ABSTRACT
Recent research has found that long noncoding RNAs (lncRNAs) were involved in various human cancers. However, the role of these lncRNAs in cervical cancer remains unexplored. Therefore, we aimed to investigate the biological function of maternally expressed gene 3 (MEG3), a cancer-related lncRNA, and its underlying mechanism in cervical cancer. In this study, MEG3 expression of 108 patients’ cervical cancer tissues and adjacent normal tissues was detected by quantitative real-time PCR analysis (qRT-PCR), and the functional effect of MEG3 was determined in vitro assays. We observed that MEG3 was downregulated in cervical cancer tissues, compared to the adjacent normal tissues, and was negatively related with FIGO stages, tumor size, lymphatic metastasis, HR-HPV infection and the expression of homo sapiens microRNA-21 (miR-21). Furthermore, we focused on the function and molecular mechanism of MEG3, finding that overexpression of MEG3 reduced the level of miR-21-5p expression, causing inhibition of proliferation and increased apoptosis in cervical cancer cells. In summary, our findings indicate that MEG3 function as a tumor suppressor by regulating miR-21-5p, resulting in the inhibition of tumor growth in cervical cancer. As a result, this study improves our understanding of the function of MEG3 in cervical cancer and will help to provide new potential target sites for cervical cancer treatment.
Abbreviations
| lncRNA, long noncoding RNA; MEG3, maternally expressed gene 3; qRT-PCR, quantitative real-time PCR analysis; miR-21, homo sapiens microRNA-21; ncRNA, noncoding RNA; NSCLC, nonsmall cell lung cancer; miRNA, MicroRNA; PDCD4, programmed cell death 4; CASC2, cancer susceptibility candidate 2; GAS5, growth arrest specific 5; LVSI, Lymphatic vascular space invasion; MDM2, mouse double minute 2 homolog; SDS-PAGE, SDS-polyacrylamide gel electrophoresis; CCK-8, Cell Counting Kit-8. | = |
Introduction
Cervical cancer is the second commonest cancer among women in the worldwide, and the majority cause of death in developing countries as well.Citation1 Reports estimate that there are approximately 500,000 new cases of cervical cancer diagnosed each year. Citation2 Treatment of cervical cancer is represented by radiotherapy or surgery for patients in FIGOI-IIa stages and concurrent chemoradiation for patients in FIGOIIb-IV stages. The prognosis of advanced patients generally remains poor and the overall 5 y survival rate is approximately 40% after conventional treatments are used.Citation3 Thus, effective therapeutic strategy is urgently needed and further exploring the mechanism underlying is urgently required.
After the discovery of noncoding RNAs (ncRNAs), which have been linked to the regulation of gene expression, a new insight to cancer etiopathogenesis occurred. LncRNA is commonly defined as a RNA molecular which is larger than 200 nucleotides and not translated into proteins.Citation4 They have important functional roles in chromatin remodeling, structural scaffolding of nuclear protein substructures, regulation of the expression and transcription genes, and posttranscriptional processing.Citation5-8
MEG3, which encodes a lncRNA, is an imprinted gene belonging to the DLK1-MEG3 locus located on chromosome 14q32.3 in humans. The loss of MEG3 expression was observed in various types of cancers, including nonsmall cell lung cancer (NSCLC), gastric cancer and gallbladder cancer. Overexpressed MEG3 could inhibit proliferation and promote apoptosis in tumor cells. Citation9-11 These studies suggest the MEG3 gene may play a role in tumor suppression. However, research on the expression and function of MEG3 in cervical cancer is still limited.
MicroRNAs (miRNAs) are a class of approximately 22 nucleotides noncoding RNAs that regulate the expression of target genes by interacting with complementary sites in the 3′ untranslated region of the target mRNAs.Citation12 Our previous study has found that miR-21 effected tumorigenesis by regulating CCL20 in cervical squamous cell.Citation13 Other studies also observe that miR-21 is an oncomiR in cervical cancer, which promotes cell proliferation by downregulating the expression of programmed cell death 4 (PDCD4),Citation14 or by mediating aberrant STAT3 signaling.Citation15 In addition, miR-21 is significantly upregulated in HR-HPV positive cervical cancer.Citation16 Taken together, it indicate that miR-21 plays a significant role in cervical cancer.
Although much ncRNAs research is focued on understanding the regulation of protein coding genes mediated by them, it has been previously suggested that ncRNAs could form a well-orchestrated regulatory interaction network.Citation17 For example, it has been reported that lncRNA cancer susceptibility candidate 2 (CASC2) and growth arrest specific 5 (GAS5) play tumor suppressive role via negative regulation of miR-21.Citation18,19 However, there is still no report about the interaction between lncRNAs and miR-21 in cervical cancer.
