ABSTRACT
Germline CDKN2A mutations have been described in 25% to 40% of melanoma families from several countries. Sicilian population is genetically different from the people of Europe and Northern Italy because of its historical background, therefore familial melanoma could be due to genes different from high-penetrance CDKN2A gene. Four hundred patients with cutaneous melanoma were observed in a 6-years period at the Plastic Surgery Unit of the University of Palermo. Forty-eight patients have met the criteria of the Italian Society of Human Genetics (SIGU) for the diagnosis of familial melanoma and were screened for CDKN2A and CDK4 mutations. Mutation testing revealed that none of the families carried mutations in CDK4 and only one patient harboured the rare CDKN2A p.R87W mutation. Unlike other studies, we have not found high mutation rate of CDKN2A in patients affected by familial melanoma or multiple melanoma. This difference could be attributed to different factors, including the genetic heterogeneity of the Sicilian population. It is likely that, as in the Australian people, the inheritance of familial melanoma in this island of the Mediterranean Sea is due to intermediate/low-penetrance susceptibility genes, which, together with environmental factors (as latitude and sun exposure), could determine the occurrence of melanoma.
Introduction
Melanoma is a high-grade, poorly differentiated malignant tumor of melanin pigment-producing cells (melanocytes) with poor prognosis in the metastatic stage, accounting for more than 70% of the skin cancer related deaths.Citation1 A familial history of melanoma is a strong predictor of melanoma development. Up to 10% of all cases of cutaneous malignant melanoma occurs in a familial setting. Familial genetic linkage studies have led to the identification of 2 high-penetrance susceptibility genes, CDKN2A and CDK4, involved in senescence and cell cycle arrest. Rare mutations or deletions in these genes confer an elevated risk of developing melanoma.Citation2,3
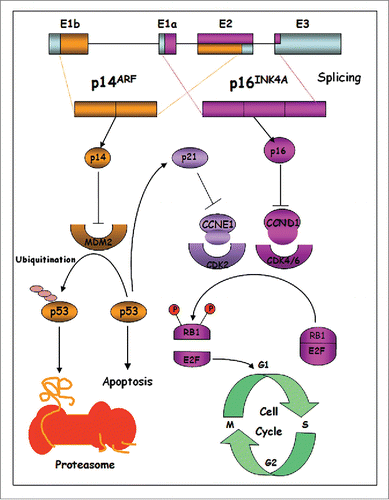
CDKN2A (cyclin-dependent kinase inhibitor 2A) is known to be a major high-risk melanoma susceptibility gene that it is transmitted according to a dominant mode if inherited in melanoma-prone families. The CDKN2A gene is located on chromosome 9p21 and includes 4 exons (1α, 1β, 2 and 3) encoding for 2 tumor-suppressor proteins, INK4A (p16INK4a) and ARF (p14ARF in humans and p19ARF in mice), which are translated from alternative spliced transcripts.Citation4,5 Most germline mutations that confer melanoma risk occur in exons 1α and 2, indicating that p16INK4a is the preferentially targeted and functionally dominant component of CDKN2A.Citation6,7 Rare deletions at the exon 1beta have also been reported, suggesting that p14ARF is a melanoma susceptibility gene independent from p16INK4a.Citation8-10 Even rarer deep intronic mutations of CDKN2A have also been described, although these account for very few cases worldwide.Citation11-14
INK4A is a cyclin-dependent kinase inhibitor that activates the pRB protein via negative regulation of Cdk4/6, promoting the progression through the G1/S transition, whereas p14ARF interacts with the MDM2 protein, whose principal function is to promote the ubiquitin-mediated degradation of the tumor-suppressor protein p53.Citation15,16 Thus, the loss of function of p16INK4a promotes CDK4 and CDK6 activation, causing hyperphosphorylation and inactivation of retinoblastoma protein (pRB), and activation of E2F1 (). Indeed, several genetic and epigenetic analyses showed that INK4A is lost in 50% of melanoma cases, inactivated by methylation of its promoter in about 10% of tumors or inactivated by point mutations in approximately 9% of melanoma cases.Citation17 The frequent and specific inactivation of p14ARF in melanoma supports its considerable role in melanomagenesis.Citation18-20 Additionally, BRAF-activating mutations and functional loss of p16INK4a and p14ARF were found in the majority of melanomas.Citation21 Binni et al. Citation22 have reported that overall frequency of p14ARF mutations in Italian familial melanoma is 3.2%.
Germline mutations in high-susceptibility CDK4 gene are rare, as reported in literature, and inhibit the regulation of the protein by p16INK4a, but maintain the interaction between CDK4 and cyclin D1, leading to constitutive activation of the complex and aberrant proliferation, through pRB inactivation and activation of the transcription factor E2F. E2F activates the transcription of S-phase genes, thereby promoting cell proliferation.Citation23 Moreover, Rane et al. reported that CDK4 mutations promote tumorigenesis and determine escape from the cellular senescence mechanism of melanocytes transplanted into nude mice.Citation24
During the last years, genome-wide association studies (GWAS) have allowed to identify common single nucleotide polymorphisms (SNPs) associated with a low risk of developing cancers, including melanoma. Therefore, the identification of genetic variants with low/intermediate allele frequency conferring a moderate risk of cancer represents an important scientific approach to discover novel melanoma-predisposing genes.Citation25,26 Among these, 2 low/intermediate-penetrance susceptibility genes, MITF (microphthalmia-associated transcription factor) and MC1R (melanocortin 1 receptor), play an important role in the development of melanoma. Genomic amplification of MITF was detected in 10% of primary tumors and 20% of metastatic melanomas and correlates with decreased 5-year overall patient survival.Citation27,28 MITF belongs to the MYC supergene family of basic helix-loop-helix transcription factors and is involved in proliferation control and survival.Citation29 A model, in which an increase in MITF expression is associated with the differentiation,Citation30 while moderate MITF levels are linked with proliferation,Citation31 and transient low MITF levels with a melanoma-initiating cell phenotype has been suggested.Citation32 MITF activity is regulated by posttranslational modifications such as phosphorylation and degradation via the ubiquitin-proteasome pathway in response to activation of the ERK pathway.Citation33 In patients with a strong family history a germline missense substitution p.E318K in MITF, conferring a fold5- increased melanoma risk, was identified.Citation34,35 This mutation confers invasive and migrative properties to melanoma cells, providing tumor progression abilities. MITF can control the expression of several genes involved in cell survival (HIF-1α, BCL-2, MET, APE-1),Citation36-38 cytoskeleton remodeling and migration,Citation39 and cell proliferation (CDK2).Citation40 Moreover, MITF activity is correlated with the resistance to UV-induced apoptosis in melanocytes.
