Abstract
microRNAs (miRNAs) are important regulators of tumor development and progression. In this study, we aimed to explore the expression and role of miR-622 in hepatocellular carcinoma (HCC). We found that miR-622 was significantly downregulated in human HCC specimens compared to adjacent noncancerous liver tissues. miR-622 downregulation was significantly associated with aggressive parameters and poor prognosis in HCC. Enforced expression of miR-622 significantly decreased the proliferation and colony formation and induced apoptosis of HCC cells. In vivo studies demonstrated that miR-622 overexpression retarded the growth of HCC xenograft tumors. Bioinformatic analysis and luciferase reporter assays revealed that miR-622 directly targeted the 3′-untranslated region (UTR) of mitogen-activated protein 4 kinase 4 (MAP4K4) mRNA. Ectopic expression of miR-622 led to a significant reduction of MAP4K4 expression in HCC cells and xenograft tumors. Overexpression of MAP4K4 partially restored cell proliferation and colony formation and reversed the induction of apoptosis in miR-622-overexpressing HCC cells. Inhibition of JNK and NF-κB signaling phenocopied the anticancer effects of miR-622 on HCC cells. Taken together, miR-622 acts as a tumor suppressor in HCC and restoration of miR-622 may provide therapeutic benefits in the treatment of HCC.
Introduction
Hepatocellular carcinoma (HCC) is the fifth most common malignancy and the second leading cause of cancer-related mortality worldwide.Citation1,2 Chronic hepatitis B virus (HBV) infection is one of the main causes of HCC, especially in Asian countries.Citation3 Despite advances in the treatment of HCC, the prognosis is still poor due to delayed diagnosis and high rate of recurrence.Citation4 The 5-year overall survival rate is only about 12%.Citation5 Therefore, identifying key regulators involved in the pathogenesis of HCC is of importance in developing effective therapeutic strategies against this disease.
microRNAs (miRNAs) are a class of endogenous, small non-coding single stranded RNAs with a length of ˜22 nucleotides.Citation6 They usually bind to the 3′-untranslated region (UTR) of target mRNAs via partial complementarity, leading to mRNA degradation or translational repression.Citation7 miRNAs play key roles in human cancers, acting as both tumor suppressors and oncogenes.Citation8 A large number of miRNAs have been identified to be involved in the development and progression of HCC.Citation9,10 Several lines of evidence indicate that miR-622 is frequently downregulated in human cancers such as gastric cancer,Citation11 colorectal cancer,Citation12 pancreatic cancerCitation13,14 and glioma Citation15. Guo et alCitation11 reported that miR-622 downregulation facilitates gastric cancer invasion and metastasis. Han et alCitation14 showed that miR-622 exhibits a growth inhibitory effect on transformed cells via targeting of K-Ras oncogene. In glioma, increased miR-622 expression has been found to interfere with cell invasion and migration.Citation15 These studies suggest that miR-622 functions as a suppressor gene in tumorigenesis. Despite these findings, relatively little is known about the role of miR-622 in HCC.
Therefore, in this study we investigated the expression and prognostic significance of miR-622 in HCC. The effects of restoration of miR-622 on HCC growth were determined in both in vitro and in vivo studies. Additionally, the potential target gene(s) of miR-622 was identified.
Results
Downregulation of miR-622 predicts poor prognosis in HCC patients
We first determined the miR-622 expression in 56 pairs of fresh HCC and adjacent noncancerous liver tissues. As illustrated in , the expression of miR-622 was significantly downregulated in HCC specimens. Next, we investigated the clinical relevance of miR-622 downregulation in HCC. Low miR-622 expression was significantly correlated with serum HBsAg positivity (P = 0.017), cirrhosis (P = 0.012), vascular invasion (P = 0.017), intrahepatic metastasis (P = 0.002), and tumor stage (P = 0.019; ). Kaplan-Meier analysis revealed that patients with low miR-622 tumors had a shorter median overall survival than those with high miR-622 tumors (32 vs. 50 months, P < 0.001; ). Multivariate survival analysis indicated that miR-622 expression was an independent predictor for overall survival of HCC patients (hazard ratio: 4.237; 95% confidence interval: 1.289–10.451; P = 0.004; ). Additionally, vascular invasion, intrahepatic metastasis, and tumor stage were also independently associated with overall survival of HCC patients ().
Figure 1. miR-622 expression in HCC tissues and its relevance to overall survival of HCC patients. (A) qRT-PCR showed that miR-622 was downregulated in HCC tissues compared with the adjacent noncancerous liver tissues. **, P < 0.01. (B) Kaplan–Meier curves for overall survival analysis by miR-622 expression in patients of HCC. P value was obtained by a log-rank test.
