation40,41 CD151 can also promote the maintenance of organized cell-cell junctions and restrain collective migration on a rigid 2D substrate.Citation42 A CD151 mutant that lacked integrin association was unable to restore organized cell-cell junctions; Citation42 however, the integrin required for CD151s pro-junction activity remained undefined. Here we present evidence that CD151s junction stabilizing activity flows through its association with α3β1 integrin. Depleting CD151 dramatically increased α3β1-dependent collective invasion in a 3D matrix, and increased tumor growth in 3D. These in vitro behaviors of CD151-silenced cells corresponded to a more invasive, rapidly growing tumor phenotype in vivo. An analysis of human cancer cases identified several examples in which CD151 expression changes are linked to better or worse clinical outcomes in different contexts, including potentially opposing roles for CD151 in hormone receptor-positive versus -negative breast cancers. These new results underscore the importance of developing a more nuanced understanding of the role of CD151-integrin complexes in cancer progression. The divergent functions of CD151 in regulating single cell vs. collective cell migration may help to explain the paradoxical pro- versus anti-invasive functions that have been ascribed to CD151-integrin complexes in different contexts.
Results
CD151-integrin interaction is required for tight cell-cell contacts
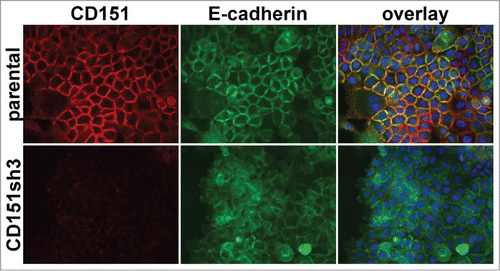
We previously observed that loss of CD151 perturbed the organization of E-cadherin-dependent cell-cell junctions.Citation42 To further explore the impact of CD151 expression and integrin association on cell-cell junctions, we examined the ultrastructure of cell junctions by electron microscopy, using monolayer cultures of our previously published set of cell lines with RNAi-mediated CD151 depletion and re-expression of CD151 mutants.Citation41 Regions of cell-cell contact contained multiple, cross-interacting filopodia-like structures that appeared less densely packed in CD151-silenced (CD151sh3) cells than in wild type parental cells, resulting in more empty space within the sh3 cell-cell contact zones (). Re-expressing a CD151 mutant, CD151-VR, which fails to interact with integrins,Citation41 failed to restore packing density at cell-cell contacts (Fig.1C). In contrast, re-expressing a CD151 mutant, CD151-Palm, which has impaired association with other tetraspanins,Citation41 was able to partially rescue the number of filopodial projections, as was re-expressing wild type CD151 ( quantified in F). Flow cytometry confirmed that CD151 was ˜97% silenced in the sh3 cells (data not shown), as previously reported,Citation40-42 (see also , below, which confirms near total silencing of CD151 in the sh3 cells). Quantification of immunoblots confirmed equivalent re-expression the CD151-VR, CD151-Palm, and CD151-WT constructs in the CD151-silenced cells (Fig. S1), as previously reported,Citation41 and we have shown that both mutant and wild type CD151 constructs are expressed on the cell surface.Citation41,42 These data revealed that loss of CD151 or CD151-integrin association reduces the tightness of cell-cell contacts, while CD151-tetraspanin association appears less important for this function.
Figure 1. Loss of CD151 or CD151-integrin interaction reduces the tightness of cell-cell junctions. (A-E) Electron micrographs of cell-cell junctions within monolayers of A431 parental cells (WT), CD151sh3 cells, CD151-VR cells, CD151-Palm cells, or CD151 WTRX cells. The rectangular area in each micrograph was zoomed and rotated to vertical to the right of each image. (F) The percentage of each junction filled with cross interacting membrane and filopodia was calculated as described in Experimental Procedures. Bars show mean with SEM for 13–17 fields per cell type. The sh3 and VR cell junctions were significantly less densely packed than the WT cell junctions, ***P ≤ 0.0006 (ANOVA with Dunnett's post-test). Bar = 1 µm.
α3β1 integrin and its ligand, laminin-332 promote the maintenance of organized cell-cell junctions
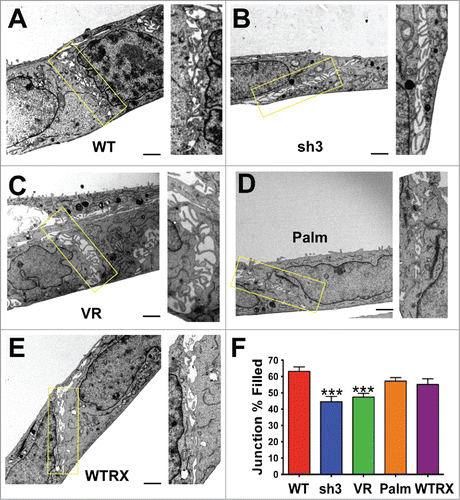
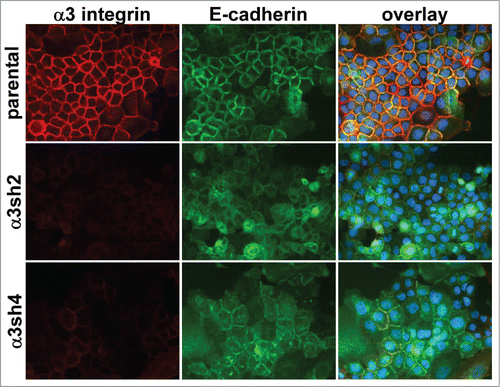
The above ultrastructural analysis, together with our previous observation that the CD151-VR integrin interaction mutant failed to sustain organized adherens junctions,Citation42 indicated that one or more of CD151s integrin partners was likely to be involved in CD151s pro-junction activity. In A431 cells, both α3β1 and α6β4 integrins are abundantly expressed, functionally relevant CD151 partners.Citation41 To test whether α3β1 or α6β4 integrin was involved in CD151s pro-junction activity, we silenced CD151, α3 integrin, or α6 integrin in cells expressing an E-cadherin-GFP fusion protein (Ecad-GFP). Total E-cadherin levels were similar in cells with E-cad-GFP expression compared to parental cells. The introduced E-cadGFP replaced ˜50% of the endogenous E-cadherin (C.S.S. unpublished data). The loss of CD151 markedly reduced the organization of cell junctions as visualized by Ecad-GFP (), confirming our previous results.Citation42 Depleting CD151 in HaCaT immortalized keratinocytes also resulted in less organized cell-cell junctions (Fig. S2), further confirming CD151s role in promoting junctional organization. Strikingly, silencing α3 integrin severely disrupted the organization of cell junctions (), while silencing α6 integrin had no obvious effect on junctional morphology (Fig. S3). Flow cytometry confirmed ˜84% silencing of α3 integrin and ˜99% silencing of α6 integrin (data not shown), similar to what we previously reported (see also and Fig. S3 for confirmation of near total silencing of α3 and α6 integrin)
Figure 3. Loss of organized cell-cell junctions in α3 integrin-silenced cells. Integrin α3 was silenced with 2 different shRNA constructs in E-cadherin-GFP-expressing A431 cells. The α3 silenced cells and parental A431 cells expressing E-cadherin-GFP (green) were fixed and stained for α3 integrin (red). DAPI-stained nuclei are shown in blue in the overlay image.
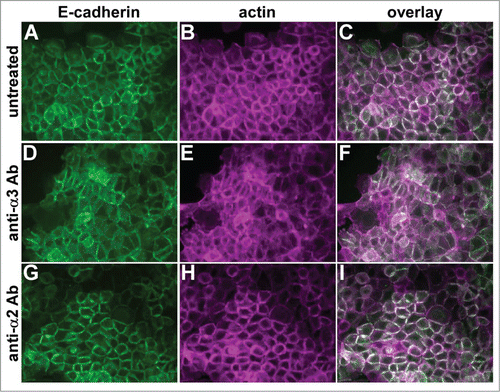
To investigate whether α3β1 integrin's junction organizing activity involves ligand binding, we treated parental cells with a function blocking anti-α3 integrin antibody that disrupts α3-dependent adhesion and migration.Citation40,41 Compared to untreated controls (), cells treated with the anti-α3 integrin antibody displayed reduced organization of E-cadherin and cortical actin at cell-cell contacts (). Treatment with a function blocking antibody recognizing α2β1 integrin, which is expressed at comparable levels to α3 integrin in A431 cells,Citation40 had no impact on the organization of cell-cell junctions ()
Figure 4. A function-blocking anti-α3 integrin antibody perturbs cell junctions. Parental A431 cells expressing E-cadherin-GFP (green) were left untreated (A–C), or treated overnight with function blocking antibodies recognizing α3 integrin (D-F) or α2 integrin (G–I). Cells were fixed and stained for actin (magenta). Antibodies used were A3-IIF5 (anti-α3) and A2-IIE10 (anti-α2).