In this study, we found a significant decrease of the MEG3 expression in cervical cancer tissues as compared to the adjacent normal tissues and MEG3 downregulation was associated with larger tumor size, advanced FIGO stage, lymph nodes metastasis and HR‑HPV positive. Moreover, we found that upregulation of MEG3 could suppress growth and enhance apoptosis of cervical cancer cells, which showed anticancer functions of MEG3 in cervical cancer. Furthermore, we sought to identify MEG3 interacting with miRNA. Expression of miR-21-5p was negatively correlated with MEG3 in cervical cancer tissues and overexpression of mir-21-5p reversed the anticancer effects of MEG3 in cervical cancer cells. Taken together, our date suggested that MEG3 may function as tumor suppressing lncRNA in cervical cancer cells at least in part by downregulating mir-21-5p.
Results
The expression level of MEG3 is significantly decreased in cervical cancer tissues
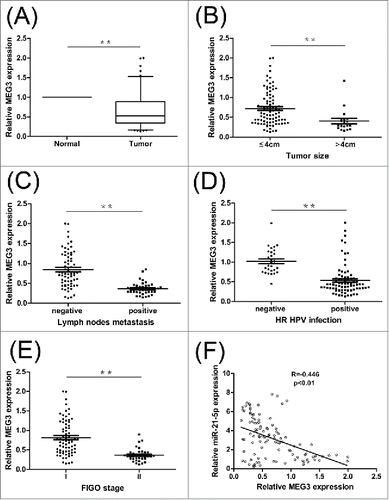
We examined the expression level of MEG3 in 108 samples of cervical cancer tissues and adjacent normal tissues. As shown in , the mean expression level of MEG3 in cervical cancer samples was significantly lower than the mean level in the adjacent normal tissues samples (P < 0.01). The results suggested that the MEG3 expression in cervical cancer were significantly downregulated compared to adjacent normal cervical tissues.
Figure 1. Relative MEG3 expression in cervical cancer tissues and its clinical significance. (A) Relative expression of MEG3 in cervical cancer tissues (n = 108) compared with adjacent normal tissues (n = 108). (B, C, D and E) MEG3 expression was significantly lower in patients with larger tumor size, lymph nodes metastasis, HR HPV positive and advanced FIGO stage. (F) Pearson's correlation coefficient analysis showed that miR-21-5p expression was negatively correlated with MEG3 in cervical cancer tissues. **P < 0.01.
Relationship between MEG3 expression and clinicopathologic features of cervical cancer
As shown in , low MEG3 expression levels were positively correlated with the tumor size, lymph node metastasis, HR‑HPV infection and FIGO stage (P < 0.01). However, there were no relationships between MEG3 expression and other parameters such as age, menopause, histology, depth of invasion, differentiation or Lymphatic vascular space invasion (LVSI) (). These data indicate that downregulation of MEG3 may relate to cervical cancer development.
Table 1. Correlation of the expression of MEG3 with clinicopathologic features.
Effect of MEG3 on cervical cancer cell prolif-eration in vitro
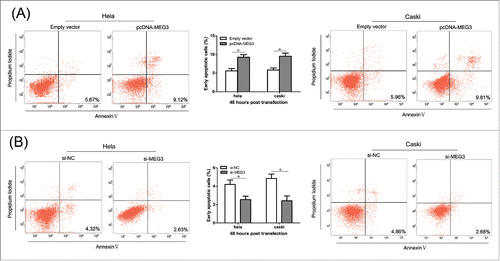
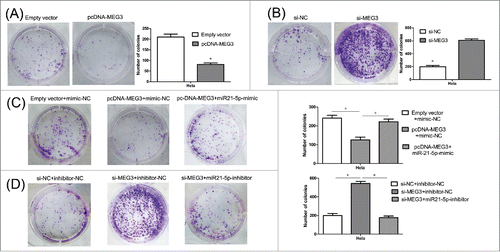
For the si-MEG3-2 demonstrated the strongest effect, it was used to perform all subsequent assays. (Result S1 and Fig. S1B) The result of CCK-8 assay showed that, compared with the negative control, the HeLa and CaSki cells transfected with pcDNA-MEG3 had lower proliferative ability (). Conversely, MEG3 knockdown significantly induced cell prolifera- (). Consistent with the CCK-8 assay, the colony numbers of HeLa and CaSki cells transfected with pcDNA3.1-MEG3 were also fewer than those of the cells transfected with negative control ( and S2A). And, as expected, MEG3 knockdown induced viability in cell proliferation ( and S2B). These results suggest that the level of MEG3 expression is significantly associated with the proliferation capacity of cervical cancer cells.