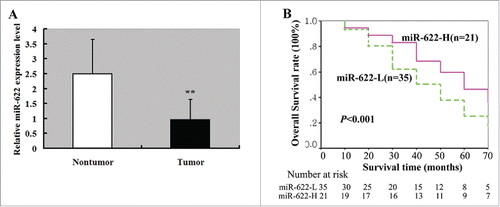
Table 1. Clinicopathologic factors and miR-622 expression in hepatocellular carcinomas (n = 56) based on qRT-PCR
Table 2. Univariate and multivariate analysis of factors associated with overall survival in patients with hepatocellular carcinoma (n = 56)
Restoration of miR-622 inhibits the growth and induces apoptosis in HCC cells
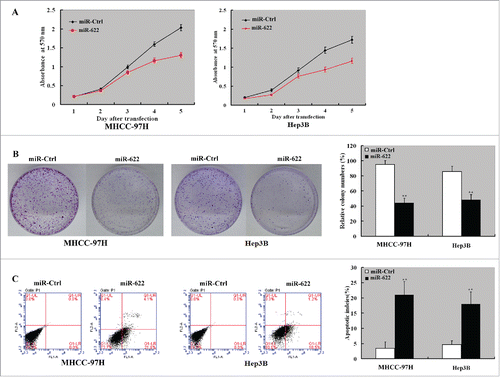
To explore the biological roles of miR-622 in HCC, we transfected miR-622 mimic into MHHC-97H and Hep3B cells and examined its impact on cell proliferation and apoptosis. The two HCC cell lines were chosen because they had relatively low abundance of miR-622 in a panel of HCC cell lines (i.e. MHHC-97H, MHHC-97L, SMMU-7721, HepG2, Hep3B, and Huh 7) (data not shown). CCK8 assay showed that enforced expression of miR-622 significantly inhibited the proliferation () and colony formation () of MHHC-97H and Hep3B cells. Apoptosis analysis revealed that the percentage of apoptotic cells was increased by 3-5 fold in miR-622-overexpressing HCC cells relative to control transfectants (P < 0.05; ).
Figure 2. miR-622 inhibits the proliferation and induces apoptosis in HCC cells. MHHC-97H and Hep3B cells were transfected with miR-622 mimic or control miRNA (NC mimic) and tested for cell proliferation and apoptosis. (A) Assessment of cell proliferation by the CCK8 assay. The proliferation of MHHC-97H and Hep3B cells was significantly inhibited by overexpression of miR-622. (B) The colony formation assay showed that overexpression of miR-622 reduced cell colony formation after 10-day incubation. Bar graph (right panel) represents quantification of colonies containing >50 cells. Data represent the means ± SD of 3 independent experiments. (C) Apoptosis detection by Annexin-V/PI staining. Left panel: Representative dot plots of flow cytometry analyses. Right panel: Quantification of apoptotic cells from 3 independent experiments. **, P < 0.01.
Overexpression of miR-622 retards HCC xenograft tumor growth
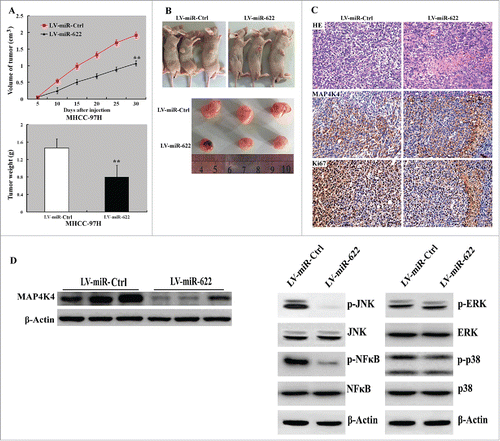
To assess the effect of miR-622 on HCC growth in vivo, we subcutaneously injected MHHC-97H cells stably expressing miR-622 or control miRNA into nude mice and tumor growth was measured. Notably, xenograft tumors formed from miR-622-overexpressing cells grew significantly slower than control xenograft tumors. At the end of the animal experiments, both the tumor volume and weight were significantly (P < 0.05) lower in the miR-622-overexpressing group than those in the control group (). Immunohistochemical analysis demonstrated that miR-622-overexpressing xenograft tumors had a marked increase in Ki-67-positive cells, compared to control tumors ().
Figure 3. Overexpression of miR-622 suppresses tumor growth in vivo. (A) (top) Tumor growth curves determined after injection of MHHC-97H cells infected with miR-622-expressing or control lentivirus into nude mice. (bottom) Tumor weight was measured at the end of the experiment. **, P < 0.01. (B) Photographs of tumor-bearing mice and resected tumors. (C) Representative tumor sections stained with H&E and anti-Ki-67 and anti-MAP4K4 antibodies. Original magnification,×400. (D) Western blot analysis of indicated proteins in miR-622-overexpressing and control xenograft tumors. Representative blots of 3 independent experiments are shown. The phosphorylation of JNK and NF-κB but not ERK1/2 or p38 was reduced by miR-622 overexpression.
miR-622 directly targets MAP4K4 in HCC
Bioinformatics analysis suggested one putative miR-622-binding site in the 3′-UTR of human MAP4K4 (). To verify the regulation of MAP4K4 expression by miR-622, we generated the luciferase reporter constructs containing a wild type or mutated MAP4K4 3′-UTR and transfected them together with miR-622 mimic or control miRNA into HCC cells. Compared to transfection of control miRNA, miR-622 significantly repressed luciferase activity of the wild-type MAP4K4 3′-UTR reporter but not that of the mutated-MAP4K4 3′-UTR reporter (). We further evaluated the effect of miR-622 overexpression on the expression of endogenous MAP4K4 in HCC cells. As shown in , ectopic expression of miR-622 significantly (P < 0.05) decreased MAP4K4 mRNA and protein levels in both the MHHC-97H and Hep3B cells. Consistent with the in vitro findings, miR-622-overexpressing HCC xenograft tumors displayed a decline in MAP4K4 protein levels, compared to control tumors (). Additionally, the phosphorylation of JNK and NF-κB but not ERK1/2 or p38 was reduced by miR-622 overexpression ().
