ABSTRACT
Malignant mesothelioma (MM) is an aggressive tumor arising from mesothelial linings of the serosal cavities. Pleural space is the most common site, accounting for about 80% of cases, while peritoneum makes up the majority of the remaining 20%. While histologically similar, tumors from these sites are epidemiologically and clinically distinct and their attribution to asbestos exposure differs. We compared DNA array-based findings from 48 epithelioid peritoneal MMs and 41 epithelioid pleural MMs to identify similarities and differences in copy number alterations (CNAs). Losses in 3p (BAP1 gene), 9p (CDKN2A) and 22q (NF2) were seen in tumors from both tumor sites, although CDKN2A and NF2 losses were seen at a higher rate in pleural disease (p<0.01). Overall, regions of copy number gain were more common in peritoneal MM, whereas losses were more common in pleural MM, with regions of loss containing known tumor suppressor genes and regions of gain encompassing genes encoding receptor tyrosine kinase pathway members. Cases with known asbestos causation (n = 32 ) were compared with those linked to radiation exposure (n = 9 ). Deletions in 6q, 14q, 17p and 22q, and gain of 17q were seen in asbestos-associated but not radiation-related cases. As reported in post-radiation sarcoma, gains outnumbered losses in radiation-associated MM. The patterns of genomic imbalances suggest overlapping and distinct molecular pathways in MM of the pleura and peritoneum, and that differences in causation (i.e., asbestos vs. radiation) may account for some of these site-dependent differences
Abbreviations
| MM | = | Malignant mesothelioma |
| SEER | = | Surveillance, Epidemiology and End Results |
| SNP | = | Single nucleotide polymorphism |
| DNA | = | DNA |
| CNV | = | copy number variation |
| CGH | = | comparative genomic hybridization |
| miRNA | = | microRNA |
| COSMIC | = | Catalog of somatic mutations in Cancer |
| TSG | = | tumor suppressor gene |
| RTK | = | receptor tyrosine kinase. |
Introduction
Malignant mesothelioma (MM) is a malignancy arising from the mesothelial cells lining serosal cavities of the thorax and abdomen. MM is a rare but aggressive cancer whose 2009 incidence was 9.6 per million in the United States and whose 5-year survival is about 8% [Surveillance, Epidemiology, and End Results (SEER) Program (www.seer.cancer.gov) Research Data 1973–2009]. Pleural MM is the most common form, accounting for roughly 80% of cases, while peritoneal MM accounts for the majority of the remaining 20%. Citation1 Other sites that can be affected include the pericardium and the tunica vaginalis of the testis. Citation2
The histology of MM is varied, but it is broadly divided into 2 patterns, epithelioid and sarcomatoid, with mixtures of each designated as biphasic. These patterns can be seen in both pleural and peritoneal sites, although the rate of biphasic and sarcomatoid disease is lower in the peritoneum. Peritoneal MM occurs in younger patients, and while pleural disease occurs much more commonly in men, peritoneal MM has a lower male-to-female ratio. Pleural MM has a relatively low 5-year survival, even for the more favorable epithelioid histology, but in contrast it has been observed that a subset of epithelioid peritoneal MM patients have prolonged survival following aggressive therapy. Collectively, these observations raise the possibility that epithelioid peritoneal MM represents a morphologically similar, but biologically distinct, disease.
Pleural MM is commonly associated with asbestos, with an attribution of 70%, including co-habitant exposure, whereas peritoneal MM has a much weaker link to asbestos (30%), with only primary exposure increasing risk. Citation3 Therefore, not all cases of MM are attributable to asbestos, and abdominal disease is less likely to be asbestos attributed. Other recognized causal factors for MM include erionite, occupational radiation exposure and medical radiation therapy. Citation4,5 Studies have shown a statistically significant increase in the number of MM cases following radiation therapy for breast cancer, testicular cancer, Hodgkin's lymphoma, and non-Hodgkin's lymphoma. Citation6-9
Based on these observations, we sought to study a group of epithelioid peritoneal MM for CNAs using high density single nucleotide polymorphism (SNP) arrays and compare these data with those obtained in a group of pleural-derived epithelioid MMs to capture possible differences in tumor biology connected with genomic imbalances (i.e., copy number gains, amplifications and deletions). In addition to site-associated differences, we examined differences related to annotated causation in these cases.