MC1R is a transmembrane G protein-coupled receptor expressed on the cell surface of epidermal melanocytes, which, upon hormonal stimulation, actives the adenylate cyclase and the cAMP/PKA/CREB pathway.Citation41 Allelic variants of MC1R are an important risk factor for melanoma, and the melanoma penetrance increases in presence of a CDKN2A mutation.Citation42 Other low-risk allelic variants are responsible for differences in pigmentation of skin, hair, and eyes, and determine variations in skin sensitivity to UV, by increasing the melanoma risk.
Sun exposure is widely considered as the critical environmental risk factor for cutaneous malignant melanoma, which originates as a consequence of deleterious interactions between ultraviolet (UV) radiations and the melanocyte genome. In fact, UV radiations may contribute to melanoma development through combined genotoxic and mitogenic effects in melanocytes.Citation43 The combination of inherited intermediate/low-penetrance allelic variants with environmental factors such as sun exposure might induce the onset of melanoma.Citation44
In this work we performed a screening of germline CDKN2A and CDK4 mutations in 48 Sicilian patients with familial malignant melanoma in order to assess the frequency and influence of these high-risk susceptibility genes in individuals belonging to this specific geographical area.
Results
Clinicopathological features of the familial melanomas in sicilian population
Forty-eight Sicilian patients who have met the selection criteria for familial melanoma genetic testing have been studied, by defining the clinical and pathological features of their melanomas. Of these patients, twenty-four were men and twenty-four were women, with a mean age at the onset of 35,8 y (range 15-60). The site of primary melanoma was the trunk in 56% of patients (27), limbs in 40% (19) and head and neck in 4% (2). In the patients enrolled in the study the most common form of melanoma was superficial spreading melanoma (52%), followed by nodular melanoma (27%), and in situ melanoma (21%). No cases of lentigo maligna melanoma (LMM) and acral lentiginous melanoma (ALM) were observed among patients who met the selection criteria for genetic counseling for familial melanoma. In only 3 cases 2 primary melanomas were diagnosed at the same time (synchronous primary melanomas). In 7 cases the second melanoma appeared in a different anatomical area than the area of primary lesion, within 5 y from diagnosis of the first melanoma. The tumor thickness measurement according to Breslow thickness highlighted that about 60% of patients presented a thin melanoma at the diagnosis with a better prognosis than other patients with a thickest melanoma. The AJCC Melanoma staging showed that almost all patients (44) had a localized melanoma at the diagnosis (stages 0-II), while only 4 patients had an advanced-stage melanoma (stages III and IV). However, the fact that most of patients showed an early-stage melanoma at diagnosis is likely due to a greater attention of individuals with family history in controlling (by means of periodic cutaneous screening) and removing suspected skin lesions.
Furthermore, in 70% of patients, in addition to melanoma, one or more dysplastic nevi have been removed ().
Table 1. Clinical and pathological features of familial melanoma patients
Screening of germline CDKN2A and CDK4 mutations
A total of 48 familial malignant cutaneous melanoma cases from Sicilian patients was screened for germline mutations in CDKN2A exons 1α, 1β, 2 and 3 and CDK4 exon 2 in order to evaluate the frequency of these high susceptibility genes in individuals belonging to this specific geographical area. Mutation analysis revealed that none of the examined families carried mutations in exon 2 of CDK4 and only one patient harboured a rare missense mutation in exon 2 of CDKN2A.Citation45 The patient was a carrier of the p.R87W mutation (c.259C > T), which causes the substitution of an arginine for a tryptophan at position 87, instead of the most common p.G101W mutation (c.301 G > T), which causes the substitution of a glycine for a tryptophan at position 101.Citation46 At the age of 49 y (first diagnosis), the mutation carrier patient had a superficial spreading melanoma (SSM) in the trunk with a Breslow thickness of 1.9 mm. The proband then developed a second metachronous melanoma at the age of 51 y and a third melanoma at the age of 54 y Both melanomas were in situ melanomas of the back and eyelid, respectively.
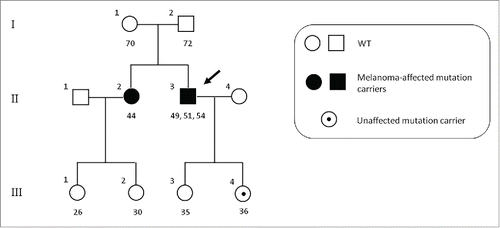
Since mutational screening of relatives of individuals harbouring germline mutations in susceptibility genes could even improve prevention and surveillance strategies of melanoma through early identification of the pigmented skin lesions at risk of malignant transformation, we performed a survey about the relatives of the CDKN2A germline mutation carrier patient. After a careful study of all the relatives of the proband, the same gene mutation was detected in his sister and his daughter, unlike other families who showed no germline mutations. This mutation has not been previously described in the literature, except in a few cases of melanoma.Citation47 Furthermore, proband's sister developed an early-stage melanoma (in situ), while his daughter was an unaffected mutation carrier ().