Figure 4. miR-622 directly targets the 3′-UTR of MAP4K4 in HCC. (A) A putative binding site for miR-622 in the 3′-UTR of MAP4K4 predicted by TargetScan software. (B) Dual-luciferase assay showed that overexpression of miR-622 repressed luciferase activity of the wild-type MAP4K4 3′-UTR reporter but not that of the mutated-MAP4K4 3′-UTR reporter. (C and D) MHHC-97H and Hep3B cells transfected with miR-622 mimic and control miRNA were tested for MAP4K4 mRNA and protein levels by real-time PCR and Western blot analysis, respectively. **, P < 0.01. (E) Pearson correlation analysis revealed a significant inverse correlation between miR-622 and MAP4K4 expression levels in human HCC specimens.
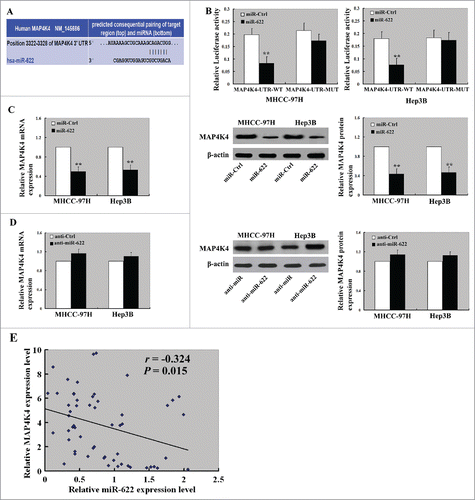
We also checked the potential associations between miR-622 and MAP4K4 expression in human HCC tissues. miR-622 and MAP4K4 expression were examined in surgically resected specimens from 56 HCC patients. Pearson correlation analysis showed that there was a significant inverse correlation between miR-622 and MAP4K4 expression levels (r = −0.324, P = 0.015; ).
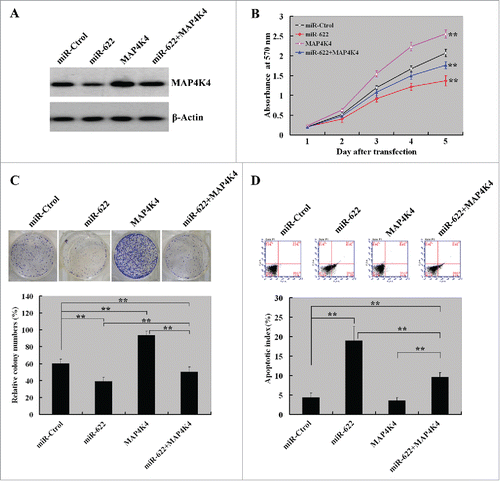
Overexpression of MAP4K4 partially reverses the growth-suppressive effects of miR-622 on HCC cells
Next, we explored whether ectopic expression of MAP4K4 reverses the growth-suppressive effects of miR-622 on HCC cells. Western blot analysis revealed that co-transfection of MAP4K4-expressing plasmid prevented the reduction of MAP4K4 expression in miR-622-expressing cells (). CCK8 assay () and colony formation assay () showed that enforced expression of MAP4K4 significantly restored, although not completely, cell proliferation and colony formation ability of miR-622-expressing HCC cells. Moreover, the pro-apoptotic activity of miR-622 was significantly (P < 0.01) compromised by co-expression of MAP4K4 ().
Figure 5. Overexpression of MAP4K4 partially reverses the growth-suppressive effects of miR-622 on HCC cells. (A) Western blot analysis of MAP4K4 protein levels in MHHC-97H cells transfected with indicated constructs at 72 h after transfection. Representative blots of 3 independent experiments are shown. (B) Cell proliferation detected by the CCK8 assay. The proliferation of MHHC-97H cells was significantly restored by co-expression of MAP4K4. (C) The colony formation assay after 10-day incubation showed that ectopic expression of MAP4K4 attenuated the suppression of colony formation by miR-622 overexpression. Bar graph (bottom panel) represents quantification of colonies containing >50 cells. Data represent the means ± SD of 3 independent experiments. (D) Apoptosis detection by Annexin-V/PI staining. Top panel: Representative dot plots of flow cytometry analyses. Bottom panel: Quantification of apoptotic cells from 3 independent experiments. **, P< 0.01.
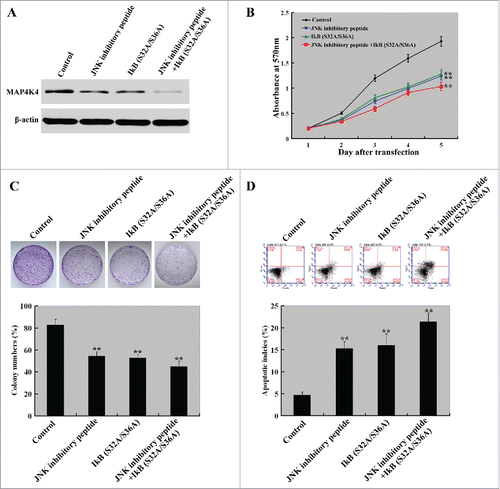
Inhibition of JNK and NF-κB signaling exerts anticancer effects against HCC cells
Finally, we sought to examine whether the anticancer effects of miR-622 are mediated through inhibition of both JNK and NF-κB signaling. To this end, we transfected a plasmid IκB (S32A/S36A) expressing a dominant negative mutant of IκB into MHHC-97H cells with or without pretreatment with a specific JNK peptide inhibitor (L-stereoisomer). Western blot analysis revealed that the reduction of MAP4K4 expression in MHHC-97H cells transfected with the plasmid IκB (S32A/S36A) or treated with L-stereoisomer was shown (). Cell proliferation assay revealed that in comparison with untreated control cells, the proliferation of cells transfected with the plasmid IκB (S32A/S36A) or treated with L-stereoisomer was significantly (P < 0.01) decreased after incubation for 2–5 d (). The combined delivery of IκB (S32A/S36A) and L-stereoisomer resulted in a greater inhibition of cell proliferation than either agent alone. Similarly, the percentage of colony formation was significantly (P < 0.01) lowered in cells with combined delivery of IκB (S32A/S36A) and L-stereoisomer (). Additionally, we observed a significantly (P < 0.01) increased apoptotic response in cells treated with IκB (S32A/S36A) and L-stereoisomer relative to untreated cells ().