Results
Demographics
Of the 48 epithelioid peritoneal MM patients seen at Columbia University, 12 were attributed to asbestos based on strong exposure history, and 7 cases were attributed to radiation exposure. Two additional post radiation epithelioid MMs of the pleura were also studied. The 9 radiation-attributed patients included 3 with prostate cancer, 2 with breast cancer, 1 each with testicular cancer, colon cancer, lymphoma or squamous carcinoma. The interval between radiation therapy and MM diagnosis was known in 7 of 9 patients and exceeded 10 y (range: 10–25 years). For comparison, we selected 38 pleural epithelioid MM cases reported by Memorial Sloan-Kettering Cancer Center (MSKCC). Of these, 20 were annotated as related to asbestos exposure. The demographic information on all 89 patients is summarized in .
Table 1. Demographics of epithelioid mesothelioma cases by tumor site and data source.
Comparison of genomic imbalances in peritoneal vs. pleural MM
The CNAs in peritoneal epithelioid MM seen in more than one third of cases are summarized in and Table S1. These regions include several Cancer Census genes, notably gains in PIK3CA, CCND1, HRAS, and EGFR and losses of 3p generally encompassing BAP1, SETD2 and PBRM1.
Figure 1. Overview of copy number alterations (CNAs) in epithelioid peritoneal MM. Peaks represented in blue to the right of the chromosome are gains, while peaks in red to the left depict losses. Areas with peaks in over 33% of cases are indicated by an asterisk (except for a broad area in chromosome 14 with a bracket), and labeled with a Cancer Census gene if present in that region.

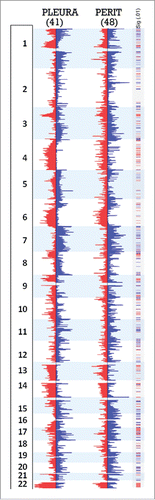
The frequency of copy number gains and losses across the genome was compared between epithelioid MMs of the peritoneum (48 cases) and pleura (41 cases; 38 from MSKCC and 3 from Columbia) (). The peritoneal and pleural tumor sets showed similar recurrent genomic imbalances, although their frequencies differed between the 2 sites of tumor origin. The general overlap in frequency of gains and losses, despite different sites and different copy number platforms, reinforces the overall similarity in epithelioid MM of the pleura and peritoneum. For example, recurrent losses in chromosome arms 3p, 9p and 22q in the genomic regions containing the tumor suppressor genes (TSGs) BAP1, CDKN2A and NF2, respectively, were seen in both tumor groups, albeit at different rates.
Figure 2. Frequency of CNAs in peritoneal and pleural MM. DNA copy number frequency output from Nexus Biodiscovery was overlapped to shows relative rates of alterations by tumor site. Regions in red and blue overlap between both conditions, while region in light blue and pink reflect higher frequency of gain or loss, respectively, in the pleural site. Areas in gray reflect regions of increased frequency of gain or loss in peritoneal site, identified as gains to the right and losses to the left of the chromosomal ideogram. Regions of increased frequency in peritoneal disease with Cancer Census genes from are marked by an asterisk.
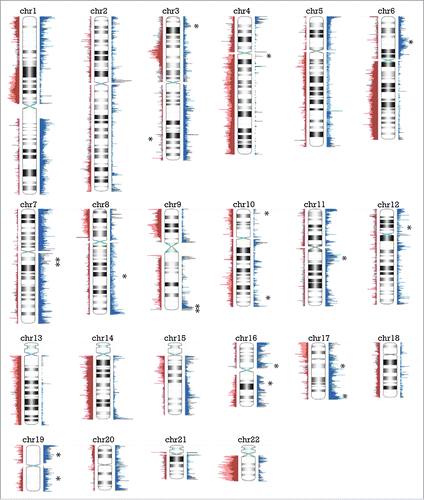
Table 2. Cancer related genes within regions of differential gains and losses in epithelioid malignant mesothelioma (MM), by tumor site.