Figure 2. Pedigree of the CDKN2A p.R87W mutation-positive family. Index case is indicated by an arrow. Numbers under symbols show the age for the relatives and age at diagnosis for the affected individuals. For patients with multiple melanomas, different ages are separated by commas. WT, non-carriers of mutation.
Overall, our data indicate that a high mutation rate of CDKN2A was not found in Sicilian patients affected by melanoma who met criteria for the diagnosis of familial or multiple melanoma, in contrast to what reported in other studies.Citation48
Discussion
Melanoma is the most serious and aggressive form of skin cancer whose incidence has been continuously increasing in the last decades, and faster than any other cancers. For patients with cutaneous melanoma, the prognosis is related to the location and depth of the primary tumor, and the presence or absence of locoregional and distant metastatic disease.Citation49
Melanoma is the most dangerous form of skin cancer in the white population, being largely resistant to conventional therapies at advanced stages. The management of patients with advanced melanoma represents a significant challenge considering that, historically, chemotherapy and immunologic therapies have produced only modest results in the treatment of metastatic melanoma.Citation50,51 Patients with metastatic melanomas have a median survival rate that typically ranges from 6 to 10 months. Currently, prevention and early detection represent the only effective strategies to reduce the incidence of this tumor. Despite improvements in early melanoma diagnosis, the 5-year survival rate remains low in advanced disease.Citation52,53
Germline mutations in genes encoding for CDK4 and CDKN2A, involved in regulation of the cell cycle, have been shown to confer a high risk of malignant melanoma.Citation54 In addition, frequent germline allelic variants in the Casp8, MTAP, MATP, MC1R, MITF and ASIP genes have been identified as low-risk susceptibility genes or as modifiers of high-risk susceptibility genes.Citation55
Germline CDKN2A mutations have been described in 25-40% of melanoma-prone families from several countries, and most of them are missense mutations mainly affecting p16INK4a. Typically, in familial melanoma there are more family members affected by melanoma with early-onset (particularly with a vertical pattern of inheritance). High number of cases of melanoma in the same family, early age of onset and presence of multiple primary melanoma (sincronous or metacronous) showed significant association with CDKN2A mutations.Citation56 Approximately from 3% to 5% of all patients with melanoma will develop additional primary melanomas in their lifetime. The prevalence of CDKN2A mutations increases with the number of primary melanoma diagnoses in the individuals. For familial cases with CDKN2A mutation carriers the overall penetrance has been estimated to be 30% by age 50, and 67% by age 80, although this risk is higher in subjects who live in sunnier climes. Melanoma risk varies by geographic area as showed by studies of families with CDKN2A mutations from North America, Europe and Australia. The reasons for these variations are not well understood, however, there may be differences in the sun exposure, other genetic or individual alterations, or a combination of these factors.Citation57,58 The probability of CDKN2A mutation in familial melanoma is higher in areas at low incidence of melanoma as Europe (57%) and North America (45%) than in areas at high incidence as Australia (20%). In these high-incidence geographical areas there may be a combined effect of moderate/low-penetrance susceptibility genes and higher sun exposure (environmental effects on susceptibility).Citation59
A high prevalence of the p.G101W germline mutation in the CDKN2A (p16INK4a) gene was detected in a large number of melanoma families worldwide from different countries such as Italy, Spain, France, United Stated, Israel, Australia.Citation46,47 Haplotype analysis suggested that this mutation is a common founder rather than a mutational hotspot.Citation60
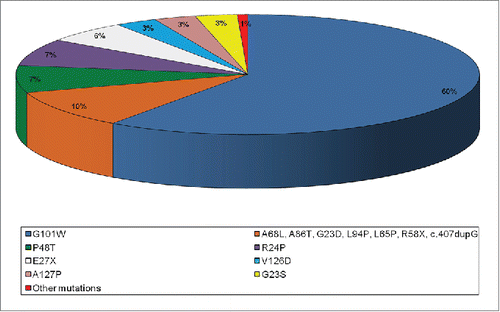
We studied 400 melanoma cases enrolled in a 6-years period at the Plastic Surgery Unit of the University of Palermo and selected 48 patients who met the criteria for familial melanoma genetic testing, in order to investigate the CDKN2A and CDK4 genes for germline mutations. The performed analyses showed that almost all patients (except one) with familial cutaneous melanoma were negative for CDKN2A mutation screening, while no variants were identified in CDK4 exon 2 of all patients, suggesting that both genes have a low impact in melanoma susceptibility of Sicilian patients. The only carrier of CDKN2A germline mutation harboured a rare missense mutation in exon 2 (p.R87W) instead of the most frequent p.G101W founder described in literature and detected in 60% of familial melanoma cases in Italy Citation61,62 (). This p.R87W rare missense mutation was found with very low frequency also in other countries belonging to Mediterranean geographical area, including Greece (Athens) and Spain (Barcelona).Citation47,63,64 In vitro experiments showed that p.R87W mutation alters both the cell cycle and reactive oxygen species (ROS) protective functions of p16INK4a.Citation45
Figure 3. Pie chart distribution of CDKN2A missense mutations detected in Italian familial melanomas.