Figure 6. Inhibition of JNK and NF-κB signaling exerts anti-growth effects on HCC cells. MHHC-97H cells were transfected with the IκB (S32A/S36A) plasmid with or without pretreatment with the JNK peptide inhibitor L-stereoisomer (10 µM) and tested for cell proliferation, colony formation, and apoptosis. (A) Western blot analysis of MAP4K4 protein levels in MHHC-97H cells transfected with the plasmid IκB (S32A/S36A) or treated with L-stereoisomer. Representative blots of 3 independent experiments are shown. (B) Cell proliferation detected by the CCK8 assay. The proliferation of MHHC-97H cells was significantly inhibited by the delivery of IκB (S32A/S36A) and L-stereoisomer, either alone or in combination. (C) Colony formation was examined after 10-day incubation. Bar graph (bottom panel) represents quantification of colonies containing >50 cells. Data represent the means ± SD of 3 independent experiments. (D) Apoptosis detection by Annexin-V/PI staining. Top panel: Representative dot plots of flow cytometry analyses. Bottom panel: Quantification of apoptotic cells from 3 independent experiments. **, P< 0.01.
Discussion
Deregulation of miRNAs is causally linked to tumor development. For instance, miR-377 is downregulated in HCC and has the ability to suppress HCC cell proliferation and invasion.Citation16 miR-622 has also been found to be downregulated in gastric cancerCitation11 and transformed human bronchial epithelial cells,Citation14 suggesting its suppressive role in tumorigenesis. Consistently, our study showed that miR-622 expression was reduced in HCC tissues compared to adjacent noncancerous liver tissues. Moreover, downregulation of miR-622 was significantly correlated with serum HBsAg positivity, cirrhosis, vascular invasion, intrahepatic metastasis, and tumor stage in HCC patients. Survival analysis further indicated that miR-622 had a positive prognostic impact on overall survival of HCC patients. These observations suggest that miR-622 may act as a tumor suppressor in HCC development and progression.
To explore the biological relevance of miR-622 downregulation in HCC, we transfected miR-622 mimic into HCC cells and examined cell proliferation and apoptosis. Our data showed that overexpression of miR-622 significantly suppressed HCC cell proliferation and colony formation and accelerated apoptotic death. Furthermore, in vivo study revealed that overexpression of miR-622 retarded HCC xenograft tumor growth in nude mice, compared to control xenograft tumors. These results argue that miR-622 functions as a tumor suppressor in HCC. Consistently, miR-622 also exerts growth suppression activity in lung cancer.Citation14 The growth-suppressive role of miR-622 offers an explanation for our clinical findings that miR-622 downregulation was significantly associated with aggressive parameters and poor prognosis in HCC.
Identification of downstream targets is of importance in deciphering the function of an miRNA. Bioinformatics analysis suggest MAP4K4 as a potential target of miR-622. MAP4K4 has been documented to be upregulated in many types of human cancers, such as HCC,Citation17 lung adenocarcinoma,Citation18 and pancreatic ductal adenocarcinoma.Citation19 Our previous work showed that MAP4K4 overexpression is correlated with poor prognosis in HCC.Citation17 Moreover, silencing of MAP4K4 using small interfering RNA technology was found to inhibit the growth of HCC cells and xenograft tumors. Likewise, Zhao et alCitation20 reported that knockdown of MAP4K4 significantly suppresses pancreatic cancer cell growth. These studies, combined with our present data, indicate that MAP4K4 knockdown and miR-622 overexpression exert similar growth suppression effects against tumor cells. To confirm the regulation of MAP4K4 by miR-622 in HCC cells, we performed MAP4K4 3′-UTR luciferase reporter assay. miR-622 was found to target MAP4K4 mRNA via directly binding to its 3′-UTR, as miR-622 failed to repress the luciferase activity of the reporter harboring mutated MAP4K4 3′-UTR. miR-622 overexpression significantly reduced the expression of endogenous MAP4K4 in HCC cells and xenograft tumors. Moreover, there was a significant inverse correlation between miR-622 and MAP4K4 expression levels in human HCC specimens. These data indicate that MAP4K4 is a bona fide target gene of miR-622 in HCC. To explore to what extent the suppressive activity of miR-622 is mediated through targeting MAP4K4, rescue experiments with overexpression of functional MAP4K4 was performed. We found that enforced expression of MAP4K4 partially restored cell proliferation and colony formation and reversed the induction of apoptosis in miR-622-overexpressing HCC cells. Taken together, miR-622 exerts its growth-suppressive effects against HCC cells largely through targeting of MAP4K4. However, a single miRNA has been found to simultaneously regulate a large number of target genes.Citation21 The partial rescue of growth suppression with MAP4K4 overexpression suggests that other relevant targets, which have not been identified yet, may be also involved in the action of miR-622 in HCC.