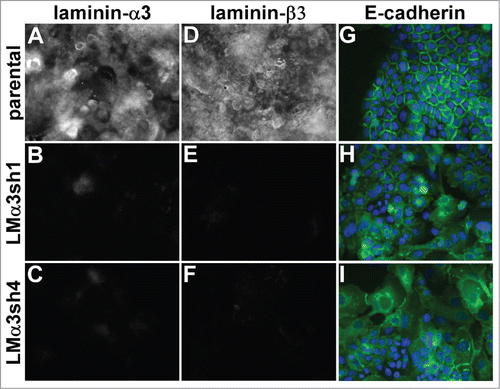
A431 cells secrete and deposit abundant laminin-332, a major α3β1 integrin ligand. To investigate whether laminin-332 contributes to the maintenance of junctional organization, we silenced the laminin-α3 subunit (LMα3) of laminin-332, the subunit that contains the α3β1 integrin binding site.Citation50 The loss of LMα3 expression dramatically reduced extracellular deposition of LM-332 as measured by staining for either LMα3 itself (), or for the laminin-β3 subunit (). An ELISA assay to detect substrate-bound LM-332 confirmed an apparent ˜80% reduction in laminin deposition in the LMα3-silenced cells (Fig. S4). Similar to the loss of α3 integrin, the loss of LM-332 deposition severely disrupted junctional organization ( and Fig. S5A-C). Organized junctions could be restored in the LMα3-silenced cells by replating them on the laminin-332 matrix deposited by wild type parental cells (Fig. S5D-F), but not on collagen I (Fig. S5G-I), fibronectin (Fig. S5J-L), or laminin-111-rich Matrigel (Fig. S5M-O), matrix proteins to which α3β1 integrin does not bind with high affinity.
Figure 5. Laminin-332 is required for the maintenance of organized cell junctions. The laminin-α3 subunit was silenced in A431 cells with 2 different shRNA constructs. (A-F) Parental cells and laminin-α3 silenced cells were extracted with Triton X-100 and the remaining extracellular matrix was stained for the α3 or β3 subunit of LM-332. (G-I) A431 parental and laminin-α3-silenced cells expressing E-cadherin-GFP (green) were fixed and photographed. DAPI-stained nuclei are shown in blue.
To explicitly test whether LM-332s junction organizing activity requires α3β1 integrin, we replated α3β1-silenced cells on the LM-332-rich extracellular matrix deposited by parental cells. Unlike LMα3-silenced cells, whose junctional organization was rescued upon replating on parental cell matrix, α3 integrin-silenced cells failed to respond to parental cell matrix by forming organized junctions (Fig. S6). Together with the data from and S5, these results strongly support a role for α3β1 integrin binding to its ligand LM-332 in promoting organized cell-cell junctions in our carcinoma cell system.
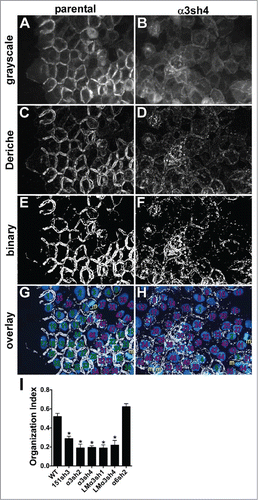
To render qualitative observations of junctional organization more quantitative, we employed an edge detection plugin for ImageJ that performs a Canny-Deriche filtering algorithm to identify edges in the image, followed by thresholding, using the Make Binary command. As shown in , well-organized junctions visualized via Ecad-GFP generally appear as bold double lines after implementation of the algorithm, while poorly organized junctions produce diffuse, thin lines that do not strongly delineate cell-cell boundaries. We overlaid the filtered images onto DAPI-stained nuclei in the same field and scored the proportion of nuclei bounded by ≥ 2 organized junctions to obtain the “organization index” of the image (). This quantitative approach confirmed that junctional organization is dramatically reduced for CD151-silenced, α3-integrin silenced, and LMα3-silenced cells, but not for α6 integrin-silenced cells.
Figure 6. Quantitative analysis of junctional organization. Junctional organization was quantified as described in Experimental Procedures. Representative fields from parental and α3-silenced A431 cells are shown. (A&B) original grayscale images; (C&D) images after Canny-Deriche edge detection filtering; (E&F) edge detection image converted to binary image; (G&H) binary image overlaid with DAPI-stained nuclei (blue). Cells scored as having ≥ 2 organized junctions are numbered in green; those with fewer than 2 organized junctions are numbered in magenta. Cells not included because they were undergoing mitosis at time of fixation are labeled with “m” in gold. (I) The proportion of cells with ≥ 2 organized junctions/cell was quantified for 8 separate fields per cell type is graphed as mean with SEM. CD151-silenced, α3 integrin-silenced, and laminin-α3-silenced cells all showed significantly reduced junctional organization compared to WT parental or α6 integrin-silenced cells, *P ≤ 0.0005 (ANOVA with Tukey post tests).
CD151 restrains collective invasion and tumor growth in a 3D matrix
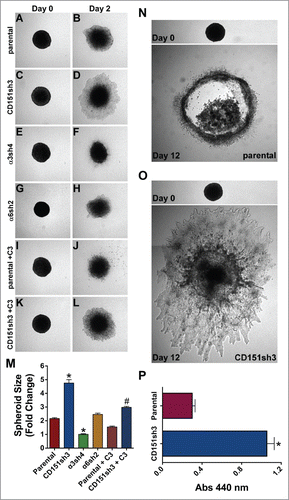
Previously, we found that CD151-depleted cells display enhanced collective migration on 2D substrates.Citation42 However, tumor invasion in vivo may be better represented by a 3D environment.Citation10 Therefore, we embedded tumor spheroids in a 3D collagen matrix to examine how loss of CD151 or its integrin partners affected 3D invasion. Compared to parental cells, the collective invasion of CD151-depleted cells was dramatically enhanced (, quantified in ). Silencing α3 integrin, but not α6 integrin, further suppressed the already low collective invasion of CD151-replete parental cells (). Thus, outright silencing of α3 integrin impaired 3D collective invasion, but loss of the α3 partner, CD151 actually enhanced invasion, suggesting that the loss of CD151 can alter the balance between α3 integrin's pro- and anti-invasive activities in intact epithelial sheets. Immunostaining confirmed that A431 cells continue to secrete abundant LM-332 in the 3D spheroid cultures, providing a potential explanation for the role of α3β1 integrin in A431 cell collective invasion(Fig. S7).
Figure 7. Loss of CD151 can promote invasion and growth in a 3D matrix. (A-H) Parental, CD151, α3, and α6 integrin-silenced cell spheroids were embedded in 3D collagen and photographed immediately (Day 0) and 2 d later (Day 2). (I-L) Parental and CD151-silenced cell spheroids were embedded in 3D collagen and grown 2 d in the presence of 1 µg/ml cell permeable C3 transferase to inhibit Rho activity. (M) Quantification of spheroid size on Day 2, plotted as fold change from size on Day 0. Bars indicate mean ± SEM for 6–12 spheroids per cell type. CD151sh3 spheroids were significantly larger and α3sh4 spheroids were significantly smaller than parental spheroids, *P < 0 .0001, and CD151sh3 spheroids treated with C3 transferase were significantly smaller than untreated CD151sh3 spheroids, #P < 0 .0001. ANOVA with Tukey post-tests. (N&O) Parental and CD151-silenced spheroids were embedded in 3D collagen and cultured for 12 d (P) Spheroids cultured for 12 d in 3D collagen were excised and relative cell number was determined by WST assay after digestion with collagenase type II. Bars indicate mean ± SEM for 12 spheroids per cell type. CD151sh3 spheroids had grown significantly larger, *P < 0.0001, unpaired t test.
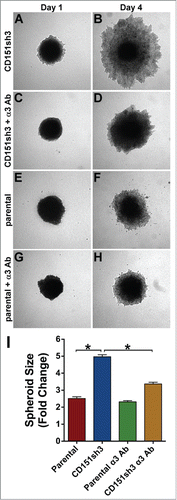
If loss of CD151 promotes α3-dependent collective invasion, then the enhanced invasion of CD151-depleted cells should be inhibited by blocking α3 integrin function. To test this hypothesis, we treated CD151-silenced and parental tumor spheroids with our function-blocking anti-α3 integrin antibody. As predicted, antibody blockade of α3 integrin significantly impaired the collective invasion of CD151-silenced cells (, quantified in I). In contrast, the anti-α3 antibody had minimal effect on the low level of invasion by parental cells (). The minimal effect of α3 integrin antibody blockade on parental cell invasion, as compared to the small but significant effect of α3 silencing quantified in , might be due to incomplete function blocking by the α3 antibody over the 3 day assay.
Figure 8. Enhanced invasion of CD151-silenced cells depends on α3β1 integrin. CD151sh3 cell spheroids were embedded in 3D collagen in the absence (A&B) or presence (C&D) of 10 µg/ml A3-IIF5 anti-α3 integrin function blocking antibody, and photographed on Day 1 and Day 4. (E-H) Parental cell spheroids were embedded in collagen with or without A3-IIF5 antibody, as for the CD151sh3 spheroids in (A-D). (I) Quantification of spheroid size on Day 4 plotted as fold change from Day 1. The untreated CD151sh3 cell spheroids were significantly larger than either parental cell spheroids or antibody-treated CD151sh3 cell spheroids, *P<0 .0001, ANOVA with Tukey post-tests.