However, differences were also noted, and these too are highlighted by . Comparison of these 2 groups revealed regions of copy number differences at a percent difference threshold of >33 % and a p value of <0 .01, as shown in and summarized in Table S2. Overall, regions of loss were more common in pleural MM than in peritoneal MM. In regions of loss common to both sites, a higher frequency of loss was seen in pleural MM, most notably in 3p, 4q, 8p, 9p, 15p, 17p and 22q. Gains, when present, were seen more frequently in peritoneal MM, including regions of 3q, 7q, 8p, 9p, 16p and 20q.
Figure 3. Significant regions of gains and losses in pleural and peritoneal MM. Frequency data of CNAs for the 41 pleural and 48 peritoneal epithelioid MMs are shown, with regions significant to p <0.01 and a difference threshold greater than 33% flagged in the column labeled Sig (.01). Red segments in the significance bar depict regions of significantly different incidence of chromosomal losses between the 2 disease sites; blue segments equal significantly different sites of chromosome gains.
Cancer related genes within regions of differential gains and losses in epithelioid MM, by tumor site, are summarized in . Losses in regions containing NF2 and MKL1 in 22q, CDKN2a in 9p and BAP1, SETD2 and PBRM1 in 3p, while identified in peritoneal cases, were seen more frequently in pleural disease. Loss of the tumor suppressor locus CDKN2A, which encodes both the cyclin dependent kinase inhibitor p16INK4A and a second tumor suppressor, p14ARF, was observed at a considerably lower frequency in peritoneal tumors than in pleural MM. Notably, however, peritoneal tumors showed frequent gains of the cyclin D1 gene, CCND1, which can act as an oncogene. Loss of CDKN2A and gain of CCND1 can have the same net effect, i.e., deregulation of the Rb pathway to promote G1 S cell cycling.
Other regions of more frequent loss in pleural MM included TSGs such as TRIM33, UBE4B, MLL3, WRN, BMPR1A, SUFU, PTPN11, SUZ12, ASXL1, SMARCB1, EP300, CHEK2, MYH9 and PATZ1. Regions of more frequent gain in peritoneal MM included the oncogenes RAF1, PIK3CA, CCND3, KIT, TFEB, EGFR, RUNX1T1, RALGDS, FGFR2, CCND1, KRAS, RARA and AKT2. Tumor site-relative losses in peritoneal MM were uncommon and did not contain any known TSGs, while relative gains in pleural disease included regions encompassing the oncogenes KDM5A and MYC and the pro-tumoral gene DVL1.
CNAs by exposure group
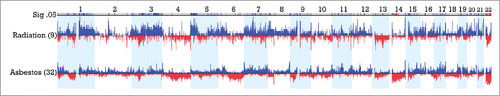
Examination of the combined dataset of epithelioid MM from Columbia and MSKCC (total 89 patients) revealed 9 patients with a documented history of medical radiation exposure for therapy of a prior malignancy and 32 patients with clearly documented asbestos exposure. Comparison of frequency of CNAs revealed several regions of significant difference between the 2 groups, with difference threshold of 33% and p<0.05, as summarized in and Table S3. Multiple regions of gain were seen in the radiation-exposed group, including regions of chromosome arms 1q, 3p, 3q and 5p, while regions of loss predominated in asbestos-related disease, including multiple regions in 14q (14q11.2, 14q12, 14q13.1-q13.2 and 14q21.1-q22.1) and 22q, as well as smaller regions in 17p and 6q (6q16.3 and 6q21-q25.1). Recurrent sites of gain in 17q21.33 and 17q22-q24.2 were also seen in the asbestos-related cases. Of note, losses in 22q11.1-q11.21, 22q13.2-q13.31, 14q, 6q21–25.1 and gains in 17q were each seen in 0% (0 of 9) radiation cases compared to 37.5%, 45.4%, 44.6%, 41.3% and 47.7% of asbestos related cases, respectively.