The tumor-promoting activity of MAP4K4 has been found to be associated with activation of JNK and NF-κB.Citation17,22 Here, we examined the effect of miR-622 overexpression on JNK and NF-κB phosphorylation in HCC xenograft tumors. Consistent with the effect of MAP4K4 silencing, overexpression of miR-622 impaired the phosphorylation of JNK and NF-κB, but not ERK1/2 or p38. JNK has been suggested to play a dual role in the development of HCC.Citation23 Pharmacological inhibition of JNK was reported to suppress the proliferation and invasionCitation24 and induce apoptosisCitation25 of HCC cells. Similarly, suppression of NF-κB activation was found to interfere with cell invasionCitation26 and trigger apoptotic deathCitation27 in HCC cells. These studies suggest a possibility that inactivation of JNK and NF-κB signaling may contribute to miR-622-mediated suppression of HCC growth. To test this possibility, we specifically inhibited the activation of JNK and NF-κB signaling and examined their effects on HCC cell growth and apoptosis. We found that specific inhibition of JNK and NF-κB signaling significantly inhibited cell proliferation and colony formation and increased apoptotic death, which mimicked the anticancer effect of overexpression of miR-622. These data suggest that miR-622-mediated growth suppression in HCC is associated with inhibition of JNK and NF-κB activity.
In conclusion, our results show that miR-622 is downregulated in HCC and has an independent prognostic impact on overall survival. Ectopic expression of miR-622 inhibits the growth of HCC cells and xenograft tumors, suggesting its tumor suppressor function. The anticancer effect of miR-622 is associated with direct targeting of MAP4K4 and inhibition of JNK and NF-κB signaling. Our data suggest that restoration of miR-622 expression may have therapeutic significance in HCC.
Materials and Methods
Cell lines and tissue specimens
Human HCC cell lines (HepG2, Huh7, HepG3B, SMMC-7721, MHHC-97H and MHHC-97L) were purchased from the Institute of Cellular Research, Chinese Academy of Science, Shanghai, China. Cells were cultured in RPMI1640 medium or Dulbecco's modified Eagle's medium with 10% fetal bovine serum (Invitrogen, Carlsbad, CA, USA), 50 U/mL penicillin, and 50 mg/mL streptomycin in a 5/ CO2 incubator at 37°C.
Fifty-six tumor specimens coupled with adjacent nontumor liver tissues were collected from patients with HCC, who underwent surgical resection for the disease at Changhai Hospital and Institute of Liver Diseases (Shanghai, China) between August 2007 and June 2009. One part of each sample was embedded in paraffin and the other was immediately snap-frozen and stored at −80°C until RNA extraction. The patients were selected on the basis of (a) distinctive pathologic diagnosis of HCC, (b) receiving curative resection, defined as macroscopically complete removal of the neoplasm, and (c) availability of detailed clinicopathologic data. Patients with preoperative anticancer treatment or with evidence of other malignancies were excluded from the study. The study protocol was approved by the Ethics Committee of Second Military Medical University (Shanghai, China), and written informed consent was obtained from each patient.
Of the 56 studied patients, 42 (75%) patients were positive for hepatitis B surface antigens (HBsAg). Liver cirrhosis was found in 38 (68%) cases. Eleven patients (20%) showed alcohol abuse and 2 (4%) had non-alcoholic steatohepatitis. All the subjects were negative for hepatitis C virus RNA. Large tumors (>2 cm) were found in 85.7% patients and intrahepatic metastasis in 67.9% patients. Tumors were staged according to the 2010 TNM Classification of Malignant Tumours.Citation28 A total of 12 (21%) patients were of stage I or II, and 44 (79%) of stage III or IV. Histologic grading of tumors was made according to the 2010 World Health Organization histologic classification of tumors of the liver and intrahepatic bile ducts (well, n = 13; moderate, n = 32; poor, n = 11). The median follow-up time was 34 months (range, 6–72 months). By the end of follow-up, 44 patients (79%) died.
Cell transfection and inhibitor treatment
miR-622 mimic and negative control miRNA were purchased from GenePharma Company (Shanghai, China). The sequences of the oligonucleotides were as follows: miR-622 mimic: 5′-ACAGUCUGCUGAGGUUGGAGC-3′ and 5′-UCCAACCUCAGCAGACUGUUU-3′; negative control miRNA: 5′-UUCUCCGAACGUGUCACGUTT-3′ and 5′-ACGUGACACGUUCGGAGAATT-3′. The MAP4K4 expression construct was obtained from JRDUN Biotechnology (Shanghai) Co., Ltd, China. A plasmid expressing the dominant negative mutant of IκB, IκB (S32A/S36A) that is unable to undergo phosphorylation and degradation was purchased from Upstate Biotechnology Inc. (Waltham, MA, USA). Transient transfections were carried out using Lipofectamine 2000, according to the manufacturer's instructions (Invitrogen). In brief, MHHC-97H cells were seeded and incubated overnight. Cells were transfected with indicated constructs and collected for gene expression analysis or for cell growth and apoptosis assays. In inhibitor experiments, we treated cells with the JNK peptide inhibitor L-stereoisomer (10 µM; Alexis Biochemicals, Carlsbad, CA, USA) 1 h before transfection with the IκB (S32A/S36A) plasmid.