Unlike individuals belonging to other European and Italian regions, the examined Sicilian families exhibited a very low frequency of the CDKN2A mutation. This difference could be attributed to different factors such as, for example, the historical background of the Sicily and its crucial geographical position in the center of Mediterranean Sea, cradle of several peoples, cultures and civilizations. In fact, the different dominations (Greeks, Phoenicians, Etruscans, Romans, Byzantines, Arabs, Normans, Aragon, Bourbons) are responsible for the genetic heterogeneity of the Sicilian people, already described for allelic variants of hemoglobin in Beta Thalassemia.Citation65 In addition, Salemi et al. Citation66 have reported that Xq27 region, containing the SPANX (sperm protein associated with the nucleus in the X chromosome) gene family, may be involved specifically in melanoma development in Sicilian male population. The latitude and higher sun exposure in our region than in other parts of Italy or Europe, probably represent the environmental factors causing the loss of heterozygosity (LOH) of moderate/low-penetrance susceptibility genes promoting the onset of melanoma. For these reasons, dermatological screening, sun exposure avoidance and self-examination are encouraged even in patients with familial melanoma negative for CDKN2A mutation screening.
Furthermore, other Italian regions belonging to Mediterranean Basin including Sardinia (Sassari) and Campania (Naples) showed a great similarity with the Sicily, exhibiting a null or very low frequency, respectively, of the CDKN2A germline mutation in melanoma families.Citation67,68
Our findings have led us to hypothesize that a very low frequency of germline CDKN2A mutations could be a population-specific genetic signature for Sicilian patients affected by familial melanoma.
A larger number of patients enrolled for genetic testing and the introduction of new genetic screenings to identify other mutations in other moderate/low-penetrance susceptibility genes involved in the genesis of familial melanoma will aid in clarifying which are the most common gene mutations and the role of environmental factors in determining this disease.
Patients and methods
Sample collection
In the period from January 2009 to December 2014, 400 patients affected by cutaneous melanoma were observed and treated in the Plastic Surgery Unit at the Department of Surgical, Oncological and Oral Sciences of the University of Palermo. Forty-eight patients have met the following criteria for familial melanoma genetic test in accordance with the recommendations of the Italian Society of Human Genetics (SIGU, www.sigu.net/show/attivita/5/1/linee%20guida?page=5 ) and Melanoma Genetics Consortium (GenoMEL, www.genomel.org) Citation69:
| - | Two or more individuals affected by melanoma in the same family (first degree relative); | ||||
| - | Multiple primary melanoma in the same patient with an early age of onset; | ||||
| - | Early-onset melanoma patient and pancreatic cancer in a member of the same family; | ||||
| - | Dysplastic nevus syndrome and a relative with a melanoma. | ||||
This data is coherent with that reported in the literature, in fact, about 10% of malignant melanoma occurs in familiar setting.
Patients with sporadic melanoma were excluded from the study. Diagnosis of melanoma was confirmed by histologic analysis. All clinical information for each enrolled patient was recorded anonymously and coded. All recruited individuals were from a Southern Italian region (Sicily, Palermo). A written informed consent was obtained from each investigated patient and the study was approved by ethical committee of the university-affiliated hospital. Patients were further informed that a negative screening for CDKN2A and CDK4 can be void of significance, as other unknown susceptibility genes may be involved.
Clinicopathological features of the familial melanoma patients were summarized in .
Mutation screening by sequencing analysis
Germline mutation screening of the CDKN2A and CDK4 genes was carried out. Genomic DNA was extracted from whole peripheral blood of patients with familial melanoma using the QIAamp Blood Kit (Qiagen, Hilden, Germany), according to the manufacturer's instructions. The DNA yields and purity were determined spectrophotometrically by measuring the absorbance of aliquots at 260 and 280 nm. All DNA samples were of sufficient quality to be genotyped. Direct sequencing of the PCR products of 4 exons (1α, 1β, 2, 3) of CDKN2A gene and exon 2 of CDK4 gene was performed using a BigDye Terminator v3.1 and then sequencing by ABI PRISM 3130 Genetic Analyzer (Applied Biosystems, Foster City, CA). Each genetic variant was confirmed by direct sequencing analysis on 2 independent peripheral blood samples. Data analysis was performed using the Sequencing Analysis 5.1.1 and Run 3100 Data Collection v2.0 softwares.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- Russo A, Rosell R, Rolfo C. Targeted therapies in melanoma. Current Clinical Pathologies. 2015; 211-27; http://dx.doi.org/10.1007/978-1-4939-2047-1_16