Figure 2. Effects of MEG3 on cervical cancer cells proliferation in vitro. (A) Upregulated MEG3 expression dramatically decreased the ability of Hela and CaSki cells proliferation by CCK-8 assay. (B) Downregulated MEG3 expression dramatically increased the ability of Hela and CaSki cells proliferation by CCK-8 assay. *P < 0.05.
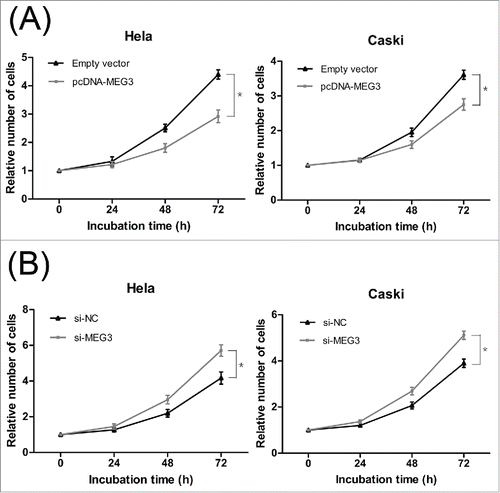
Figure 3. Colony formation assay was performed to determine the colony formation ability of transfected Hela cells. (A, B) The colony number of pcDNA-MEG3 or si-MEG3 transfected Hela cells. (C, D) The colony number of Hela cells transfected with pcDNA- MEG3 and miR-21-5p mimic or transfected with si-MEG3 and miR-21-5p inhibitor. *P < 0.05.
Effect of MEG3 on cervical cancer cell apoptosis in vitro
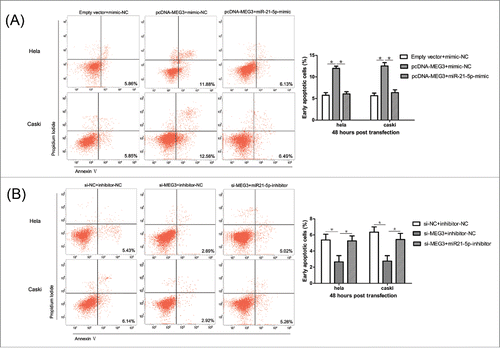
To further determine the physiological role of MEG3 in cervical cancer cell apoptosis, HeLa and CaSki cells were transfected with pcDNA- MEG3 or si-MEG3. The results revealed that more apoptotic cells in pcDNA- MEG3-transfected groups than control groups (). In contrast, MEG3 knockdown significantly reduced apoptosis in HeLa and CaSki cells (). These results confirmed that MEG3 induced cell apoptosis in cervical cancer cells.
MEG3 affects the levels of P53 and caspase 3 in cervical cancer cell
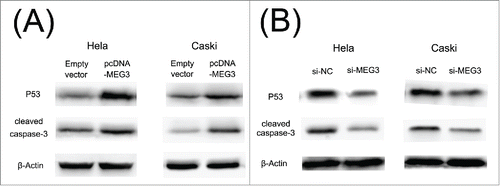
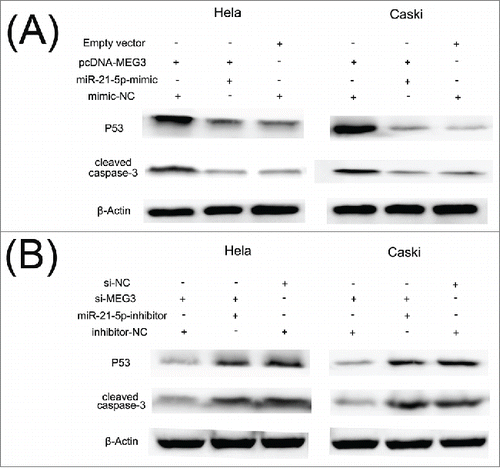
shows that the expression of p53 and cleaved caspase 3 proteins was significantly increased in HeLa and CaSki cells transfected with pcDNA- MEG3 compared with the control cells. As anticipated, we also found that knockdown of MEG3 by si-MEG3 in HeLa and CaSki cells significantly decreased p53 and cleaved caspase 3 protein expression (). These data confirm that MEG3 functions as a tumor suppressor gene by affecting p53 and caspase 3 activations in cervical cancer cell.