RNA isolation and quantitative real-time RT-PCR
Total RNA, containing miRNA, was extracted from either tissue samples or transfected cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) a according to the manufacturer's instructions. The reverse transcription was conducted by using a reverse transcription kit (Invitrogen). The expression of mature miRNAs and potential target genes were measured by quantitative real-time PCR (qRT-PCR) with SYBR Green PCR Master Mix (Life Technologies Corporation, Foster City, CA, USA)on an ABI-7500 Sequence Detection System (Applied Biosystems, Foster City, CA, USA) . For evaluating miRNA expression, human U6 RNA was amplified as an internal control. For the detection of target gene mRNA, humanβ-actin transcripts was used as a control. Primer sequences were shown in Table S1. All samples were run in triplicate, and the relative mRNA or miRNA levels were calculated according to the 2−ΔΔCt method.Citation29
Western blot analysis
Total protein was lysed in lysis buffer (50 mmol/L Tris–HCl, pH 7.4; 150 mmol/L NaCl; 1% NP-40, and 0.5% sodium deoxycholate) with proteinase inhibitors (Roche Diagnostics, Mannheim, Germany). Equal amounts of protein (50 µg) was loaded, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and transferred onto polyvinylidene difluoride membranes. After blocking, the membranes were probed with primary antibody overnight at 4°C, followed by incubation with appropriate secondary antibody for 1 h. Details of the primary antibodies used are shown in Table S2. Blots were developed using an enhanced chemiluminescence kit from Santa Cruz Biotechnology. The intensities of immunoreactive bands were measured by computerized image analysis (QuantityOne-software, Bio-Rad, Hercules, CA, USA) and normalized to β-actin levels.
Immunohistochemistry
Immunohistochemistry was performed by DAKO Envision+ Reagent (DakoCytomation, Carpinteria, CA, USA) as previously described.Citation17 Briefly, paraffin sections (4-µm thick) were deparaffinized with xylene, rehydrated, and heated for 10 min in a steamer containing 10 mM of sodium citrate (pH 6.0) to retrieve antigen. Sections were incubated with primary antibodies against rabbit polyclonal anti-mitogen-activated protein 4 kinase 4 (MAP4K4) (HGK, sc-25738; 1:50) for 1 h, followed by the secondary reaction with DAKO Envision+ Reagent (DakoCytomation, Carpinteria, CA, USA). Negative controls were included by omitting the primary antibody, and a known positive control was included with each batch. The staining results were blindly evaluated by 2 experienced pathologists without known the information of the patients.
Cell proliferation and colony formation assay
Cell counting kit-8 assays (Dojindo, Japan) were used to measure cell viability according to the manufacturer's instructions. MHHC-97H and HepG3B cells (3,000 cells/well) were seeded in 96-well plates and transfected with miR-622 mimic, control miRNA or together with MAP4K4-expressing plasmid. After incubation for 1 to 5 days, each well was added 10 μl of CCK8 solution and the plates were continued to be incubated for another 4 h at 37°C. The absorbance was measured at 450 nm using a microplate reader (Bio-Rad). The CCK8 assay was repeated 3 times with 6 replicates.
The colony formation was performed as follows: At 24 h posttransfection with miR-622 mimic, control miRNA or together with MAP4K4-expressing plasmid, 1,000 cells were seeded into each well of a 6-well plate and then incubated for another 10 d. Culture media were replaced on day 5. At the end of the experiment, the wells were washed, fixed with 4% paraformaldehyde, and stained with 0.1% crystal violet. Colonies that consisted of >50 cells were scored. Each experiment was repeated 3 times in duplicate.
Apoptosis analysis
At 72 h posttransfection, cells were collected and stained with the Annexin V-FITC/Propidium Iodide Kit (LHK601–100, MBI) according to the manufacturer's instructions. Data were analyzed with Multicycle software (Phoenix Flow Systems, San Diego, CA, USA). All experiments were repeated 3 times.
miRNA target prediction
To identify putative targets for the indicated miRNAs, we used 4 miRNA databases including miRBase (http://www.mirbase.org/), TargetScan (y), PicTar (http://pictar.bio.nyu.edu/) and miRDB (http://mirdb.org/miRDB/), and predicted targets were converged and overlapped for further analysis.
Dual luciferase assay
To check whether MAP4K4 was a target of miR-622, the pMIR-REPORT assay was applied. Briefly, MHHC-97H were seeded in 96-well plates (5× 103 cells per well) and cotransfected with 100 ng pMIR-REPORT Luciferase vector (Shanghai JRDUN Biotechnology Co., Ltd) containing MAP4K4 3′UTR (3,441 bp, termed wt-MAP4K4) or mutated forms (termed mt-MAP4K4) with or without 50 nmol/L miR-622 mimic or control miRNA. After incubation for 48 h, luciferase activity was determined using the Dual-Luciferase Reporter Assay system according to the manufacturer's instructions (Promega, Madison, WI, USA). All experiments were performed in triplicate and relative luciferase activity was normalized to that of Firefly luciferase.
Tumorigenicity in nude mice
Tumor formation was studied by establishing a mouse xenograft model as described previously.Citation17,30 Lentiviral vectors (Shanghai JRDUN Biotechnology Co., Ltd) expressing miR-622 (LV-miR-622) or control miRNA were used to infect MHHC-97H cells according to the manufacturer's instructions. BALB/c female nude mice (4 weeks old) were purchased from Experimental Animal Center of Shanghai, Shanghai, China. Mice were randomly divided into 2 groups with 3 mice in each group. Infected MHHC-97H cells (5×106 cells/mouse) suspended in PBS solution were injected subcutaneously into the mice. Tumor volumes were measured every 5 d using calipers along 2 major axes, and calculated according to the formula V = 0.5 × L (length) × W2 (width). At 30 d after cell inoculation, mice were sacrificed. All excised tumors and lungs of the mice were evaluated for volume and weight, and fixed in 10/ formalin before paraffin embedding, then were sectioned and stained in hematoxylin and eosin (H&E). The expression of MAP4K4 in tumors was detected by Western blot and immunohistochemical staining. All experimental manipulations were undertaken in accordance with the guidelines for Institutional and Animal Care and Use Committees, with the approval of the Scientific Investigation Board of the Shanghai Jiaotong University, Shanghai, China.