In our previous study, the enhanced collective migration of CD151-silenced cells in 2D was dependent on RhoA signaling.Citation42 In agreement with these earlier results, inhibiting Rho activity with cell permeable C3 transferase suppressed the collective 3D invasion of the CD151-silenced cells, while the inhibitor had a more modest effect on invasion of the parental cells (). These data suggest that Rho signaling may be at least partially responsible for the enhanced invasiveness of the CD151-silenced cells.
To evaluate the longer-term consequences of CD151 silencing in 3D cultures, we examined cultures of spheroids grown for 12 d post-embedding. By this time point, the less invasive parental spheroids showed necrotic centers, while the more invasive CD151-silenced spheroids showed continued expansion (). To quantify differences in growth, we excised individual spheroids, digested the matrix with collagenase, and measured relative cell numbers by WST assay. This analysis revealed an apparent fold3- increase in the number of CD151-silenced cells compared to parental controls (). The enhanced growth of the CD151-silenced cells in 3D is not due to an artifact of CD151-silencing that renders the cells intrinsically more proliferative. In standard proliferation assays in 2D cultures, parental and CD151-silenced cells showed identical proliferation on collagen I, and the CD151-silenced cells actually showed modestly impaired proliferation on laminin-332 (Fig. S8). Thus, the loss of CD151 specifically enhanced growth under 3D conditions.
CD151-depleted cells appear more invasive and aggressive in vivo
To explore how the enhanced collective invasion of CD151-depleted cells in vitro may translate to invasiveness in vivo, we implanted spheroids of parental or CD151-depleted cells intradermally in nude mice. The intradermal site was chosen because of the squamous epidermoid carcinoma origin of the A431 cell line. Six days post-implantation, one set of spheroids was recovered and sectioned for morphological analysis. To quantify differences in tumor morphology we adopted the metric of numerical roundness, which has been applied to differences in cellular morphology.Citation51 Numerical roundness, defined by the equation (Area × 4π)/(perimeter),Citation2 is equal to exactly 1.0 for a perfect circle. The more complex the perimeter, the lower the numerical roundness. Analysis of tumor sections taken near the midpoint of the tumors at day 6 after implantation revealed that sh3 cell tumors appeared more locally invasive, with complex perimeters resulting from chords of cells emanating from the tumor centers (). The mean numerical roundness of CD151-silenced tumors was less than one third that of the parental cell tumors, which had smooth perimeters encircled by layers of stromal cells (0.27 ± 0.006 vs. 0.84 ± 0.07, P = 0.0017; ). Thus, the enhanced collective migration of the CD151-silenced cells in vitro appears to correspond to a more invasive tumor morphology in vivo.
Figure 9. Loss of CD151 can promote a more invasive and rapidly growing tumor phenotype in vivo. (A) CD151-silenced cell spheroids (left) and parental spheroids (right) were recovered 6 d after intradermal implantation, sectioned, and stained with hematoxylin and eosin (H&E). Tumors (labeled “t”) are surrounded by lightly stained stromal cells. Fat and muscle are visible in some sections. Silhouettes of CD151-silenced and parental tumor sections are displayed below the H&E panels, along with the numerical roundness of each section. The more tortuous the perimeter, the lower the roundness value. The mean roundness of the CD151-silenced spheroids (0.27 ± 0.01) was significantly less than the mean roundness of the parental spheroids (0.84 ± 0.07), n = 3 spheroids/cell type, P = 0.0017, unpaired t test. (B) In vivo growth of parental and CD151-silenced spheroids was monitored by bioluminescence imaging. By day 20, CD151sh3 spheroids were significantly larger than parental spheroids, *P = 0.0188, unpaired t test, n = 3 spheroids/cell type. (C) H&E staining of day 20 parental and CD151-silenced spheroids. Tumors (“t”) and epidermis (“e”) are labeled.
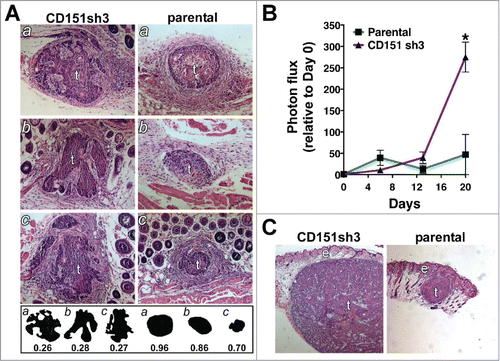
We used bioluminescence imaging (BLI) to follow the growth of implanted spheroids over time. At 6 d post-implantation, when differences in local invasion were apparent, there was no obvious difference between the parental and CD151-silenced spheroids. However by 20 d post-implantation, CD151-silenced spheroids were over fold5- larger, as measured by BLI () and confirmed by H&E staining of dissected tumors (). The enhanced growth of the CD151-silenced cells in vivo thus recapitulated their growth phenotype in our 3D in vitro assay.
Expression of CD151 in human cancer cases
Our in depth functional analyses of CD151-integrin complexes in the A431 squamous cell carcinoma indicates that the role of CD151 and its integrin partners in regulating invasive behavior is context dependent. For example, CD151 promotes rapid, single cell migration,Citation40,41 but restrains slower, collective migration of intact epithelial sheets (ref. Citation42 and this study). To search for potential correlates between results from our squamous cell carcinoma model and human cancer cases, we queried the TCGA database using the cBioPortal interface.Citation45,46
In lung squamous cell carcinoma, a substantial fraction of cases display alterations in genes involved in regulating squamous differentiation. When we applied a previously published set of gene alterations,Citation47 to the cases accrued in the provisional TCGA Lung Squamous Cell Carcinoma database, we found that 69% of cases displayed amplification or upregulation of SOX2 or TP63, and/or mutations in NOTCH1, NOTCH2, ASCL4, or FOXP1 genes (Fig. S9A). SOX2, which was upregulated or amplified in 60% of the cases, has been linked to a poorly differentiated, stem-like phenotype and epithelial-mesenchymal transitions (EMTs).Citation52,53 Within these 69% of cases predicted to display disrupted squamous differentiation, reduced CD151 mRNA expression was associated with better overall survival (, P = 0.0079), consistent with the possibility that CD151 could contribute to invasion of poorly differentiated squamous lung cancers. In the remaining 31% of cases lacking alterations in the squamous differentiation gene set, there were not enough cases with changes in CD151 expression to reveal any statistically significant relationships. Likewise, there were too few cases in the TCGA Esophageal Squamous Cell Carcinoma and Cervical Squamous Cell Carcinoma databases with altered CD151 expression to reveal significant associations (data not shown).
Figure 10. The relationship of CD151 expression to clinical outcomes in human cancers. (A-E) Analysis of CD151 expression by RNAseq in TCGA Lung Squamous Cell Carcinoma and Invasive Breast Carcinoma databases. Survival curves are shown for CD151-low cases (Z-score < −1; shown in green) or CD151-high cases (Z-score > 1; shown in red) compared to cases in which CD151 expression was unaltered (shown in black). (A) Reduced CD151 expression was associated with increased overall survival in lung squamous cell carcinomas harboring changes in genes that regulate squamous cell differentiation. (B&C) In less aggressive ESR1+/GATA3+ breast cancers (in which estrogen receptor α (ESR1) or GATA3 were not reduced; see Fig. S8), reduced CD151 expression was associated with reduced survival, while increased CD151 expression was associated with increased survival. (D&E) In the more aggressive ESR1low/GATA3low breast cancer cases, CD151-low cases showed a trend toward increased survival, while CD151-high cases showed reduced overall survival. (F-I) Analysis of CD151 expression in different breast cancer molecular subtypes using KM-plotter to query Affymetrix microarray data. (F) In luminal A breast cancers, CD151-high cases (red) showed increased relapse-free survival compared to CD151-low cases (green). (G) In luminal B breast cancers, no association between CD151 expression and relapse-free survival was observed. (H&I) in Her2+ and basal-like breast cancers, CD151-high cases showed reduced relapse-free survival. In all panels, P values were calculated by the logrank test. More detailed information about the number of cases represented in each panel is found in Table S1.