The tumor suppressive effects of MEG3 may through regulation of miR-21-5p in cervical cancer cells
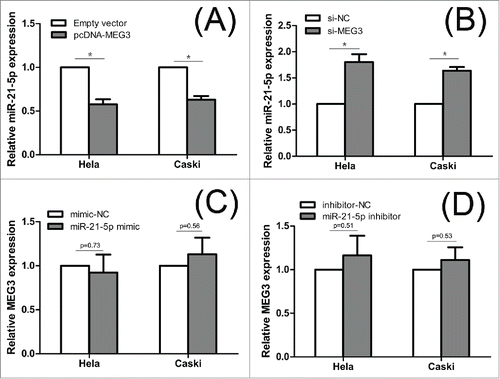
To explore the underlying mechanism responsible for MEG3, we tested whether MEG3 could regulate miR-21-5p expression. First of all, Pearson's correlation showed that expression of miR-21-5p was negatively correlated with MEG3 level in cervical cancer tissues (). Subsequently, we detected the miR-21-5p level after overexpression or knockdown MEG3. shows that the miR-21-5p expression level was significantly decreased in cells transfected with pcDNA- MEG3 comparing with control cells. In contrast, knockdown the expression of MEG3 resulted significant upregulation of miR-21-5p expression in HeLa and CaSki cells (). These results show that MEG3 is regulate the expression levels of the miR-21-5p in HeLa and CaSki cells.
Figure 6. The interaction between MEG3 and miR-21-5p. (A, B) Hela and CaSki cells were transfected with pcDNA-MEG3 or si-MEG3, and qRT-PCR was used to detect the miR-21-5p levels compared with controls. (C, D) Hela and CaSki cells were transfected with miR-21-5p mimic or inhibitor, and qRT-PCR was used to detect the MEG3 levels compared with controls. *P < 0.05.
To confirm that the MEG3 functions were indeed due to regulation of miR-21-5p, HeLa and CaSki cells were transfected together with plasmids encoding MEG3 and miR-21-5p mimic or with si-MEG3 and miR-21-5p inhibitor. CCK-8 assay and colony formation assay revealed that the proliferation of cells in pcDNA- MEG3 + miR-21-5p mimics groups was increased, compared with the cells in pcDNA- MEG3 groups (, and S2C). Furthermore, we found that the apoptotic rates of cells in pcDNA- MEG3 + miR-21-5p mimic groups were decreased compared with the cells in pcDNA- MEG3 groups (). The functional assays were also performed to monitor the effect of co-transfection of si-MEG3 and miR-21-5p inhibitor. As shown in , 3D, S2D and 8B, the effects of si-MEG3 could be partly eliminated after decreasing the expression of miR-21-5p. In summary, we suggest that miR-21-5p could rescue the effect on both cell proliferation and apoptosis which induced by MEG3 in HeLa and CaSki cells.
Figure 7. MiR-21-5p mediated the effects of MEG3 on Hela and CaSki cells proliferation. (A) CCK8 assay to evaluate the effect of MEG3 overexpression and miR-21-5p stimulation on Hela and CaSki cells proliferation. (B) CCK8 assay to evaluate the effect of MEG3 knockdown and miR-21-5p inhibition on Hela and CaSki cells proliferation. *P < 0.05.
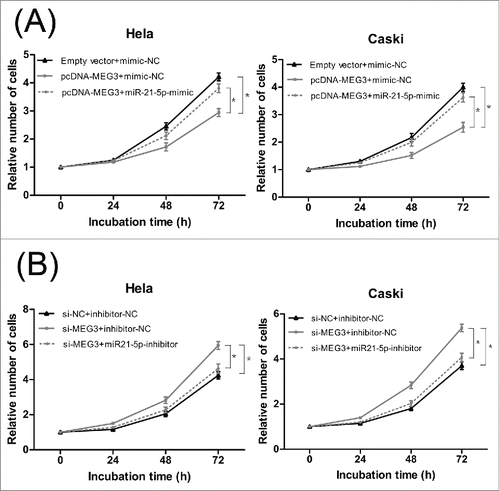
Figure 8. MiR-21-5p mediated the effects of MEG3 on Hela and CaSki cells apoptosis. (A) Effect of co-transfection of pcDNA- MEG3 and miR-21-5p mimic on apoptosis of Hela and CaSki cells. (B) Effect of co-transfection of si-MEG3 and miR-21-5p inhibitor on apoptosis of Hela and CaSki cells. *P < 0.05.