Statistical analysis
Unless otherwise stated, data are expressed as mean ± standard deviation (SD) and were analyzed using the SPSS 13.0 software (SPSS, Chicago, IL, USA). Differences between groups were assessed using the Student t test and Fisher exact test. The relationship between the expression of MAP4K4 and miR-622 in tissues or cell lines was analyzed with Pearson correlation. The relationship between miR-622 expression and clinicopathologic features of pancreatic cancer was analyzed using the Pearson χ2 test. Kaplan–Meier method was used to calculate overall survival of 2 patient groups, and differences were analyzed by log-rank test. The survival data were evaluated using a multivariate Cox regression analysis. P < 0.05 was considered statistically significant.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Supplemental_Material.doc
Download MS Word (37 KB)Funding
This work was supported in part by grants from National Nature Science Foundation of China (No. 81072020 and 81172311 to S.H. Zhang, and No. 30973458 to A.M. Xu) and from '085' and “Integrated Traditional Chinese and Western Medicine” first-class discipline construction of science and technology innovation in Shanghai University of Traditional Chinese Medicine (No. 085ZY1220 to S.H. Zhang).
References
- Boyle P, Levin B. World cancer report 2008. WHO Press, Avenue,Geneva, Switzerland 2009:pp350
- Venook AP, Papandreou C, Furuse J, de Guevara LL. The incidence and epidemiology of hepatocellular carcinoma: a global and regional perspective. Oncologist 2010; 15 Suppl 4:5-13; PMID:21115576; http://dx.doi.org/10.1634/theoncologist.2010-S4-05
- Nagaoki Y, Hyogo H, Aikata H, Tanaka M, Naeshiro N, Nakahara T, Honda Y, Miyaki D, Kawaoka T, Takaki S, et al. Recent trend of clinical features in patients with hepatocellular carcinoma. Hepatol Res 2012; 42:368-75; PMID:22151896; http://dx.doi.org/10.1111/j.1872-034X.2011.00929.x
- Tinkle CL, Haas-Kogan D. Hepatocellular carcinoma: natural history, current management, and emerging tools. Biologics 2012; 6:207-19; PMID:22904613
- El-Serag HB. Hepatocellular carcinoma. N Engl J Med 2011; 365:1118-27; PMID:21992124; http://dx.doi.org/10.1056/NEJMra1001683
- Hermeking H. MicroRNAs in the p53 network: micromanagement of tumour suppression. Nat Rev Cancer 2012; 12:613-26; PMID:22898542; http://dx.doi.org/10.1038/nrc3318
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116:281-97; PMID:14744438; http://dx.doi.org/10.1016/S0092-8674(04)00045-5
- Taylor MA, Schiemann WP. Therapeutic Opportunities for Targeting microRNAs in Cancer. Mol Cell Ther 2014; 2:1-13; PMID:25717380; http://dx.doi.org/10.1186/2052-8426-2-30
- Zhang Z, Zhang Y, Sun XX, Ma X, Chen ZN. microRNA-146a inhibits cancer metastasis by downregulating VEGF through dual pathways in hepatocellular carcinoma. Mol Cancer 2015; 14:5; PMID:25608619; http://dx.doi.org/10.1186/1476-4598-14-5
- Ni F, Zhao H, Cui H, Wu Z, Chen L, Hu Z, Guo C, Liu Y, Chen Z, Wang X, Chen D, Wei H, Wang S. MicroRNA-362-5p promotes tumor growth and metastasis by targeting CYLD in hepatocellular carcinoma. Cancer Lett 2015; 356:809-18; PMID:25449782; http://dx.doi.org/10.1016/j.canlet.2014.10.041
- Guo XB, Jing CQ, Li LP, Zhang L, Shi YL, Wang JS, Liu JL, Li CS. Down-regulation of miR-622 in gastric cancer promotes cellular invasion and tumor metastasis by targeting ING1 gene. World J Gastroenterol 2011; 17:1895-902; PMID:21528065
- Balaguer F, Moreira L, Lozano JJ, Link A, Ramirez G, Shen Y, Cuatrecasas M, Arnold M, Meltzer SJ, Syngal S, et al. Colorectal cancers with microsatellite instability display unique miRNA profiles. Clin Cancer Res 2011; 17:6239-49; PMID:21844009; http://dx.doi.org/10.1158/1078-0432.CCR-11-1424
- Schultz NA, Werner J, Willenbrock H, Roslind A, Giese N, Horn T, Wojdemann M, Johansen JS. MicroRNA expression profiles associated with pancreatic adenocarcinoma and ampullary adenocarcinoma. Mod Pathol 2012; 25:1609-22; PMID:22878649; http://dx.doi.org/10.1038/modpathol.2012.122
- Han Z, Yang Q, Liu B, Wu J, Li Y, Yang C, Jiang Y. MicroRNA-622 functions as a tumor suppressor by targeting K-Ras and enhancing the anticarcinogenic effect of resveratrol. Carcinogenesis 2012; 33:131-9; PMID:22016468; http://dx.doi.org/10.1093/carcin/bgr226