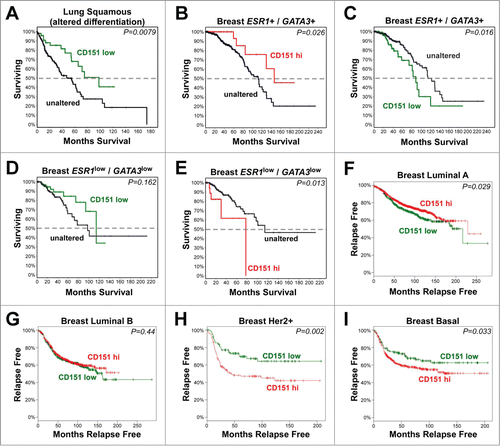
To further explore the relationship between CD151 expression and clinical outcomes in human cancer, we turned to the TCGA Breast Invasive Carcinoma provisional database, which contains RNAseq gene expression data for over 1000 breast cancer cases. Initial analyses of CD151 expression in breast cancer cases revealed no obvious relationship between CD151 up or downregulation and clinical outcomes (data not shown). Therefore, we subdivided breast cancers on the basis of the expression of the estrogen receptor (ESR1) and its co-regulator, GATA3,Citation48 which are associated with a luminal cell phenotype. Cases with reduced ESR1 and/or GATA3 mRNA (hereafter referred to as ESR1low/GATA3low cases) accounted for ˜26% (288 out of 1098) of the total breast cancer cases and were associated with loss of other luminal markers, reduced disease-free survival, higher expression of cyclins B1 and E1, increased p53 mutation rate, and upregulation of P-cadherin (CDH3), a marker of aggressive, basal-like breast cancers (Fig. S9B-E).Citation54 Interestingly, in the less aggressive ESR1+/GATA3+ breast cancer cases, loss of CD151 was associated with reduced survival time (P = 0.016; ), while elevated CD151 was associated with increased survival time (P = 0.026; ). In contrast, in the more aggressive ESR1low/GATA3low cancers, cases with loss of CD151 showed a trend toward increased survival time (), while cases with elevated CD151 showed a significant reduction in survival time (P = 0.013; ). These results are consistent with CD151 contributing to the maintenance of a less aggressive phenotype in the ESR1+/GATA3+ cases, but helping to promote a more aggressive phenotype in ESR1low/GATA3low cases.
For a second, independent assessment of the relationship between CD151 expression and breast cancer outcomes, we evaluated Affymetrix microarray data on CD151 expression in the different molecular subtypes of breast cancer, using the KM-plotter program.Citation49 In luminal A breast cancers, higher CD151 expression was associated with a modest but significant increase in relapse-free survival (, P = 0.029). In contrast, in the more aggressive Her2+ and basal breast cancers, higher CD151 expression was associated with significant reductions in relapse-free survival (, P = 0.002, and P = 0.033 respectively). In the intermediate luminal B breast cancers, there was no significant association between CD151 expression and outcomes (). Thus, data from Affymetrix arrays evaluated by KM-plotter supported the RNAseq data from the TCGA database in that both datasets revealed better outcomes for CD151 high cases in less aggressive breast cancer subtypes and worse outcomes for CD151 high cases in more aggressive breast cancers. Collectively, these data suggest that CD151 can make both positive and negative contributions to disease progression in different settings, highlighting the importance of understanding the diversity of mechanisms by which CD151 regulates tumor invasion and motility in different contexts. As discussed below, the divergent roles of CD151 in regulating single cell vs. collective invasion may help to explain why CD151 can have seemingly opposing roles in the progression of different types of cancer.
Discussion
The role of CD151-α3β1 in the maintenance of organized cell-cell junctions and the regulation of collective cell invasion and migration.
Several earlier studies pointed to a role for tetraspanin CD151 in the maintenance of organized cell-cell junctions. Over-expressing CD151 in A431 cells enhanced cell-cell contact formation and reduced the rate of collective migration.Citation55 Conversely, an anti-CD151 antibody capable of dissociating CD151 from its integrin partners perturbed cell-cell junctions and enhanced migration of HaCaT immortalized keratinocytes.Citation56 We previously showed that silencing CD151 disrupted the organization of E-cadherin-based cell-cell junctions, and a CD151 mutant incapable of interacting with integrins failed to restore organized junctions upon re-expression in CD151-silenced cells.Citation42 CD151 also promoted VE-cadherin-dependent endothelial cell-cell adhesion.Citation57
An earlier study with α3-null murine kidney epithelial cells implicated α3 integrin as a CD151 partner involved in promoting junctional stability.Citation58 However, the proposed mechanism in the kidney epithelial cell system, which involved CD151-α3β1 regulation of β-catenin phosphorylation, α-actinin-cadherin association, and PTPµ expression, appears not to operate in our human carcinoma cells.Citation42 An important recent study revealed that silencing CD151 in ovarian carcinoma cells can perturb cell-cell junctions by a mechanism that may involve upregulation of β-catenin and induction of epithelial-mesenchymal transition (EMT)-like changes, including loss of E-cadherin.Citation35 However, we previously observed no changes in β-catenin or E-cadherin expression level upon loss of CD151 in our squamous skin cell carcinoma system.Citation42 Depleting CD151 in endothelial cells also results in loss of organized junctions, but the proposed mechanism for this phenomenon was suggested to be independent of CD151-association with laminin-binding integrins.Citation57 In contrast to its role in promoting organized junctions in many settings, in lung airway epithelial cells, α3β1 integrin has been reported to interact with E-cadherin to promote an EMT in response to TGFβ or bleomycin induced injury.Citation59,60 However, we could not detect a α3β1-E-cadherin interaction in our carcinoma cells.Citation42 Given all of these contrasting results, which, if any, of CD151s integrin partners are responsible for CD151s pro-junction activity in human carcinoma cells had remained unclear.
We now show that CD151s ability to promote organized junctions in our carcinoma cell system strongly depends on α3β1 integrin. The CD151-VR mutant lacking integrin association fails to promote tight cell-cell contacts as visualized by electron microscopy (this study) or organized junctions as visualized by E-cadherin localization.Citation42 Depletion of α3 integrin or the α3β1 integrin ligand, laminin-332, disrupted junctional organization to at least as great an extent as depletion of CD151. In contrast, near total depletion of the laminin-332-binding α6 integrin had no obvious impact on the organization of cell-cell junctions. Blocking α3 function with an antibody also disrupted junctional organization, and replating α3-silenced cells on parental cell matrix failed to restore organized junctions. Collectively, our new data support a model in which α3β1 integrin binding to its ligand, laminin-332, generates a cue or a signal that, in a CD151-dependent manner, promotes the formation or maintenance of organized cell-cell junctions.
The mechanism by which the CD151-α3β1 complex promotes the stability of cell junctions appears to involve moderating Rho signaling. Inhibiting Rho with cell permeable C3 transferase suppressed 3D collective invasion in the present study. This is consistent with our earlier study, in which we observed that RhoA activity is elevated in CD151-silenced cells, and that suppressing RhoA partially restored the stability of cell-cell junctions,Citation42 and with the deregulation of RhoA signaling that results from conditional deletion of α3 integrin in mammary myoepithelium.Citation61 In endothelial cells, CD151 depletion also results in aberrant RhoA activation, by a mechanism that may involve reduced cAMP levels and the loss of an inhibitory phosphorylation of RhoA mediated by protein kinase A,Citation57 while in hippocampal neurons, the loss of α3 integrin triggers a collapse of dendritic spines due to increased RhoA activation.Citation62 In this case elevated RhoA activity results from a loss of α3β1 integrin signaling to p190RhoGAP via the Arg/ABL2 tyrosine kinase.Citation62 It will be important to determine which if either of these mechanisms is responsible for CD151-α3β1 regulation of RhoA in our squamous cell carcinoma model.
In our carcinoma cell model, an important functional effect of CD151 depletion is enhanced collective migration and invasion in 2D and 3D settings. Depleting α3 integrin itself also disrupted cell junctions, but actually suppressed collective invasion, probably because α3β1 is required as a motility receptor during invasion in 3D. In support of this possibility, much of the increased invasion resulting from CD151 depletion can be blocked with an anti-α3 integrin antibody. Thus, the loss of CD151, which disrupts junction stability but preserves α3 cell surface expression,Citation40,42 might shift the balance between α3β1s junction-stabilizing activity and α3β1s motility functions, leaving enough of the latter intact to allow α3β1 to participate in invasion. The loss of CD151 might also shift the balance between adhesion and traction, which governs migration velocity in both 2D and 3D settings,Citation63,64 to a ratio that better facilitates the coordinated migration of cell sheets.
A potential corollary of enhanced invasion of CD151-silenced cells at early time points is their increased growth by later time points, observed both in vitro and in vivo. Conceivably, ongoing local invasion might facilitate more rapid growth by alleviating competition for limiting nutrients in the cell-dense core of a growing tumor or spheroid. Determining whether enhanced invasion and more rapid growth are linked or can be uncoupled will be an important future direction.