Discussion
Several studies reported that MEG3 acted as a tumor suppressing lncRNA in various tumors. The MEG3 expression levels were found to be lower in cancer tissues than the adjacent normal tissues in NSCLC,Citation9, gastric cancerCitation10 and gallbladder cancer.Citation11 So, we hypothesized that MEG3 also had similar impact on the progression of cervical cancer. As other studies, we found that the levels of MEG3 in cervical cancer tissues were significantly lower than those in the matched adjacent normal tissues. According to other reports, MEG3 was mainly correlated with tumor size.Citation9,10 Similarly, we found that MEG3 was significantly lower in patients with large tumors. By now, there is no report about the relationship between HR‑HPV infection and MEG3 expression in cervical cancer. Our study found that the MEG3 levels in HR-HPV negative patients are higher than positive patients. More important, we found that low expression of MEG3 was significantly associated with advanced FIGO stages and lymph nodes metastasis. And as we know, patients with advanced FIGO stages or lymph nodes metastasis always tend to have shorter overall survival time. So MEG3 may represent a novel indicator of prognosis in cervical cancer. Taken together, these results suggested that loss expression of MEG3 may associate with progression and poor prognosis of cervical cancer.
Qin et al. demonstrated that MEG3 inhibits proliferation and induces apoptosis in human cervical cancer cells.Citation20 Consistent with it, we also found that MEG3 overexpression can dramatically induce apoptosis and growth arrest in vitro. P53 and caspase 3 are apoptosis-related proteins. Many studies have demonstrated that MEG3 may play an important role in regulating cell growth via the p53 and caspase 3.Citation9,20 In our study, we found as expected that MEG3 can activate p53 and caspase 3. It suggested that MEG3 can indirectly or directly target p53 and caspase 3 to control proliferation and apoptosis of cervical cancer cells.
Recently, increasing evidences have demonstrated the interplay between lncRNAs and miRNAs. Some lncRNAs are degraded by miRNAsCitation21 and some can serve as sponges or decoys for miRNAs.Citation22 Moreover, Several lncRNAs also have been reported that can compete with miRNAs for binding to mRNAsCitation23 or produce miRNAs and other small RNAs.Citation24 Although MEG3 has been reported involved in the development of cervical cancer,Citation20 the interaction between MEG3 and miRNAs in cervical cancer is still not verified. In our previous study, we found that miR-21-5p was correlated with lymph nodes metastasis, HR‑HPV infection, cell proliferation and apoptosis in cervical cancer,Citation13 which is similar to MEG3. Therefore, we would like to explore the interaction between MEG3 and miR-21-5p in cervical cancer. First, we found that miR-21-5p level was inversely correlated with MEG3 level in cervical cancer tissues. So, we speculated that miR-21-5p is under the regulation of MEG3. Then we found that overexpression of MEG3 contributed to downregulation of miR-21-5p, and knockdown of MEG3 contributed to upregulation of miR-21-5p. In addition, miR-21-5p mimic was shown to rescue the effects which induced by MEG3 and miR-21-5p inhibitor could partly eliminate the effect of si-MEG3 in HeLa and CaSki cells. These provided additional evidences to such a regulatory network. Interestingly, miR-21-5p overexpression or knockdown had no significant effect on the expression level of MEG3 (). This finding indicated that the MEG3 may negative regulated the expression of miR-21-5p but they may not form a reciprocal repression feedback loop. Here, we report for the first time that MEG3 can inhibit cervical cancer cells proliferation and induce apoptosis through regulating miR-21-5p. However, the exact molecular mechanisms of interaction between MEG3 and miR-21-5p require further study.
It has recently been reported that mouse double minute 2 homolog (MDM2) expression is repressed by MEG3, suggesting that down-regulation of MDM2 protein levels contributes to p53 activation by MEG3.Citation25 However, the molecular mechanisms of MEG3 activates p53 remain elusive, especially whether miRNAs is involved in it. In our study, we revealed that expression level of p53 protein was significantly decreased in pcDNA- MEG3 + miR-21-5p mimic group and it was dramatically increase in si-MEG3 + miR-21-5p inhibitor groups, compared with the cells in pcDNA- MEG3 groups or si-MEG3 groups, respectively (). This result indicated that miR-21-5p suppression contributes at least in part to p53 accumulation induced by MEG3. Although there is no miR-21 binding site in the p53 3′ untranslated region in human, there are several reports that can support our results. A recent study reports that downregulation of miR-21 in glioblastoma cells leads to elevated expression of p63 and activators of the p53 pathway.Citation26 In breast cancer cells, miR-21 inhibition induces the expression of several genes regulated by p53 at the mRNA level, suggesting that miR-21 over-expression could impair the tumor-suppressive function of the p53 pathway.Citation27 However, more studies should be applied to elucidate the molecular mechanisms of interaction among MEG3, miR-21 and p53 in the progression of cervical cancer.