- Soufir N, Avril MF, Chompret A, Demenais F, Bombled J, Spatz A, Stoppa-Lyonnet D, Benard J, Bressac-de Paillerets B. Prevalence of p16 and CDK4 germline mutations in 48 melanoma-prone families in France. The French Familial Melanoma Study Group. Hum Mol Genet 1998; 7:209-16; PMID:9425228; http://dx.doi.org/10.1093/hmg/7.2.209
- Nelson AA, Tsao H. Melanoma and genetics. Clin Dermatol 2009; 27:46-52; PMID:19095153; http://dx.doi.org/10.1016/j.clindermatol.2008.09.005
- Chin L, Garraway LA, Fisher DE. Malignant melanoma: genetics and therapeutics in the genomic era. Genes Dev 2006; 20:2149-82; PMID:16912270; http://dx.doi.org/10.1101/gad.1437206
- Kamb A, Shattuck-Eidens D, Eeles R, Liu Q, Gruis NA, Ding W, Hussey C, Tran T, Miki Y, Weaver-Feldhaus J, et al. Analysis of the p16 gene (CDKN2) as a candidate for the chromosome 9p melanoma susceptibility locus. Nat Genet 1994; 8:23-6; PMID:7987388; http://dx.doi.org/10.1038/ng0994-22
- Begg CB, Orlow I, Hummer AJ, Armstrong BK, Kricker A, Marrett LD, Millikan RC, Gruber SB, Anton-Culver H, Zanetti R, et al. Lifetime risk of melanoma in CDKN2A mutation carriers in a population-based sample. J Natl Cancer Inst 2005; 97:1507-15; PMID:16234564; http://dx.doi.org/10.1093/jnci/dji312
- Hayward NK. Genetics of melanoma predisposition. Oncogene 2003; 22:3053-62; PMID:12789280; http://dx.doi.org/10.1038/sj.onc.1206445
- Harland M, Taylor CF, Chambers PA, Kukalizch K, Randerson-Moor JA, Gruis NA, de Snoo FA, ter Huurne JA, Goldstein AM, Tucker MA, et al. A mutation hotspot at the p14ARF splice site. Oncogene 2005; 24:4604-8; PMID:15856016; http://dx.doi.org/10.1038/sj.onc.1208678
- Hewitt C, Lee Wu C, Evans G, Howell A, Elles RG, Jordan R, Sloan P, Read AP, Thakker N. Germline mutation of ARF in a melanoma kindred. Hum Mol Genet 2002; 11:1273-9; PMID:12019208; http://dx.doi.org/10.1093/hmg/11.11.1273
- Garcia-Casado Z, Nagore E, Fernandez-Serra A, Botella-Estrada R, Lopez-Guerrero JA. A germline mutation of p14/ARF in a melanoma kindred. Melanoma Res 2009; 19:335-7; PMID:19741424; http://dx.doi.org/10.1097/CMR.0b013e32832dd2d4
- Balogh K, Szell M, Polyanka H, Pagani F, Bussani E, Kemeny L, Olah J. Detection of a rare CDKN2A intronic mutation in a Hungarian melanoma-prone family and its role in splicing regulation. Br J Dermatol 2012; 167:131-3; PMID:22292911; http://dx.doi.org/10.1111/j.1365-2133.2012.10864.x
- Djursby M, Wadt K, Lorentzen H, Borg A, Gerdes AM, Krogh L.; CDKN2A-mutation in a family with hereditary malignant melanoma.. Ugeskr Laeger 2014; 176:pii: V10130587; PMID:25294512
- Veinalde R, Ozola A, Azarjana K, Molven A, Akslen LA, Donina S, Proboka G, Cema I, Baginskis A, Pjanova D. Analysis of Latvian familial melanoma patients shows novel variants in the noncoding regions of CDKN2A and that the CDK4 mutation R24H is a founder mutation. Melanoma Res 2013; 23:221-6; PMID:23546221; http://dx.doi.org/10.1097/CMR.0b013e3283608695
- Harland M, Mistry S, Bishop DT, Bishop JA. A deep intronic mutation in CDKN2A is associated with disease in a subset of melanoma pedigrees. Hum Mol Genet 2001; 10:2679-86; PMID:11726555; http://dx.doi.org/10.1093/hmg/10.23.2679
- Laud K, Marian C, Avril MF, Barrois M, Chompret A, Goldstein AM, Tucker MA, Clark PA, Peters G, Chaudru V, et al. Comprehensive analysis of CDKN2A (p16INK4A/p14ARF) and CDKN2B genes in 53 melanoma index cases considered to be at heightened risk of melanoma. J Med Genet 2006; 43:39-47; PMID:15937071; http://dx.doi.org/10.1136/jmg.2005.033498
- Pavletich NP. Mechanisms of cyclin-dependent kinase regulation: structures of Cdks, their cyclin activators, and Cip and INK4 inhibitors. J Mol Biol 1999; 287:821-8; PMID:10222191; http://dx.doi.org/10.1006/jmbi.1999.2640
- Bennett DC. How to make a melanoma: what do we know of the primary clonal events? Pigment Cell Melanoma Res 2008; 21:27-38; PMID:18353141; http://dx.doi.org/10.1111/j.1755-148X.2007.00433.x
- Sharpless NE, Kannan K, Xu J, Bosenberg MW, Chin L. Both products of the mouse Ink4a/Arf locus suppress melanoma formation in vivo. Oncogene 2003; 22:5055-9; PMID:12902988; http://dx.doi.org/10.1038/sj.onc.1206809