Implications for tumor progression
The loss of organized cell-cell junctions and the loss of stable cell-substrate attachments in the form of hemidesmosomes have long been recognized as hallmarks of progression toward invasive carcinoma. Destabilization of cell-cell and cell-matrix attachments can result from EMT-like events in which epithelial proteins such as E-cadherin and β4 integrin are downregulated, and mesenchymal proteins such as N-cadherin and vimentin are upregulated. However, outright EMT, in which cohesive epithelial cells convert to solitary, invasive mesenchymal cells, is at one end of a spectrum of altered cell-cell and cell-substrate interactions that can lead from stable epithelial structure to locally invasive, collectively migrating carcinoma cells, to distant metastases.Citation10,65
Where CD151 has been positively linked to tumor formation, progression or metastasis, it may function by promoting migration, invasion, survival, and proliferation mediated through its laminin-binding integrin partners,Citation25-28,30,66 or by integrin-independent mechanisms that remain to be defined.Citation34 These pro-metastatic CD151 functions may sometimes be more readily apparent in cells with a mesenchymal phenotype, which might help to explain CD151s pro-metastatic role in the TRAMP prostate carcinoma model,Citation31 which features a loss of epithelial phenotype at advanced stages.Citation31,67 Aggressive, Her2+ and basal-like breast cancers or Sox2-positive squamous lung carcinomas, in which CD151 may contribute to progression and poor clinical outcomes, may be more prone to display EMT-like phenotypes.Citation52,53,68-70
In contrast, where CD151 has been negatively linked to invasion and metastasis,Citation35-38 it may function by helping to enforce integrin-dependent epithelial phenotypes, such as stable cell-cell and cell-matrix contacts, which are retained to a greater extent in some cancers. Hormone receptor-positive, GATA3-positive, or luminal A breast cancers, in which we found that CD151 expression was associated with better clinical outcomes, feature a more differentiated, epithelial phenotype, with retention of E-cadherin expression.Citation71 Potentially, the loss of CD151 might contribute to destabilization of epithelial structure and increased collective invasion in E-cadherin-positive breast cancers, similar to what we observed in our squamous skin cell carcinoma model. These potential anti-invasive CD151 functions are also reflected in CD151s role in maintaining kidney and lung epithelial integrity.Citation72-75 Thus, the role of CD151 in regulating metastasis may depend, at least in some cases, on where the tumor cells lie on the spectrum of epithelial to mesenchymal phenotype. Given the complexities arising from intra-tumoral heterogeneity,Citation76 the role of CD151 might also vary from cell to cell in a primary tumor or change dynamically during different stages of tumor progression.
Materials and Methods
Antibodies and extracellular Matrix Proteins
Monoclonal antibodies used in this study included anti- E-cadherin (BD Biosciences, clone 36/E-cadherin), P3H9, anti-laminin-α3 (Santa Cruz Biotechnology), and 6F12 anti-laminin-β3.Citation43 The 5C11 anti-CD151, A2-IIE10 anti-2 integrin, A3-X8 and A3-IIF5 anti-α3 integrin, and A6-ELE anti-α6 integrin antibodies have been previously referenced.Citation40 Alexa Fluor 488 and 594 goat anti-mouse IgG secondary antibodies were from Life Technologies. Horseradish peroxidase (HRP)-conjugated goat anti-mouse secondary antibody was from Jackson ImmunoResearch. Rat tail collagen I, human plasma fibronectin, and Matrigel were from BD Biosciences. Laminin-332 was purified from SCC-25 squamous carcinoma cell conditioned medium as previously described.Citation40
Cell Lines and RNA interference (RNAi)
A431 squamous cell carcinoma lines and HaCaT cells were cultured in DMEM supplemented with 10% fetal bovine serum, 2 mM L-glutamine, 100 units/ml penicillin, and 100 µg/ml streptomycin. The retroviral RNAi-mediated silencing of CD151, α3 integrin, and α6 integrin, and the reconstitution of CD151-silenced cells with wild type and mutant CD151 constructs has been previously described.Citation40,41 Targeting sequences used in the current study were as follows: CD151 sh3, 5′-AGTACCTGCTGTTTACCTACA-3′; α3 integrin sh2, 5′-GGCCAGCATTGGTGACATCAA-3′; α3 integrin sh4, 5′-GGATGACTGTGAGCGGATG-AA-3′; α6 integrin sh2, 5′-GTATGTAACAG-CAACCTTAAA-3′; laminin-α3 sh1, 5′-GTC-CGACTGCCAAATACC-3′; and laminin-α3 sh4, 5′-GGAGGTCAT-GTCGTCTTGG’-3′.
Ultrastructural analysis of cell junctions
Cells growing in 35 mm tissue culture dishes were fixed overnight with 2.5% glutaraldehyde in 0.1 M cacodylate buffer, and post-fixed for 1 hour at room temperature with a buffered 1% osmium tetroxide solution reduced with 1.5% potassium ferrocyanide. Samples were stained en bloc with 2.5% uranyl acetate for 20 minutes, then rinsed and dehydrated using gradually increasing concentrations of ethanol to 100%. Infiltration of EPONate 12 (Ted Pella, Redding, CA) epoxy resin and ethanol were performed over several hours to 100% resin and cured 48 hours in a 60°C oven. Sections of 80 nm thickness were cut using a Leica UC-6 ultramicrotome and collected on formvar-coated 2×1 slot copper grids. Samples were counterstained with 5% uranyl acetate for 2 minutes and Reynold's lead citrate for 2 minutes, and then imaged at 1200X using a JEOL 1230 transmission electron microscope at 120 KV.
For morphometric analysis of cell junctions, images were converted to binary using the Threshold function in ImageJ. Then, the pencil tool was used to demarcate the boundaries of the cell-cell junctions, and, where necessary, the brush tool was used to correct gaps in the filopodial projections. The boundary of a cell-cell junction was defined as the space between 2 cells where only 2 cells could be distinguished at the junction and 2 distinct connections could be discerned. Cells that were stacked vertically instead of side-to-side, and cells that were not connected at the tops of the monolayer were not included in the analysis. The percentage of each junction filled by filopodial projections was calculated using ImageJ's Analyze Particles function.
Immunostaining and Enzyme-linked Immunoassay (ELISA)
Cells cultured on acid-washed glass coverslips were fixed with 10% formalin in HEPES-buffered saline with 4% sucrose and 1 mM MgCl2, rinsed with TBS, and blocked with 10% goat serum in PBS prior to exposure to primary antibodies. To examine laminin-332 deposition, cells were extracted with 1% Triton X-100 prior to immunostaining. Primary antibodies were detected with Alexa Fluor 488 or 594-labeled secondary antibodies, and coverslips were mounted in ProLong Gold (Life Technologies) for fluorescence microscopy. In some experiments, cells were permeabilized with 0.1% NP-40 detergent and counterstained with Alexa-594 phalloidin (Life Technologies) to reveal F-actin. To quantify substrate-bound laminin-332 by ELISA, 1.5 × 104 cells/well growing in 96 well plates were extracted with 1% Triton X-100, and the exposed substrates were blocked with 10% heat-inactivated BSA in PBS. Wells were then stained with the 6F12 anti-laminin-β3 antibody, followed by an HRP-linked goat-anti-mouse secondary antibody. The wells were developed using the Super AquaBlue ELISA substrate (eBioscience, Inc.) followed by quantification using a plate reader (Molecular Devices).
Quantification of Cell Junction Organization
Using ImageJ software,Citation44 images of E-cadherin localization were converted to 8 bit grayscale and then the Image Edge plugin (http://imagejdocu.tudor.lu/doku.php?id = plugin:filter:edge_detection:start) was used to perform a Canny-Deriche filtering for edge detection. The smoothing parameter α was left at the default value of 1.0. The plug in generates 2 images, a normal image and a second image with non-maximal suppression to generate thinner edges. For the purposes of our analysis, th49. normal image produced more robust results. The Canny-Deriche filtered image was converted to binary and then overlaid with a 4′,6-diamidino-2-phenylindole (DAPI)-stained image of the cell nuclei from the same field. Junctions with prominent E-cadherin localization generally produce bold double lines at the cell-cell boundaries using this protocol, while junctions lacking prominent E-cadherin tend not to survive the filtering process. De-identified overlaid images were blindly scored for the percentage of nuclei bounded by 2 or more distinct junctions. Eight separate fields per cell type were analyzed.
3D Collective Invasion Assay
To create spheroids for the invasion assay, 10,000 cells per spheroid were plated in 96 well V-bottom plates that had been coated twice by air-drying 25 µl of 20 mg/ml polyhydroxyethylmethacrylate (poly-HEMA) in 95% ethanol in each well. Plates were spun briefly in a tabletop centrifuge to settle the cells and then cultured overnight in standard growth medium to allow spheroid formation. To create collagen gels for the invasion assay, 700 µl per well of 0.8 mg/ml rat tail collagen I in DMEM was allowed to polymerize for 1 h at 37°C in a 12 well plate. The plate was then placed on ice, and the spaces between the wells were filled with ice cold PBS to maintain temperature during the transfer of spheroids. A second 700 µl of ice cold 0.8 mg/ml collagen I was overlaid on the cushion of pre-polymerized collagen I, and 6 spheroids per cell type were transferred to a collagen-containing well. To transfer, spheroids were recovered from the V-bottom wells by pipeting with a yellow tip to dislodge and capture the spheroids, and then spheroids were allowed to settle by gravity out of the tip and into the collagen, without ejecting any culture medium. After transfer, the collagen was polymerized for 1 h at 37°C, and each spheroid was photographed with a 4X objective and the cultures were overlaid with serum-free DMEM with or without 1 µg/ml cell permeable C3-transferase (Cytoskeleton, Inc.). After 2 d, spheroids were photographed again, and the change in area was measured with Image J. In one set of experiments, the A3-IIF5 anti-α3 integrin antibody was included in both collagen layers and the overlying medium at a concentration of 10 µg/ml. In another set of experiments, spheroids were cultured for 12 d after implantation. In these longer-term experiments, the relative number of viable cells was determined by WST assay (Roche Life Science) after excising the spheroids from the collagen gel with the aid of a dissecting microscope and dissolving the collagen matrix with 200 U/ml type II collagenase (Worthington Biochemical Corp.).