Figure 9. MiR-21-5p mediated the effects of MEG3 on regulating P53 and cleaved caspase 3 protein levels. (A) The level of p53 and cleaved caspase 3 protein was decreased in pcDNA- MEG3 + miR-21-5p mimic group. (B) The level of p53 and cleaved caspase 3 protein was increased in si-MEG3 + miR-21-5p inhibitor groups.
In conclusion, our data suggests that MEG3 may play crucial role during cervical cancer progression, and MEG3 may interact with miR-21-5p to link miRNAs and the post-transcriptional network. Targeting the MEG3/miR-21-5p interaction may help establish new strategies for therapy and improve therapeutic effect of cervical cancer.
Material and methods
Clinical samples
Paired cervical cancer and adjacent normal tissues were obtained from 108 patients (FIGOI-IIa) who underwent primary surgery in the Second Affiliated Hospital of Sun Yat-sen University (Guangzhou, China), between November 2014 and Jun 2015. No patients had received any form of tumor specific therapy before diagnosis. Informed consent was obtained from each patient recruited, and the study protocol was approved by the institutional ethics committee of the Second Affiliated Hospital of Sun Yat-sen University. All specimens were immediately frozen in tubes containing RNAlater preservation liquid after removal and stored at -80°C until RNA extraction. Tissue sections from each patient were reviewed and classified by a pathologist. The clinical characteristics of all patients are shown in .
Cell culture
Human cervical cancer cell lines (HeLa, CaSki) were purchased from the Cell Bank of the Chinese Academy of Sciences. HeLa was grown in DMEM (Thermo Scientific) containing high glucose. CaSki was grown in RPMI 1640 medium (Thermo Scientific). Medium was supplemented with 10 % fetal bovine serum (Gibco). All cells were grown in at 37°C in a humidified 5 % CO2 atmosphere.
RNA isolation, Reverse transcription, and qRT-PCR
Total RNA of tissues and cells was isolated with Trizol reagent (TaKaRa). RNAs were reverse transcribed into cDNAs using the RT-PCR kit purchased from TaKaRa according to the manufacturer's protocol. SYBR Premix Ex Taq (TaKaRa) was used to detect MEG3 and mir-21 according to the manufacturer's instructions, and β-Actin and U6 was used as an internal control. QRT-PCR and data collection were performed on CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, California, US). The results were calculated by the 2−ΔΔCt method. All results were expressed as the mean ± SEM of 3 independent experiments. Primers were synthesized by Generay Biotech and TaKaRa. The primer sequences were listed in Table S1.
Western blot
Fifty micrograms of protein were separated by 10% SDS-PAGE (SDS-PAGE) and transferred to PVDF membrane (Roche). The membrane was blocked at room temperature for 1 hour in tris-buffered saline (TBS) containing 0.1% Tween-20 with 5% skim-milk and subsequently incubated with specific primary antibodies at 4°C for 12 hours. Then the membranes were washed 3 times in TBST. Following incubation with HRP Goat-anti-Rabbit (1:2000; Santa Cruz Biotechnology) at room temperature for 2 hours, bound proteins were visualized using ECL (Thermo Scientific). Primary antibody for caspases 3 was purchased from Cell Signaling Technology (1:1,000) and P53 were obtained from Santa Cruz Biotechnology (1:500). Results were normalized to the expression of β-Actin (1:1,000; Cell Signaling Technology)
Transfection of cervical cancer cell lines
Cervical cancer cell lines were transfected with plasmids, siRNA oligonucleotides, miRNA mimic and inhibitor using Lipofectamine 2000 (Invitrogen), according to the manufacturer's protocol. The full-length MEG3 sequence was cloned into pcDNA3.1 (+) expression vector (GenePharma). For the RNAi-mediated knockdown of MEG3, GenePharma provided 3 different siRNAs against MEG3 (Table S2). MiRNA mimic and inhibitor for miR-21-5p were purchased from GenePharma too. Cells were harvested 48 hours after transfection then subjected to qRT-PCR analyses, western blot or functional assays.
Colony formation assay
For the colony formation assay, a total of 1000 cells were plated into each well of a 6-well plate and maintained in proper media containing 10 %FBS for 7 days, during which the medium was replaced every 3 d. Colonies were then fixed with 10 % formaldehyde for 15 minutes and stained with 1.0 % crystal violet for 5 minutes. The colony formation was determined by counting the number of stained colonies. Triplicate wells were measured in each treatment group.
Cell proliferation assay
Cell proliferation was assayed by the Cell Counting Kit-8 (CCK-8) assay (Dojindo) according to the manufacturer's protocol. A total of approximately 5 × 103 transfected cells were plated in 96-well plates in triplicate and cultured in 90μL medium containing 10% FBS. Then 10 μL CCK-8 solution was added to each well and incubated for 2 hours at 37°C at each indicated time points. Absorbance was then recorded at 450 nm using Multifunctional microplate reader SpectraMax M5 (Molecular Devise, California, USA). Each experiment was repeated 3 times, and the data represent the mean of all measurements. The cell proliferation curves were plotted using the absorbance at each time point.