- Ha L, Ichikawa T, Anver M, Dickins R, Lowe S, Sharpless NE, Krimpenfort P, Depinho RA, Bennett DC, Sviderskaya EV, et al. ARF functions as a melanoma tumor suppressor by inducing p53-independent senescence. Proc Natl Acad Sci U S A 2007; 104:10968-73; PMID:17576930; http://dx.doi.org/10.1073/pnas.0611638104
- Freedberg DE, Rigas SH, Russak J, Gai W, Kaplow M, Osman I, Turner F, Randerson-Moor JA, Houghton A, Busam K, et al. Frequent p16-independent inactivation of p14ARF in human melanoma. J Natl Cancer Inst 2008; 100:784-95; PMID:18505964; http://dx.doi.org/10.1093/jnci/djn157
- Daniotti M, Oggionni M, Ranzani T, Vallacchi V, Campi V, Di Stasi D, Torre GD, Perrone F, Luoni C, Suardi S, et al. BRAF alterations are associated with complex mutational profiles in malignant melanoma. Oncogene 2004; 23:5968-77; PMID:15195137; http://dx.doi.org/10.1038/sj.onc.1207780
- Binni F, Antigoni I, De Simone P, Majore S, Silipo V, Crisi A, Amantea A, Pacchiarini D, Castori M, De Bernardo C, et al. Novel and recurrent p14 mutations in Italian familial melanoma. Clin Genet 2010; 77:581-6; PMID:20132244; http://dx.doi.org/10.1111/j.1399-0004.2009.01298.x
- Zuo L, Weger J, Yang Q, Goldstein AM, Tucker MA, Walker GJ, Hayward N, Dracopoli NC. Germline mutations in the p16INK4a binding domain of CDK4 in familial melanoma. Nat Genet 1996; 12:97-9; PMID:8528263; http://dx.doi.org/10.1038/ng0196-97
- Rane SG, Cosenza SC, Mettus RV, Reddy EP. Germ line transmission of the Cdk4(R24C) mutation facilitates tumorigenesis and escape from cellular senescence. Mol Cell Biol 2002; 22:644-56; PMID:11756559; http://dx.doi.org/10.1128/MCB.22.2.644-656.2002
- Fanale D, Amodeo V, Corsini LR, Rizzo S, Bazan V, Russo A. Breast cancer genome-wide association studies: there is strength in numbers. Oncogene 2012; 31:2121-8; PMID:21996731; http://dx.doi.org/10.1038/onc.2011.408
- Barrett JH, Taylor JC, Bright C, Harland M, Dunning AM, Akslen LA, Andresen PA, Avril MF, Azizi E, Bianchi Scarra G, et al. Fine mapping of genetic susceptibility loci for melanoma reveals a mixture of single variant and multiple variant regions. Int J Cancer 2015; 136:1351-60; PMID:25077817; http://dx.doi.org/10.1002/ijc.29099
- Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, Beroukhim R, Milner DA, Granter SR, Du J, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature 2005; 436:117-22; PMID:16001072; http://dx.doi.org/10.1038/nature03664
- Ugurel S, Houben R, Schrama D, Voigt H, Zapatka M, Schadendorf D, Brocker EB, Becker JC. Microphthalmia-associated transcription factor gene amplification in metastatic melanoma is a prognostic marker for patient survival, but not a predictive marker for chemosensitivity and chemotherapy response. Clin Cancer Res 2007; 13:6344-50; PMID:17975146; http://dx.doi.org/10.1158/1078-0432.CCR-06-2682
- Cheli Y, Ohanna M, Ballotti R, Bertolotto C. Fifteen-year quest for microphthalmia-associated transcription factor target genes. Pigment Cell Melanoma Res 2010; 23:27-40; PMID:19995375; http://dx.doi.org/10.1111/j.1755-148X.2009.00653.x
- Bertolotto C, Abbe P, Hemesath TJ, Bille K, Fisher DE, Ortonne JP, Ballotti R. Microphthalmia gene product as a signal transducer in cAMP-induced differentiation of melanocytes. J Cell Biol 1998; 142:827-35; PMID:9700169; http://dx.doi.org/10.1083/jcb.142.3.827
- Carreira S, Goodall J, Aksan I, La Rocca SA, Galibert MD, Denat L, Larue L, Goding CR. Mitf cooperates with Rb1 and activates p21Cip1 expression to regulate cell cycle progression. Nature 2005; 433:764-9; PMID:15716956; http://dx.doi.org/10.1038/nature03269
- Cheli Y, Giuliano S, Botton T, Rocchi S, Hofman V, Hofman P, Bahadoran P, Bertolotto C, Ballotti R. Mitf is the key molecular switch between mouse or human melanoma initiating cells and their differentiated progeny. Oncogene 2011; 30:2307-18; PMID:21278797; http://dx.doi.org/10.1038/onc.2010.598
- Xu W, Gong L, Haddad MM, Bischof O, Campisi J, Yeh ET, Medrano EE. Regulation of microphthalmia-associated transcription factor MITF protein levels by association with the ubiquitin-conjugating enzyme hUBC9. Exp Cell Res 2000; 255:135-43; PMID:10694430; http://dx.doi.org/10.1006/excr.2000.4803
- Yokoyama S, Woods SL, Boyle GM, Aoude LG, MacGregor S, Zismann V, Gartside M, Cust AE, Haq R, Harland M, et al. A novel recurrent mutation in MITF predisposes to familial and sporadic melanoma. Nature 2011; 480:99-103; PMID:22080950; http://dx.doi.org/10.1038/nature10630
- Bertolotto C, Lesueur F, Giuliano S, Strub T, de Lichy M, Bille K, Dessen P, d'Hayer B, Mohamdi H, Remenieras A, et al. A SUMOylation-defective MITF germline mutation predisposes to melanoma and renal carcinoma. Nature 2011; 480:94-8; PMID:22012259; http://dx.doi.org/10.1038/nature10539