Intradermal Implantation Assay
All animal experiments were approved by the University of Iowa Institutional Animal Care and Use Committee (Approval #1202041). Spheroids were created, using the protocol described above, except starting with 5,000 cells per spheroid instead of 10,000. Cells were transduced with a luciferase retroviral vector to aid in confirming successful implantation and assist in subsequent dissection. Spheroids were recovered, rinsed with a 50:50 mixture of PBS and OptiMEM medium (Life Technologies). A 27 gauge needle was used to create small intradermal spaces in the periscapular skin of nude mice, and individual spheroids were implanted under a dissecting scope using a drawn glass pipet fitted with mouth transfer pipet tubing. After 6 or 20 days, skin containing the spheroids was dissected, formalin fixed, and paraffin-embedded for sectioning. For morphometric analysis of the tumor spheroids, tumor boundaries were traced, and perimeters and enclosed areas were calculated using Image J software. Tumor roundness was calculated with the equation, roundness = (Area × 4π)/perimeter,Citation2 which yields a value of exactly 1.0 for a perfect circle. Tumor growth was monitored during the assay by bioluminescence imaging using an IVIS100 instrument (Caliper Life Sciences).
Analysis of data from the Cancer Genome Atlas (TCGA) and KM-plotter
Analyses of human cancer data utilized only de-identified, publicly available databases, and thus were in accordance with Helsinki Declaration of 1975. The cBioportal website.Citation45,46 at the Memorial Sloan-Kettering Cancer Center was used to query the TCGA Provisional RNA seq databases (http://cancergenome.nih.gov/). Expression data for CD151 were retrieved, with reduced expression defined as a Z-score of < −1, and increased expression defined as a Z-score of > 1. Clinical outcome data (overall survival or disease-free survival) associated with altered CD151 expression were also retrieved. For lung squamous cell carcinoma cases, we identified a subset with predicted loss of squamous differentiation using a previously defined gene set containing SOX2, TP63, NOTCH1, NOTCH2, ASCL4 and FOXP1.Citation47 For invasive breast carcinoma, no relationship between CD151 expression and clinical outcomes was observed when all cases were considered together. Therefore, cases were divided based on the expression level of estrogen receptor (ESR1) and its co-regulator GATA3.Citation48 Cases with ESR1 or GATA3 mRNA downregulation (defined as a Z-score < −1) were retrieved using the Download tab of the cBioportal interface and analyzed separately from cases in which ESR1 and GATA3 were not downregulated. Additional characteristics of the ESR1low/GATA3low cases were retrieved from the reverse phase protein array (RPPA) data set and by analyzing the prevalence of TP53 mutations using cBioportal. To analyze CD151 expression in breast cancer subtypes, luminal A, luminal B, Her2+, and basal, we used the KM Plotter online tool,Citation49 which queries aggregated Affymetrix microarray data. The “auto select best cutoff” function was used to separate CD151-high and CD151-low cases.
2D proliferation assay
Cells were plated in serum-free DMEM on wells coated with 20 µg/ml rat tail collagen I or 1 µg/ml laminin-332 in duplicate 96 well plates, 6 wells per cell type. On subsequent days, plates were developed by refeeding with fresh medium containing WST reagent (Roche Life Sciences) and developed for 60 minutes before reading absorbance at 440 nm using a plate reader.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Supplemental Material
Download PDF (1.3 MB)Acknowledgments
We thank Moussa Aboisa for assistance with data quantification, Will Hacker for critical reading of the manuscript, the Flow Cytometry Core Facility at the Holden Comprehensive Cancer Center in the University of Iowa Carver College of Medicine for cell sorting, and the Central Microscopy Research Core Facility for sectioning and H&E staining.
Funding
This work was supported by NIH R01 grants CA13664 to CSS, and CA130916 to MDH. Core facilities used in these studies were supported by NIH grant P30 CA086862.
References
- Bjerke MA, Dzamba BJ, Wang C, DeSimone DW. FAK is required for tension-dependent organization of collective cell movements in Xenopus mesendoderm. Dev Biol 2014; 394:340-56; PMID:25127991; http://dx.doi.org/10.1016/j.ydbio.2014.07.023
- Cai D, Chen SC, Prasad M, He L, Wang X, Choesmel-Cadamuro V, Sawyer JK, Danuser G, Montell DJ. Mechanical feedback through E-cadherin promotes direction sensing during collective cell migration. Cell 2014; 157:1146-59; PMID:24855950; http://dx.doi.org/10.1016/j.cell.2014.03.045
- Aman A, Piotrowski T. Cell-cell signaling interactions coordinate multiple cell behaviors that drive morphogenesis of the lateral line. Cell Adh Migr 2011; 5:499-508; PMID:22274715; http://dx.doi.org/10.4161/cam.5.6.19113
- Arima S, Nishiyama K, Ko T, Arima Y, Hakozaki Y, Sugihara K, Koseki H, Uchijima Y, Kurihara Y, Kurihara H. Angiogenic morphogenesis driven by dynamic and heterogeneous collective endothelial cell movement. Development 2011; 138:4763-76; PMID:21965612; http://dx.doi.org/10.1242/dev.068023
- Theveneau E, Mayor R. Neural crest delamination and migration: from epithelium-to-mesenchyme transition to collective cell migration. Dev Biol 2012; 366:34-54; PMID:22261150; http://dx.doi.org/10.1016/j.ydbio.2011.12.041
- Einenkel J, Braumann U-D, Horn L-C, Kuska J-P, Höckel M. Three-D analysis of the invasion front in squamous cell carcinoma of the uterine cervix: histopathologic evidence for collective invasion per continuitatem. Anal Quant Cytol Histol 2007; 29:279-90; PMID:17987808
- Patsialou A, Bravo-Cordero JJ, Wang Y, Entenberg D, Liu H, Clarke M, Condeelis JS. Intravital multiphoton imaging reveals multicellular streaming as a crucial component of in vivo cell migration in human breast tumors. Intravital 2013; 2:e25294; PMID:25013744; http://dx.doi.org/10.4161/intv.25294
- Cheung KJ, Gabrielson E, Werb Z, Ewald AJ. Collective invasion in breast cancer requires a conserved basal epithelial program. Cell 2013; 155:1639-51; PMID:24332913; http://dx.doi.org/10.1016/j.cell.2013.11.029
- Nguyen-Ngoc KV, Cheung KJ, Brenot A, Shamir ER, Gray RS, Hines WC, Yaswen P, Werb Z, Ewald AJ. ECM microenvironment regulates collective migration and local dissemination in normal and malignant mammary epithelium. Proc Natl Acad Sci USA 2012; 109:E2595-604; PMID:22923691; http://dx.doi.org/10.1073/pnas.1212834109
- Friedl P, Locker J, Sahai E, Segall JE. Classifying collective cancer cell invasion. Nat Cell Biol 2012; 14:777-83; PMID:22854810; http://dx.doi.org/10.1038/ncb2548
- Cheung KJ, Ewald AJ. Illuminating breast cancer invasion: diverse roles for cell-cell interactions. Curr Opin Cell Biol 2014; 30C:99-111; ; http://dx.doi.org/10.1016/j.ceb.2014.07.003
- Macpherson IR, Hooper S, Serrels A, McGarry L, Ozanne BW, Harrington K, Frame MC, Sahai E, Brunton VG. p120-catenin is required for the collective invasion of squamous cell carcinoma cells via a phosphorylation-independent mechanism. Oncogene 2007; 26:5214-28; PMID:17334396; http://dx.doi.org/10.1038/sj.onc.1210334
- Canel M, Serrels A, Miller D, Timpson P, Serrels B, Frame MC, Brunton VG. Quantitative in vivo imaging of the effects of inhibiting integrin signaling via Src and FAK on cancer cell movement: effects on E-cadherin dynamics. Cancer Res 2010; 70:9413-22; PMID:21045155; http://dx.doi.org/10.1158/0008-5472.CAN-10-1454