Apoptosis analysis
At 48 hours post transfection, cells were harvested by trypsinization and washed with PBS twice. Then the cells were resuspended in binding buffer and stained with Annexin V and PI (Annexin V-FITC Apoptosis Detection Kit, eBioscience) for 15 min in the dark at room temperature, according to the manufacturer's recommendations. The stained cells were examined by BD FACSCalibur flow cytometer (BD Biosciences, California, USA) equipped with Cell Quest software (BD Biosciences). The cells were categorized into early apoptotic cells, late apoptotic cells, dead cells, and viable cells. The relative ratio of early apoptotic cells were counted for further comparisons. All of the samples assayed were in triplicates.
Statistical analyses
Statistical analysis was performed using the SPSS software package (version 13.0) and GraphPad Prism Software. The significance of differences between 2 groups was estimated using the Student's t-test. The data is shown as the mean ± SEM from at least 3 independent experiments. All of the p-values were 2-sided and p < 0.05 was considered to be statistically significant.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
KCBT_S_1108496.zip
Download Zip (2.2 MB)Acknowledgments
The Project Supported by National Natural Science Foundation of China (Grant No.81572575), Key Project of Guangdong Natural Science Foundation (Grant No.S2012020010913), Guangdong Natural Science Foundation (Grant No.2014A030313109) and The PhD Program Foundation of the China Education Ministry (Grant No.20120171110097).
References
- Forouzanfar MH, Foreman KJ, Delossantos AM, Lozano R, Lopez AD, Murray CJ, Naghavi M. Breast and cervical cancer in 187 countries between 1980 and 2010: a systematic analysis. Lancet 2011; 378:1461–84; PMID:21924486; http://dx.doi.org/10.1016/S0140-6736(11)61351-2
- Tewari KS, Sill MW, Long HJ 3rd, Penson RT, Huang H, Ramondetta LM, Landrum LM, Oaknin A, Reid TJ, Leitao MM, et al. Improved survival with bevacizumab in advanced cervical cancer. N Engl J Med 2014; 370(8):734–43; PMID:24552320; http://dx.doi.org/10.1056/NEJMoa1309748
- Zhao YB, Wang JH, Chen XX, Wu YZ, Wu Q. Values of three different preoperative regimens in comprehensive treatment for young patients with stage Ib2 cervical cancer. Asian Pac J Cancer Prev 2012; 13(4):1487–9; PMID:22799353; http://dx.doi.org/10.7314/APJCP.2012.13.4.1487
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell 2009; 136(4):629–41; PMID:19239885; http://dx.doi.org/10.1016/j.cell.2009.02.006
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009; 458(7235):223–7; PMID:19182780; http://dx.doi.org/10.1038/nature07672
- Lujambio A, Portela A, Liz J, Melo SA, Rossi S, Spizzo R, Croce CM, Calin GA, Esteller M. CpG island hypermethylation-associated silencing of non-coding RNAs transcribed from ultraconserved regions in human cancer. Oncogene 2010; 29(48):6390–401; PMID:20802525; http://dx.doi.org/10.1038/onc.2010.361
- Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev 2009; 23(13):1494–504; PMID:19571179; http://dx.doi.org/10.1101/gad.1800909
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A 2009; 106(28):11667–72; PMID:19571010; http://dx.doi.org/10.1073/pnas.0904715106
- Lu KH, Li W, Liu XH, Sun M, Zhang ML, Wu WQ, Xie WP, Hou YY. Long non-coding RNA MEG3 inhibits NSCLC cells proliferation and induces apoptosis by affecting p53 expression. BMC Cancer 2013; 13:461; PMID:24098911; http://dx.doi.org/10.1186/1471-2407-13-461
- Sun M, Xia R, Jin F, Xu T, Liu Z, De W, Liu X. Downregulated long noncoding RNA MEG3 is associated with poor prognosis and promotes cell proliferation in gastric cancer. Tumor Biol 2014; 35(2):1065–73; PMID:24006224; http://dx.doi.org/10.1007/s13277-013-1142-z