Figure 4. Significant regions of gains (blue) and losses (red) in radiation- associated versus asbestos-associated epithelioid MM. CNAs between 9 post-radiation MM cases and 32 post-asbestos cases are displayed with regions significant to p <0.05 and a difference threshold greater than 33% flagged in the upper row labeled Sig .05. Note the elevated proportion of radiation-exposed MMs with extended regions of chromosomal gain in 1q, 3q and 5p relative to asbestos-exposed tumors. Additionally, there is an elevated proportion of asbestos-associated MMs with extended or multiple regions of chromosomal loss in 6q, proximal 14q and 22q relative to that observed in post-radiation cases of MM.
summarizes the Cancer Census genes in the areas of differential CNAs between radiation- and asbestos-related cases. The region of 17q gain seen in nearly half of the asbestos-related cases contains COL1A1 and the deleted region in 22q contains the TSG EP300. The region of recurrent loss in 6q contains LATS1, a TSG previously shown to be mutated in some pleural MMs,Citation10 as well as several other putative TSGs, including GRIK2, CDK19, AMD1, and LRP11. The region in 14q shows no cancer census genes, but contains HECTD1, a proposed TSG as well as NFKBIA (14q13.2), a putative TSG whose product inhibits signaling of the
Table 3. Cancer related genes within regions of differential gains and losses in epithelioid MM, by radiation and asbestos causality.
Discussion
Malignant mesothelioma is a tumor that arises from the mesothelial lining of the pleura, peritoneum, tunica vaginalis and pericardium. Pleural disease is the most common, followed by peritoneal disease. While there are a variety of growth patterns in MM, these can be broadly characterized into epithelioid, biphasic and sarcomatoid, which are seen in all sites of origin of MM.
While tumors arising from different sites can have similar morphology, peritoneal MM has some unique characteristics. In a review of published series, peritoneal MM patients had a median age of 50 and male-to-female ratio of 1.3 to 1.Citation12 This is a younger average age and a larger proportion of female patients than in pleural disease, which has an age peak closer to age 70Citation13 and male-to-female ratio of 4:1.Citation14,15 Epithelioid histology is the most common subtype in peritoneal disease, as in pleural disease, but the rate of biphasic and sarcomatoid disease is lower in the peritoneal site (~15%), with few pure sarcomatoid tumors.Citation12 Another major difference is the attribution to asbestos exposure, which in pleural disease is nearly 80%Citation16; in peritoneal disease, the rate is lower, closer to 30%. One of the most important differences is the median survival of 54 months and 5-year survival of 47% in peritoneal MM. In contrast, the 5-year survival of pleural MM is just under 10%, with a median survival of about 1 y.Citation17
These demographic and biologic differences have not been previously explored at the DNA copy number level, as the focus of large CNA series has been on pleural disease. Since some differences in CNA distribution may be attributed to histologic differences (which are known to impact survival in MM), we focused on epithelioid MM only. First summarizing data in peritoneal MM, we found losses in NF2, BAP1 and CDKN2A, but only BAP1 loss was seen in more than 33% of cases. The finding of frequent co-deletion of BAP1, SETD2 and PBRM1 is of uncertain significance given the proximity of these genes to each other in chromosome band 3p21; however the mutational profile of these genes in renal clear cell carcinoma and their impact on chromosome remodeling in renal carcinogenesis suggest a potential parallel in peritoneal MM.Citation18 In addition, increases in copy number of oncogenes such as EGFRCitation19 and PIK3CACitation20 may be reflected in increased activation of associated pathways implicated in MM previously, although in one series, EGFR copy number was not significantly associated with EGFR immunoreactivity.Citation21
Direct comparison of peritoneal MM and pleural MM revealed extensive areas of overlapping recurrent genomic gains and losses, which underscores the CNA similarities between these 2 tumor sites. The top 3 encountered alterations in cancer census genes in pleural MM, when examined using the Catalog of Somatic Mutations in Cancer (COSMIC) database, are CDKN2A, NF2 and BAP1. For all 3 genes, the alteration most commonly consists of loss of part or all of the affected gene; however, for NF2 and BAP1, a significant proportion of cases involve point mutations and indels. While genomic losses encompassing these genes are also seen in peritoneal disease, the rate of loss is lower, and this difference is significant for CDKN2A and NF2, but not for BAP1.