- Ng MR, Besser A, Danuser G, Brugge JS. Substrate stiffness regulates cadherin-dependent collective migration through myosin-II contractility. J Cell Biol 2012; 199:545-63; PMID:23091067; http://dx.doi.org/10.1083/jcb.201207148
- Martinez-Rico C, Pincet F, Thiery JP, Dufour S. Integrins stimulate E-cadherin-mediated intercellular adhesion by regulating Src-kinase activation and actomyosin contractility. J Cell Sci 2010; 123:712-22; PMID:20144995; http://dx.doi.org/10.1242/jcs.047878
- Pirraglia C, Walters J, Ahn N, Myat MM. Rac1 GTPase acts downstream of αPS1βPS integrin to control collective migration and lumen size in the Drosophila salivary gland. Dev Biol 2013; 377:21-32; PMID:23500171; http://dx.doi.org/10.1016/j.ydbio.2013.02.020
- Canel M, Serrels A, Frame MC, Brunton VG. E-cadherin-integrin crosstalk in cancer invasion and metastasis. J Cell Sci 2013; 126:393-401; PMID:23525005; http://dx.doi.org/10.1242/jcs.100115
- Stipp CS. Laminin-binding integrins and their tetraspanin partners as potential antimetastatic targets. Expert Rev Mol Med 2010; 12:e3; PMID:20078909; http://dx.doi.org/10.1017/S1462399409001355
- Romanska HM, Berditchevski F. Tetraspanins in human epithelial malignancies. J Pathol 2011; 223:4-14; PMID:20938929; http://dx.doi.org/10.1002/path.2779
- Charrin S, Jouannet S, Boucheix C, Rubinstein E. Tetraspanins at a glance. J Cell Sci 2014; 127:3641-8; PMID:25128561; http://dx.doi.org/10.1242/jcs.154906
- Hemler ME. Tetraspanin proteins promote multiple cancer stages. Nat Rev Cancer 2014; 14:49-60; PMID:24505619; http://dx.doi.org/10.1038/nrc3640
- Wang HX, Li Q, Sharma C, Knoblich K, Hemler ME. Tetraspanin protein contributions to cancer. Biochem Soc Trans 2011; 39:547-52; PMID:21428937; http://dx.doi.org/10.1042/BST0390547
- Zöller M. Tetraspanins: push and pull in suppressing and promoting metastasis. Nat Rev Cancer 2009; 9:40-55; PMID:19078974; http://dx.doi.org/10.1038/nrc2543
- Sadej R, Grudowska A, Turczyk L, Kordek R, Romanska HM. CD151 in cancer progression and metastasis: a complex scenario. Lab Invest 2013; 94(1):41-51; PMID:24247563
- Roselli S, Kahl RGS, Copeland BT, Naylor MJ, Weidenhofer J, Muller WJ, Ashman LK. Deletion of Cd151 reduces mammary tumorigenesis in the MMTV/PyMT mouse model. BMC Cancer 2014; 14:509; PMID:25012362; http://dx.doi.org/10.1186/1471-2407-14-509
- Deng X, Li Q, Hoff J, Novak M, Yang H, Jin H, Erfani SF, Sharma C, Zhou P, Rabinovitz I, et al. Integrin-Associated CD151 Drives ErbB2-Evoked Mammary Tumor Onset and Metastasis. Neoplasia 2012; 14:678-89; PMID:22952421; http://dx.doi.org/10.1593/neo.12922
- Sadej R, Romanska H, Kavanagh D, Baldwin G, Takahashi T, Kalia N, Berditchevski F. Tetraspanin CD151 regulates transforming growth factor β signaling: implication in tumor metastasis. Cancer Res 2010; 70:6059-70; PMID:20570898; http://dx.doi.org/10.1158/0008-5472.CAN-09-3497
- Sadej R, Romanska H, Baldwin G, Gkirtzimanaki K, Novitskaya V, Filer AD, Krcova Z, Kusinska R, Ehrmann J, Buckley CD, et al. CD151 regulates tumorigenesis by modulating the communication between tumor cells and endothelium. Mol Cancer Res 2009; 7:787-98; PMID:19531562; http://dx.doi.org/10.1158/1541-7786.MCR-08-0574
- Yang XH, Richardson AL, Torres-Arzayus MI, Zhou P, Sharma C, Kazarov AR, Andzelm MM, Strominger JL, Brown M, Hemler ME. CD151 accelerates breast cancer by regulating α 6 integrin function, signaling, and molecular organization. Cancer Res 2008; 68:3204-13; PMID:18451146; http://dx.doi.org/10.1158/0008-5472.CAN-07-2949
- Li Q, Yang XH, Xu F, Sharma C, Wang HX, Knoblich K, Rabinovitz I, Granter SR, Hemler ME. Tetraspanin CD151 plays a key role in skin squamous cell carcinoma. Oncogene 2012; 32(14):1772-83
- Copeland BT, Bowman MJ, Ashman LK. Genetic ablation of the tetraspanin CD151 reduces spontaneous metastatic spread of prostate cancer in the TRAMP model. Mol Cancer Res 2013; 11:95-105; PMID:23131993; http://dx.doi.org/10.1158/1541-7786.MCR-12-0468
- Yue S, Mu W, Zöller M. Tspan8 and CD151 promote metastasis by distinct mechanisms. Eur J Cancer 2013; 49:2934-48; PMID:23683890; http://dx.doi.org/10.1016/j.ejca.2013.03.032
- Novitskaya V, Romanska H, Kordek R, Potemski P, Kusińska R, Parsons M, Odintsova E, Berditchevski F. Integrin α3β1-CD151 complex regulates dimerization of ErbB2 via RhoA. Oncogene 2013; 33(21):2779-89; PMID:23792450
- Palmer TD, Martinez CH, Vasquez C, Hebron K, Jones-Paris C, Arnold SA, Chan SM, Chalasani V, Gomez-Lemus JA, Williams AK, et al. Integrin-free tetraspanin CD151 can inhibit tumor cell motility upon clustering and is a clinical indicator of prostate cancer progression. Cancer Res 2013; 74(1):173-87
- Baldwin LA, Hoff JT, Lefringhouse J, Zhang M, Jia C, Liu Z, Erfani S, Jin H, Xu M, She QB, et al. CD151-α3β1 integrin complexes suppress ovarian tumor growth by repressing slug-mediated EMT and canonical wnt signaling. Oncotarget 2014; 5(23):12203-17; PMID:25356755
- Voss MA, Gordon N, Maloney S, Ganesan R, Ludeman L, McCarthy K, Gornall R, Schaller G, Wei W, Berditchevski F, et al. Tetraspanin CD151 is a novel prognostic marker in poor outcome endometrial cancer. Br J Cancer 2011; 104:1611-8; PMID:21505452; http://dx.doi.org/10.1038/bjc.2011.80
- Romanska HM, Potemski P, Collins SI, Williams H, Parmar S, Berditchevski F. Loss of CD151/Tspan24 from the complex with integrin α3β1 in invasive front of the tumour is a negative predictor of disease-free survival in oral squamous cell carcinoma. Oral Oncol 2013; 49:224-9; PMID:23099281; http://dx.doi.org/10.1016/j.oraloncology.2012.09.013
- Chien CW, Lin SC, Lai YY, Lin BW, Lin SC, Lee JC, Tsai SJ. Regulation of CD151 by hypoxia controls cell adhesion and metastasis in colorectal cancer. Clin Cancer Res 2008; 14:8043-51; PMID:19073968; http://dx.doi.org/10.1158/1078-0432.CCR-08-1651
- Sawada S, Yoshimoto M, Odintsova E, Hotchin NA, Berditchevski F. The tetraspanin CD151 functions as a negative regulator in the adhesion-dependent activation of Ras. J Biol Chem 2003; 278:26323-6; PMID:12782641; http://dx.doi.org/10.1074/jbc.C300210200
- Winterwood NE, Varzavand A, Meland MN, Ashman LK, Stipp CS. A critical role for tetraspanin CD151 in alpha3beta1 and alpha6beta4 integrin-dependent tumor cell functions on laminin-5. Mol Biol Cell 2006; 17:2707-21; PMID:16571677; http://dx.doi.org/10.1091/mbc.E05-11-1042
- Zevian S, Winterwood NE, Stipp CS. Structure-function analysis of tetraspanin CD151 reveals distinct requirements for tumor cell behaviors mediated by α3β1 versus α6β4 integrin. J Biol Chem 2011; 286:7496-506; PMID:21193415; http://dx.doi.org/10.1074/jbc.M110.173583
- Johnson JL, Winterwood N, DeMali KA, Stipp CS. Tetraspanin CD151 regulates RhoA activation and the dynamic stability of carcinoma cell-cell contacts. J Cell Sci 2009; 122:2263-73; PMID:19509057; http://dx.doi.org/10.1242/jcs.045997
- Marinkovich MP, Verrando P, Keene DR, Meneguzzi G, Lunstrum GP, Ortonne JP, Burgeson RE. Basement membrane proteins kalinin and nicein are structurally and immunologically identical. Lab Invest 1993; 69:295-9; PMID:8377472
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 2012; 9:671-5; PMID:22930834; http://dx.doi.org/10.1038/nmeth.2089
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012; 2:401-4; PMID:22588877; http://dx.doi.org/10.1158/2159-8290.CD-12-0095
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013; 6:pl1