- Zhang R, Luo H, Wang S, Chen Z, Hua L, Wang HW, Chen W, Yuan Y, Zhou X, Li D, et al. miR-622 suppresses proliferation, invasion and migration by directly targeting activating transcription factor 2 in glioma cells. J Neurooncol 2015; 121:63-72; PMID:25258251; http://dx.doi.org/10.1007/s11060-014-1607-y
- Chen G, Lu L, Liu C, Shan L, Yuan D. MicroRNA-377 Suppresses Cell Proliferation and Invasion by Inhibiting TIAM1 Expression in Hepatocellular Carcinoma. PLoS One 2015; 10:e0117714; PMID:25739101; http://dx.doi.org/10.1371/journal.pone.0117714
- Liu AW, Cai J, Zhao XL, Jiang TH, He TF, Fu HQ, Zhu MH, Zhang SH. ShRNA-targeted MAP4K4 inhibits hepatocellular carcinoma growth. Clin Cancer Res 2011; 17:710-20; PMID:21196414; http://dx.doi.org/10.1158/1078-0432.CCR-10-0331
- Qiu MH, Qian YM, Zhao XL, Wang SM, Feng XJ, Chen XF, Zhang SH. Expression and prognostic significance of MAP4K4 in lung adenocarcinoma. Pathol Res Pract 2012; 208:541-8; PMID:22824148; http://dx.doi.org/10.1016/j.prp.2012.06.001
- Liang JJ, Wang H, Rashid A, Tan TH, Hwang RF, Hamilton SR, Abbruzzese JL, Evans DB, Wang H. Expression of MAP4K4 is associated with worse prognosis in patients with stage II pancreatic ductal adenocarcinoma. Clin Cancer Res 2008; 14:7043-9; PMID:18981001; http://dx.doi.org/10.1158/1078-0432.CCR-08-0381
- Zhao G, Wang B, Liu Y, Zhang JG, Deng SC, Qin Q, Tian K, Li X, Zhu S, Niu Y, et al. miRNA-141, downregulated in pancreatic cancer, inhibits cell proliferation and invasion by directly targeting MAP4K4. Mol Cancer Ther 2013; 12:2569-80; PMID:24013097; http://dx.doi.org/10.1158/1535-7163.MCT-13-0296
- Gaken J, Mohamedali AM, Jiang J, Malik F, Stangl D, Smith AE, Chronis C, Kulasekararaj AG, Thomas NS, Farzaneh F, et al. A functional assay for microRNA target identification and validation. Nucleic Acids Res 2012; 40:e75; PMID:22323518; http://dx.doi.org/10.1093/nar/gks145
- Collins CS, Hong J, Sapinoso L, Zhou Y, Liu Z, Micklash K, Schultz PG, Hampton GM. A small interfering RNA screen for modulators of tumor cell motility identifies MAP4K4 as a promigratory kinase. Proc Natl Acad Sci U S A 2006; 103:3775-80; PMID:16537454; http://dx.doi.org/10.1073/pnas.0600040103
- Das M, Garlick DS, Greiner DL, Davis RJ. The role of JNK in the development of hepatocellular carcinoma. Genes Dev 2011; 25:634-45; PMID:21406557; http://dx.doi.org/10.1101/gad.1989311
- Lee EK, Kim HJ, Lee KJ, Lee HJ, Lee JS, Kim DG, Hong SW, Yoon Y, Kim JS. Inhibition of the proliferation and invasion of hepatocellular carcinoma cells by lipocalin 2 through blockade of JNK and PI3K/Akt signaling. Int J Oncol 2011; 38:325-33; PMID:21132267
- Lai JP, Sandhu DS, Yu C, Moser CD, Hu C, Shire AM, Aderca I, Murphy LM, Adjei AA, Sanderson S, et al. Sulfatase 2 protects hepatocellular carcinoma cells against apoptosis induced by the PI3K inhibitor LY294002 and ERK and JNK kinase inhibitors. Liver Int 2010; 30:1522-8; PMID:21040406; http://dx.doi.org/10.1111/j.1478-3231.2010.02336.x
- Song R, Song H, Liang Y, Yin D, Zhang H, Zheng T, Wang J, Lu Z, Song X, Pei T, et al. Reciprocal activation between ATPase inhibitory factor 1 and NF-kappaB drives hepatocellular carcinoma angiogenesis and metastasis. Hepatology 2014; 60:1659-73; PMID:25042864; http://dx.doi.org/10.1002/hep.27312
- Wu LF, Li GP, Su JD, Pu ZJ, Feng JL, Ye YQ, Wei BL. Involvement of NF-kappaB activation in the apoptosis induced by extracellular adenosine in human hepatocellular carcinoma HepG2 cells. Biochem Cell Biol 2010; 88:705-14; PMID:20651843; http://dx.doi.org/10.1139/O10-008
- Sobin L, Gospodarowicz M, Wittekind C. TNM Classification of Malignant Tumours. International Union against Cancer: Wiley-Blackwell, 2010
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001; 25:402-8; PMID:11846609; http://dx.doi.org/10.1006/meth.2001.1262
- Wang Y, Shek FH, Wong KF, Liu LX, Zhang XQ, Yuan Y, Khin E, Hu MY, Wang JH, Poon RT, et al. Anti-cadherin-17 antibody modulates β-catenin signaling and tumorigenicity of hepatocellular carcinoma. PLoS One 2013; 8:e72386; PMID:24039755; http://dx.doi.org/10.1371/journal.pone.0072386