- Beuret L, Flori E, Denoyelle C, Bille K, Busca R, Picardo M, Bertolotto C, Ballotti R. Up-regulation of MET expression by alpha-melanocyte-stimulating hormone and MITF allows hepatocyte growth factor to protect melanocytes and melanoma cells from apoptosis. J Biol Chem 2007; 282:14140-7; PMID:17371876; http://dx.doi.org/10.1074/jbc.M611563200
- Busca R, Berra E, Gaggioli C, Khaled M, Bille K, Marchetti B, Thyss R, Fitsialos G, Larribere L, Bertolotto C, et al. Hypoxia-inducible factor 1{alpha} is a new target of microphthalmia-associated transcription factor (MITF) in melanoma cells. J Cell Biol 2005; 170:49-59; PMID:15983061; http://dx.doi.org/10.1083/jcb.200501067
- Liu F, Fu Y, Meyskens FL, Jr. MiTF regulates cellular response to reactive oxygen species through transcriptional regulation of APE-1/Ref-1. J Invest Dermatol 2009; 129:422-31; PMID:18971960; http://dx.doi.org/10.1038/jid.2008.255
- Carreira S, Goodall J, Denat L, Rodriguez M, Nuciforo P, Hoek KS, Testori A, Larue L, Goding CR. Mitf regulation of Dia1 controls melanoma proliferation and invasiveness. Genes Dev 2006; 20:3426-39; PMID:17182868; http://dx.doi.org/10.1101/gad.406406
- Du J, Widlund HR, Horstmann MA, Ramaswamy S, Ross K, Huber WE, Nishimura EK, Golub TR, Fisher DE. Critical role of CDK2 for melanoma growth linked to its melanocyte-specific transcriptional regulation by MITF. Cancer Cell 2004; 6:565-76; PMID:15607961; http://dx.doi.org/10.1016/j.ccr.2004.10.014
- Sturm RA. Skin colour and skin cancer - MC1R, the genetic link. Melanoma Res 2002; 12:405-16; PMID:12394181; http://dx.doi.org/10.1097/00008390-200209000-00001
- Box NF, Duffy DL, Chen W, Stark M, Martin NG, Sturm RA, Hayward NK. MC1R genotype modifies risk of melanoma in families segregating CDKN2A mutations. Am J Hum Genet 2001; 69:765-73; PMID:11500805; http://dx.doi.org/10.1086/323412
- Jhappan C, Noonan FP, Merlino G. Ultraviolet radiation and cutaneous malignant melanoma. Oncogene 2003; 22:3099-112; PMID:12789287; http://dx.doi.org/10.1038/sj.onc.1206450
- Demenais F, Mohamdi H, Chaudru V, Goldstein AM, Newton Bishop JA, Bishop DT, Kanetsky PA, Hayward NK, Gillanders E, Elder DE, et al. Association of MC1R variants and host phenotypes with melanoma risk in CDKN2A mutation carriers: a GenoMEL study. J Natl Cancer Inst 2010; 102:1568-83; PMID:20876876; http://dx.doi.org/10.1093/jnci/djq363
- Jenkins NC, Jung J, Liu T, Wilde M, Holmen SL, Grossman D. Familial melanoma-associated mutations in p16 uncouple its tumor-suppressor functions. J Invest Dermatol 2013; 133:1043-51; PMID:23190892; http://dx.doi.org/10.1038/jid.2012.401
- Mantelli M, Barile M, Ciotti P, Ghiorzo P, Lantieri F, Pastorino L, Catricala C, Torre GD, Folco U, Grammatico P, et al. High prevalence of the G101W germline mutation in the CDKN2A (P16(ink4a)) gene in 62 Italian malignant melanoma families. Am J Med Genet 2002; 107:214-21; PMID:11807902; http://dx.doi.org/10.1002/ajmg.10137
- Puig S, Malvehy J, Badenas C, Ruiz A, Jimenez D, Cuellar F, Azon A, Gonzalez U, Castel T, Campoy A, et al. Role of the CDKN2A locus in patients with multiple primary melanomas. J Clin Oncol 2005; 23:3043-51; PMID:15860862; http://dx.doi.org/10.1200/JCO.2005.08.034
- Hashemi J, Platz A, Ueno T, Stierner U, Ringborg U, Hansson J. CDKN2A germ-line mutations in individuals with multiple cutaneous melanomas. Cancer Res 2000; 60:6864-7; PMID:11156381
- Tsao H, Chin L, Garraway LA, Fisher DE. Melanoma: from mutations to medicine. Genes Dev 2012; 26:1131-55; PMID:22661227; http://dx.doi.org/10.1101/gad.191999.112
- Bishop JN, Harland M, Randerson-Moor J, Bishop DT. Management of familial melanoma. Lancet Oncol 2007; 8:46-54; PMID:17196510; http://dx.doi.org/10.1016/S1470-2045(06)71010-5
- Massey PR, Prasad V, Figg WD, Fojo T. Multiplying therapies and reducing toxicity in metastatic melanoma. Cancer Biol Ther 2015; 16(7):1014-8:1-5
- Bhatia S, Emdad L, Das SK, Hamed H, Dent P, Sarkar D, Fisher PB. Non-BRAF targeted therapies for melanoma: protein kinase inhibitors in Phase II clinical trials. Expert Opin Investig Drugs 2014; 23:489-500; PMID:24502370; http://dx.doi.org/10.1517/13543784.2014.884558
- Wyluda EJ, Cheng J, Schell TD, Haley JS, Mallon C, Neves RI, Robertson G, Sivik J, Mackley H, Talamo G, et al. Durable complete responses off all treatment in patients with metastatic malignant melanoma after sequential immunotherapy followed by a finite course of BRAF inhibitor therapy. Cancer Biol Ther 2015; 16:662-70; PMID:25806780; http://dx.doi.org/10.1080/15384047.2015.1026507