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012; 489:519-25; PMID:22960745; http://dx.doi.org/10.1038/nature11404
- Eeckhoute J, Keeton EK, Lupien M, Krum SA, Carroll JS, Brown M. Positive cross-regulatory loop ties GATA-3 to estrogen receptor α expression in breast cancer. Cancer Res 2007; 67:6477-83; PMID:17616709; http://dx.doi.org/10.1158/0008-5472.CAN-07-0746
- Győrffy B, Lánczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat 2010; 123:725-31; PMID:20020197; http://dx.doi.org/10.1007/s10549-009-0674-9
- Shang M, Koshikawa N, Schenk S, Quaranta V. The LG3 module of laminin-5 harbors a binding site for integrin alpha3beta1 that promotes cell adhesion, spreading, and migration. J Biol Chem 2001; 276:33045-53; PMID:11395486; http://dx.doi.org/10.1074/jbc.M100798200
- Falk DL, Wessels D, Jenkins L, Pham T, Kuhl S, Titus MA, Soll DR. Shared, unique and redundant functions of three members of the class I myosins (MyoA, MyoB and MyoF) in motility and chemotaxis in Dictyostelium. J Cell Sci 2003; 116:3985-99; PMID:12953059; http://dx.doi.org/10.1242/jcs.00696
- Lee SH, Oh SY, Do SI, Lee HJ, Kang HJ, Rho YS, Bae WJ, Lim YC. SOX2 regulates self-renewal and tumorigenicity of stem-like cells of head and neck squamous cell carcinoma. Br J Cancer 2014; 111:2122-30; PMID:25321191; http://dx.doi.org/10.1038/bjc.2014.528
- Chen C, Wei Y, Hummel M, Hoffmann TK, Gross M, Kaufmann AM, Albers AE. Evidence for epithelial-mesenchymal transition in cancer stem cells of head and neck squamous cell carcinoma. PLoS One 2011; 6:e16466; PMID:21304586; http://dx.doi.org/10.1371/journal.pone.0016466
- Ribeiro AS, Paredes J. P-Cadherin Linking Breast Cancer Stem Cells and Invasion: A promising marker to Identify an “Intermediate/Metastable” EMT State. Front Oncol 2014; 4:371; PMID:25601904; http://dx.doi.org/10.3389/fonc.2014.00045
- Shigeta M, Sanzen N, Ozawa M, Gu J, Hasegawa H, Sekiguchi K. CD151 regulates epithelial cell-cell adhesion through PKC- and Cdc42-dependent actin cytoskeletal reorganization. J Cell Biol 2003; 163:165-76; PMID:14557253; http://dx.doi.org/10.1083/jcb.200301075
- Chometon G, Zhang ZG, Rubinstein E, Boucheix C, Mauch C, Aumailley M. Dissociation of the complex between CD151 and laminin-binding integrins permits migration of epithelial cells. Exp Cell Res 2006; 312:983-95; PMID:16490193; http://dx.doi.org/10.1016/j.yexcr.2005.12.034
- Zhang F, Michaelson JE, Moshiach S, Sachs N, Zhao W, Sun Y, Sonnenberg A, Lahti JM, Huang H, Zhang XA. Tetraspanin CD151 maintains vascular stability by balancing the forces of cell adhesion and cytoskeletal tension. Blood 2011; 118:4274-84; PMID:21832275; http://dx.doi.org/10.1182/blood-2011-03-339531
- Chattopadhyay N, Wang Z, Ashman LK, Brady-Kalnay SM, Kreidberg JA. alpha3beta1 integrin-CD151, a component of the cadherin-catenin complex, regulates PTPmu expression and cell-cell adhesion. J Cell Biol 2003; 163:1351-62; PMID:14691142; http://dx.doi.org/10.1083/jcb.200306067
- Kim KK, Wei Y, Szekeres C, Kugler MC, Wolters PJ, Hill ML, Frank JA, Brumwell AN, Wheeler SE, Kreidberg JA, et al. Epithelial cell alpha3beta1 integrin links β-catenin and Smad signaling to promote myofibroblast formation and pulmonary fibrosis. J Clin Invest 2009; 119:213-24; PMID:19104148
- Kim Y, Kugler MC, Wei Y, Kim KK, Li X, Brumwell AN, Chapman HA. Integrin alpha3beta1-dependent β-catenin phosphorylation links epithelial Smad signaling to cell contacts. J Cell Biol 2009; 184:309-22; PMID:19171760; http://dx.doi.org/10.1083/jcb.200806067
- Raymond K, Cagnet S, Kreft M, Janssen H, Sonnenberg A, Glukhova MA. Control of mammary myoepithelial cell contractile function by α3β1 integrin signalling. EMBO J 2011; 30:1896-906; PMID:21487391; http://dx.doi.org/10.1038/emboj.2011.113
- Kerrisk ME, Greer CA, Koleske AJ. Integrin α3 is required for late postnatal stability of dendrite arbors, dendritic spines and synapses, and mouse behavior. J Neurosci 2013; 33:6742-52; PMID:23595732; http://dx.doi.org/10.1523/JNEUROSCI.0528-13.2013
- Palecek SP, Loftus JC, Ginsberg MH, Lauffenburger DA, Horwitz AF. Integrin-ligand binding properties govern cell migration speed through cell-substratum adhesiveness. Nature 1997; 385:537-40; PMID:9020360; http://dx.doi.org/10.1038/385537a0
- Zaman MH, Trapani LM, Sieminski AL, Siemeski A, Mackellar D, Gong H, Kamm RD, Wells A, Lauffenburger DA, Matsudaira P. Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis. Proc Natl Acad Sci USA 2006; 103:10889-94; PMID:16832052; http://dx.doi.org/10.1073/pnas.0604460103
- Friedl P, Alexander S. Cancer invasion and the microenvironment: plasticity and reciprocity. Cell 2011; 147:992-1009; PMID:22118458; http://dx.doi.org/10.1016/j.cell.2011.11.016
- Yang XH, Flores LM, Li Q, Zhou P, Xu F, Krop IE, Hemler ME. Disruption of laminin-integrin-CD151-focal adhesion kinase axis sensitizes breast cancer cells to ErbB2 antagonists. Cancer Res 2010; 70:2256-63; PMID:20197472; http://dx.doi.org/10.1158/0008-5472.CAN-09-4032
- Kaplan-Lefko PJ, Chen TM, Ittmann MM, Barrios RJ, Ayala GE, Huss WJ, Maddison LA, Foster BA, Greenberg NM. Pathobiology of autochthonous prostate cancer in a pre-clinical transgenic mouse model. Prostate 2003; 55:219-37; PMID:12692788; http://dx.doi.org/10.1002/pros.10215
- Sarrió D, Rodriguez-Pinilla SM, Hardisson D, Cano A, Moreno-Bueno G, Palacios J. Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res 2008; 68:989-97; http://dx.doi.org/10.1158/0008-5472.CAN-07-2017
- Giordano A, Gao H, Anfossi S, Cohen E, Mego M, Lee BN, Tin S, De Laurentiis M, Parker CA, Alvarez RH, et al. Epithelial-mesenchymal transition and stem cell markers in patients with HER2-positive metastatic breast cancer. Mol Cancer Ther 2012; 11:2526-34; PMID:22973057; http://dx.doi.org/10.1158/1535-7163.MCT-12-0460
- Chung SS, Giehl N, Wu Y, Vadgama JV. STAT3 activation in HER2-overexpressing breast cancer promotes epithelial-mesenchymal transition and cancer stem cell traits. Int J Oncol 2014; 44:403-11; PMID:24297508
- Yan W, Cao QJ, Arenas RB, Bentley B, Shao R. GATA3 inhibits breast cancer metastasis through the reversal of epithelial-mesenchymal transition. J Biol Chem 2010; 285:14042-51; PMID:20189993; http://dx.doi.org/10.1074/jbc.M110.105262
- Sachs N, Claessen N, Aten J, Kreft M, Teske GJD, Koeman A, Zuurbier CJ, Janssen H, Sonnenberg A. Blood pressure influences end-stage renal disease of Cd151 knockout mice. J Clin Invest 2012; 122:348-58; PMID:22201679; http://dx.doi.org/10.1172/JCI58878
- Sachs N, Kreft M, van den Bergh Weerman MA, Beynon AJ, Peters TA, Weening JJ, Sonnenberg A. Kidney failure in mice lacking the tetraspanin CD151. J Cell Biol 2006; 175:33-9; PMID:17015618; http://dx.doi.org/10.1083/jcb.200603073
- Baleato RM, Guthrie PL, Gubler MC, Ashman LK, Roselli S. Deletion of CD151 results in a strain-dependent glomerular disease due to severe alterations of the glomerular basement membrane. Am J Pathol 2008; 173:927-37; PMID:18787104; http://dx.doi.org/10.2353/ajpath.2008.071149
- Tsujino K, Takeda Y, Arai T, Shintani Y, Inagaki R, Saiga H, Iwasaki T, Tetsumoto S, Jin Y, Ihara S, et al. Tetraspanin CD151 Protects against Pulmonary Fibrosis by maintaining epithelial integrity. Am J Respir Crit Care Med 2012; 186:170-80; PMID:22592804; http://dx.doi.org/10.1164/rccm.201201-0117OC
- Marusyk A, Almendro V, Polyak K. Intra-tumour heterogeneity: a looking glass for cancer? Nat Rev Cancer 2012; 12:323-34; PMID:22513401; http://dx.doi.org/10.1038/nrc3261