- Wu XS, Wang XA, Wu WG, Hu YP, Li ML, Ding Q, Weng H, Shu YJ, Liu TY, Jiang L, et al. MALAT1 promotes the proliferation and metastasis of gallbladder cancer cells by activating the ERK/MAPK pathway. Cancer Biol Ther 2014; 15(6):806–14; PMID:24658096; http://dx.doi.org/10.4161/cbt.28584
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116(2):281–97; PMID:14744438; http://dx.doi.org/10.1016/S0092-8674(04)00045-5
- Yao T, Lin Z. MiR-21 is involved in cervical squamous cell tumorigenesis and regulates CCL20. Biochim Biophys Acta 2012; 1822(2):248–60; PMID:22001440; http://dx.doi.org/10.1016/j.bbadis.2011.09.018
- Yao Q, Xu H, Zhang QQ, Zhou H, Qu LH. MicroRNA-21 promotes cell proliferation and down-regulates the expression of programmed cell death 4 (PDCD4) in HeLa cervical carcinoma cells. Biochem Biophys Res Commun 2009; 388(3):539–42; PMID:19682430; http://dx.doi.org/10.1016/j.bbrc.2009.08.044
- Shishodia G, Verma G, Srivastava Y, Mehrotra R, Das BC, Bharti AC. Deregulation of microRNAs Let-7a and miR-21 mediate aberrant STAT3 signaling during human papillomavirus-induced cervical carcinogenesis: role of E6 oncoprotein. BMC Cancer 2014; 14:996; PMID:25539644; http://dx.doi.org/10.1186/1471-2407-14-996
- Liu S, Song L, Zhang L, Zeng S, Gao F. miR-21 modulates resistance of HR-HPV positive cervical cancer cells to radiation through targeting LATS1. Biochem Biophys Res Commun 2015; 459(4):679–85; PMID:25769949; http://dx.doi.org/10.1016/j.bbrc.2015.03.004
- Collins LJ. The RNA infrastructure: an introduction to ncRNA networks. Adv Exp Med Biol 2011; 722: 1–19; PMID:21915779; http://dx.doi.org/10.1007/978-1-4614-0332-6_1
- Wang P, Liu YH, Yao YL, Li Z, Li ZQ, Ma J, Xue YX. Long non-coding RNA CASC2 suppresses malignancy in human gliomas by miR-21. Cell Signal 2015; 27(2):275–82; PMID:25446261; http://dx.doi.org/10.1016/j.cellsig.2014.11.011
- Zhang Z, Zhu Z, Watabe K, Zhang X, Bai C, Xu M, Wu F, Mo YY. Negative regulation of lncRNA GAS5 by miR-21. Cell Death Differ 2013; 20(11):1558–68; PMID:23933812; http://dx.doi.org/10.1038/cdd.2013.110
- Qin R, Chen Z, Ding Y, Hao J, Hu J, Guo F. Long non-coding RNA MEG3 inhibits the proliferation of cervical carcinoma cells through the induction of cell cycle arrest and apoptosis. Neoplasma 2013; 60(5):486–92; PMID:23790166; http://dx.doi.org/10.4149/neo_2013_063
- Leucci E, Patella F, Waage J, Holmstrøm K, Lindow M, Porse B, Kauppinen S, Lund AH. microRNA-9 targets the long non-coding RNA MALAT1 for degradation in the nucleus. Sci Rep 2013; 3:2535; PMID:23985560; http://dx.doi.org/10.1038/srep02535
- Wang Y, Xu Z, Jiang J, Xu C, Kang J, Xiao L, Wu M, Xiong J, Guo X, Liu H. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev Cell 2013; 25(1):69–80; PMID:23541921; http://dx.doi.org/10.1016/j.devcel.2013.03.002
- Faghihi MA, Zhang M, Huang J, Modarresi F, Van der Brug MP, Nalls MA, Cookson MR, St-Laurent G 3rd, Wahlestedt C. Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol 2010; 11(5):R56; PMID:20507594; http://dx.doi.org/10.1186/gb-2010-11-5-r56
- Keniry A, Oxley D, Monnier P, Kyba M, Dandolo L, Smits G, Reik W. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol 2012; 14(7):659–65; PMID:22684254; http://dx.doi.org/10.1038/ncb2521
- Zhou Y, Zhong Y, Wang Y, Zhang X, Batista DL, Gejman R, Ansell PJ, Zhao J, Weng C, Klibanski A. Activation of p53 by meg3 non-coding RNA. J Biol Chem 2007; 282(34):24731–42; PMID:17569660; http://dx.doi.org/10.1074/jbc.M702029200
- Papagiannakopoulos T, Shapiro A, Kosik KS. MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res 2008; 68(19)8164–72; PMID:18829576; http://dx.doi.org/10.1158/0008-5472.CAN-08-1305
- Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem 2008; 283(2)1026–33; PMID:17991735; http://dx.doi.org/10.1074/jbc.M707224200