- Della Torre G, Pasini B, Frigerio S, Donghi R, Rovini D, Delia D, Peters G, Huot TJ, Bianchi-Scarra G, Lantieri F, et al. CDKN2A and CDK4 mutation analysis in Italian melanoma-prone families: functional characterization of a novel CDKN2A germ line mutation. Br J Cancer 2001; 85:836-44; PMID:11556834; http://dx.doi.org/10.1054/bjoc.2001.1991
- Udayakumar D, Mahato B, Gabree M, Tsao H. Genetic determinants of cutaneous melanoma predisposition. Semin Cutan Med Surg 2010; 29:190-5; PMID:21051013; http://dx.doi.org/10.1016/j.sder.2010.06.002
- Tsao H, Zhang X, Kwitkiwski K, Finkelstein DM, Sober AJ, Haluska FG. Low prevalence of germline CDKN2A and CDK4 mutations in patients with early-onset melanoma. Arch Dermatol 2000; 136:1118-22; PMID:10987867; http://dx.doi.org/10.1001/archderm.136.9.1118
- Bishop DT, Demenais F, Goldstein AM, Bergman W, Bishop JN, Bressac-de Paillerets B, Chompret A, Ghiorzo P, Gruis N, Hansson J, et al. Geographical variation in the penetrance of CDKN2A mutations for melanoma. J Natl Cancer Inst 2002; 94:894-903; PMID:12072543; http://dx.doi.org/10.1093/jnci/94.12.894
- Fallah M, Pukkala E, Sundquist K, Tretli S, Olsen JH, Tryggvadottir L, Hemminki K. Familial melanoma by histology and age: joint data from five Nordic countries. Eur J Cancer 2014; 50:1176-83; PMID:24461199; http://dx.doi.org/10.1016/j.ejca.2013.12.023
- Goldstein AM, Chan M, Harland M, Hayward NK, Demenais F, Bishop DT, Azizi E, Bergman W, Bianchi-Scarra G, Bruno W, et al. Features associated with germline CDKN2A mutations: a GenoMEL study of melanoma-prone families from three continents. J Med Genet 2007; 44:99-106; PMID:16905682; http://dx.doi.org/10.1136/jmg.2006.043802
- Ciotti P, Struewing JP, Mantelli M, Chompret A, Avril MF, Santi PL, Tucker MA, Bianchi-Scarra G, Bressac-de Paillerets B, Goldstein AM. A single genetic origin for the G101W CDKN2A mutation in 20 melanoma-prone families. Am J Hum Genet 2000; 67:311-9; PMID:10869234; http://dx.doi.org/10.1086/303001
- Bruno W, Ghiorzo P, Battistuzzi L, Ascierto PA, Barile M, Gargiulo S, Gensini F, Gliori S, Guida M, Lombardo M, et al. Clinical genetic testing for familial melanoma in Italy: a cooperative study. J Am Acad Dermatol 2009; 61:775-82; PMID:19500876; http://dx.doi.org/10.1016/j.jaad.2009.03.039
- Mantelli M, Pastorino L, Ghiorzo P, Barile M, Bruno W, Gargiulo S, Sormani MP, Gliori S, Vecchio S, Ciotti P, et al. Early onset may predict G101W CDKN2A founder mutation carrier status in Ligurian melanoma patients. Melanoma Res 2004; 14:443-8; PMID:15577313; http://dx.doi.org/10.1097/00008390-200412000-00002
- Harland M, Cust AE, Badenas C, Chang YM, Holland EA, Aguilera P, Aitken JF, Armstrong BK, Barrett JH, Carrera C, et al. Prevalence and predictors of germline CDKN2A mutations for melanoma cases from Australia, Spain and the United Kingdom. Hered Cancer Clin Pract 2014; 12:20; PMID:25780468; http://dx.doi.org/10.1186/1897-4287-12-20
- Nikolaou V, Kang X, Stratigos A, Gogas H, Latorre MC, Gabree M, Plaka M, Njauw CN, Kypreou K, Mirmigi I, et al. Comprehensive mutational analysis of CDKN2A and CDK4 in Greek patients with cutaneous melanoma. Br J Dermatol 2011; 165:1219-22; PMID:21801156; http://dx.doi.org/10.1111/j.1365-2133.2011.10551.x
- Giambona A, Vinciguerra M, Cannata M, Cassara F, Fiorentino G, Leto F, Gioco PL, Renda D, Passarello C, Maggio A. The genetic heterogeneity of beta-globin gene defects in Sicily reflects the historic population migrations of the island. Blood Cells Mol Dis 2011; 46:282-7; PMID:21353607; http://dx.doi.org/10.1016/j.bcmd.2011.01.006
- Salemi M, Bosco P, Cali F, Calogero AE, Soma PF, Galia A, Lanzafame M, Romano C, Vicari E, Grasso G, et al. SPANX-B and SPANX-C (Xq27 region) gene dosage analysis in Sicilian patients with melanoma. Melanoma Res 2008; 18:295-9; PMID:18626316; http://dx.doi.org/10.1097/CMR.0b013e32830aaa90
- Casula M, Colombino M, Satta MP, Cossu A, Lissia A, Budroni M, Simeone E, Calemma R, Loddo C, Caraco C, et al. Factors predicting the occurrence of germline mutations in candidate genes among patients with cutaneous malignant melanoma from South Italy. Eur J Cancer 2007; 43:137-43; PMID:17055252; http://dx.doi.org/10.1016/j.ejca.2006.07.017
- Casula M, Muggiano A, Cossu A, Budroni M, Caraco C, Ascierto PA, Pagani E, Stanganelli I, Canzanella S, Sini M, et al. Role of key-regulator genes in melanoma susceptibility and pathogenesis among patients from South Italy. BMC Cancer 2009; 9:352; PMID:19799798; http://dx.doi.org/10.1186/1471-2407-9-352
- Leachman SA, Carucci J, Kohlmann W, Banks KC, Asgari MM, Bergman W, Bianchi-Scarra G, Brentnall T, Bressac-de Paillerets B, Bruno W, et al. Selection criteria for genetic assessment of patients with familial melanoma. J Am Acad Dermatol 2009; 61:677 e1-14; PMID:19751883; http://dx.doi.org/10.1016/j.jaad.2009.03.016