Beyond the similarities, there were significant differences as well. Overall, pleural MMs had more frequent rates of genomic loss, whereas peritoneal MM had more regions of chromosomal gain. Examination of regions of loss in pleural disease revealed many sites of known and putative TSGs, while the few regions of loss in peritoneal disease did not. However, recurrent sites of chromosomal gain in peritoneal MM contained several genes known to encode components of important receptor tyrosine kinase (RTK) pathways, which in prior reports have been shown to be activated in MM. In addition to the previously mentioned EGFR and PIK3CA, the importance of KRAS and AKT2 expression has been reported, albeit in only certain MM subsets.Citation22,23 While fewer genomic gains were seen in pleural disease, one site of gain included the MYC gene, which has been reported to have increased copy number and pro-tumoral activity in pleural MM.Citation24
Examination of CNA profiles in connection with disease causation revealed further differences within MM subgroups. Post-radiation MMs, when compared to asbestos-induced MMs, showed multiple regions of copy number gain. The asbestos-induced tumors frequently showed losses in 6q, 14q, 17p and 22q as well as gains in 17q, and this was seen in both pleural and peritoneal tumors. This supports the hypothesis that a subset of peritoneal MMs is genetically similar to their pleural counterparts, and this may correspond to the asbestos causation group.
Our analysis included 32 pleural and peritoneal MM patients with asbestos-attributed disease and 9 with radiation-attributed disease. A comparison of DNA copy number imbalances between these 2 groups revealed regions of loss at 14q11.2–13.3 and 14q21.1–22.1 in up to 50% of MM patients with asbestos exposure compared to 0% of patients whose disease was thought to be connected with radiation exposure. Similar regions, 14q11.2–13.2 and 14q22.3–24.3, have been previously documented in a loss of heterozygosity (LOH) analysis of 14q in pleural MM.Citation25 Recurrent LOH at 14q11-q13 has also been reported in head and neck cancer, lung carcinoma, and nasopharyngeal carcinoma,Citation26-28 leading us to hypothesize that this region harbors one or more TSGs. The putative TSG NFKBIA, located at 14q13.2, is an intriguing possibility.Citation11 Deletion, rather than mutation, of NFKBIA promotes tumorigenesis in glioblastomas that do not have alterations of EGFR. Moreover, restoration of the expression of NFKBIA attenuated the malignant phenotype and increased the vulnerability to chemotherapy of glioblastoma cells cultured from tumors with NFKBIA deletion. It also reduced the viability of glioblastoma cells with EGFR amplification but not of cells with normal gene dosages of both NFKBIA and EGFR. A promising candidate in the 14q22.3–24.3 region is HIF1A, which has been implicated in renal cell carcinoma.Citation29 This gene showed no copy number losses in the radiation-related MM cases, but was seen in 17% of the asbestos-related MM cases, a difference that did not meet our difference threshold or p-value cutoff. Loss of 14q is also seen in breast carcinomas that harbor BRCA2 mutations,Citation30 suggesting that progressive loss of tumor suppressor function involving 14q requires more than one locus for cancer progression; this may be a necessary oncogenic event in some tumors but not a sufficient one.
The regions of loss in chromosome arms 6q and14q in asbestos-related MM overlap in peritoneal and pleural tumors and, thus, are independent of disease site and may represent a common link between asbestos exposure and MM. In part this may explain some of the aforementioned differences seen in pleural and peritoneal disease in that only the asbestos-attributed cases have genetic similarity. If correct, this indicates that a subset of peritoneal MMs has similar causation and behavior as pleural tumors, but that a higher proportion of non-asbestos-related peritoneal cases results in the observed epidemiologic and molecular features that are distinct from those of pleural MM.
Likewise, it may be that a subset of pleural MM are genetically similar to peritoneal disease when radiation is thought to be the cause. Our series has only 2 post-radiation pleural MMs, and neither of these cases showed losses of 6q or 14q. In addition, it has been noted that patients with pleural MM secondary to radiation are younger and have a better overall survival than other pleural MM,Citation31 clinical features also observed in peritoneal MM.
A variety of post-radiation human tumors have been shown to exhibit differences in copy number when compared to counterparts not connected with such an exposure. In survivors of atomic bomb irradiation, significant differences in genomic instability have been described in subsequent breast carcinomas, with post-radiation tumors tending to show more copy number gains than losses, when compared to a control group of breast cancers.Citation32 In another series, characterization of genomic alterations in radiation-associated breast cancer among 32 childhood cancer survivors revealed that 23 (72%) showed an amplifier phenotype,Citation33 with the remainder showing either simple alterations or complex mixtures of gains and losses. In post-radiation sarcomas, increased CNAs, mostly gains, have been described in most subtypes, except osteosarcoma. Specifically, gains of chromosomal regions 7q11.2-q21, 7q22, 7p15-pter and 8q23-qter were associated with radiation exposure and a poor prognosis.Citation34 In thyroid cancer, largely studied in post-radiation Chernobyl cohorts, gains again exceeded losses,Citation35-37 particularly in regions of chromosomes 1, 2, 5, 6p, 7, 9p, 12q, 13, 14, 16p, 17q, 20q, 21 and 22. RET translocations and amplifications were also described. Thus, like the post-radiation MMs in our investigation, most post-radiation tumor types are characterized by an increased prevalence of DNA copy number gains.
Radiation exposure has been associated with induction of CNAs in various cell types. For example, in vitro studies of fibroblasts have shown an induction of CNAs after radiation exposure,Citation38 and amplification of MYC was observed in rat skin tumors induced by radiation.Citation39 When lymphoma cells are exposed to radiation in vitro, amplification of the MCL1 gene and a preponderance of copy number gains in 1q, 2p, 3q, 6p, 7, 11q, 12p and 16 have been reported.Citation40
In summary, peritoneal and pleural MMs show a similar overall distribution of CNAs, although peritoneal tumors display a higher frequency of gains and lower frequency of losses than their pleural counterparts. Losses at sites of TSGs are proportionately more common in pleural MM, whereas gains tend to occur more frequently in regions encompassing genes encoding RTK pathway members in peritoneal MM. More striking regional chromosomal differences are seen when focusing on causation, with deletions in chromosome arms 6q, 14q, 17p and 22q and gains in 17q seen in connection with asbestos causality regardless of disease site.
Materials and methods
Columbia university MM samples
We identified 48 cases of snap frozen, epithelioid peritoneal MM tissue and 3 cases of pleural MM tissue from the Columbia University Cancer Center Tissue Bank with Columbia IRB approval. Frozen sections were cut at 8-micron thickness, and manual needle dissection of tumor-rich areas was performed with an 18-gauge needle on an Olympus microscope at 40X magnification and collected by vacuum suction into a pipette tip. For each sample, a total of 500 ng of DNA was collected and subjected to DNA copy number analysis using an Affymetrix Genome Wide Human SNP Array 6.0, which includes more than 906,600 SNP probes and >946,000 oligonucleotide probes for the detection of copy number variation (CNV) (www.affymetrix.com). Sample preparation, hybridization and scanning were performed using GeneChip® Instrument System hardware according to the manufacturer's specifications (Affymetrix, Santa Clara, CA). The intensities of both SNP and CNV probes were used to determine segments that varied in copy number. Data analysis was performed using Nexus Copy Number Software, Version 6.0 (BioDiscovery, Hawthorne, CA). The FASST2 Segmentation algorithm was used to make copy number calls (a Hidden Markov Model approach). The significance threshold for segmentation was set at 5 × 10−7, also requiring a minimum of 3 probes per segment and a maximum probe spacing of 1000 between adjacent probes before breaking a segment. The log ratio thresholds for single copy gain and single copy loss were set at +0.2 and −0.15, respectively. The log ratio thresholds for gain of 2 or more copies and for a homozygous loss were set at +0.7 and −1.1, respectively. Array results were analyzed using Nexus Copy Number 6.0 software for subgroup comparisons studies.
Memorial sloan-kettering cancer center (MSKCC) data
Copy number data were obtained from 53 pleural MM cases previously published by Bott et al. Citation10 (http://cbio.mskcc.org/Public/Ladanyi_lab_mesothelioma_data sets). Specifically, annotated data was downloaded along with Agilent 244K comparative genomic hybridization arrays and imported into a Nexus Copy Number 6.0 experiment using FASST2 Segmentation protocol with a significance threshold of 1 × 10−4, maximum contiguous spacing of 1000 and a minimum of 3 probes per segment. The log ratio thresholds for gain of 2 or more copies and for homozygous loss were set at +0.7 and −1.1, respectively. Altogether, 38 cases of epithelioid type pleural MM were then selected, and the annotated data included asbestos exposure history in 20 cases.
All analyses were performed using Nexus software analysis tools. Factor aggregates based on site of disease (pleural versus peritoneal) and for cases with annotation of suspected causation (radiation vs. asbestos) were displayed and paired comparisons for CNAs were performed. Tracks for genes, microRNA and Cancer Census genesCitation41 were displayed. Cancer Census genes are an ongoing effort by COSMIC/Wellcome Trust Sanger Institute to catalog cancer associated genes.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplemental_Datas.zip
Download Zip (77.1 KB)Funding
The work was supported in part by the Mesothelioma Center at Columbia University Medical (JC salary, 20%), which received support from the Simmons employee foundation. JP and JRT are supported in part by the Fox Chase Cancer Center CCSG Genomics Facility (CA-06927 from the NCI). Work by JRT related to BAP1 is also supported by NIH grant CA-175691
References
- Yang H, Testa JR, Carbone M. Mesothelioma epidemiology, carcinogenesis, and pathogenesis. Curr Treat Options Oncol 2008; 9:147-57; PMID:18709470; http://dx.doi.org/10.1007/s11864-008-0067-z
- Musti M, Kettunen E, Dragonieri S, Lindholm P, Cavone D, Serio G, Knuutila S. Cytogenetic and molecular genetic changes in malignant mesothelioma. Cancer Genet Cytogenet 2006; 170:9-15; PMID:16965949; http://dx.doi.org/10.1016/j.cancergencyto.2006.04.011
- Spirtas R, Heineman EF, Bernstein L, Beebe GW, Keehn RJ, Stark A, Harlow BL, Benichou J. Malignant mesothelioma: attributable risk of asbestos exposure. Occup Environ Med 1994; 51:804-11; PMID:7849863; http://dx.doi.org/10.1136/oem.51.12.804
- Cavazza A, Travis LB, Travis WD, Wolfe JT, 3rd, Foo ML, Gillespie DJ, Weidner N, Colby TV. Post-irradiation malignant mesothelioma. Cancer 1996; 77:1379-85; PMID:8608519; http://dx.doi.org/10.1002/(SICI)1097-0142(19960401)77:7%3c1379::AID-CNCR24%3e3.0.CO;2-Y
- Jasani B, Gibbs A. Mesothelioma not associated with asbestos exposure. Arch Pathol Lab Med 2012; 136:262-7; PMID:22372902; http://dx.doi.org/10.5858/arpa.2011-0039-RA
- Tward JD, Wendland MM, Shrieve DC, Szabo A, Gaffney DK. The risk of secondary malignancies over 30 years after the treatment of non-Hodgkin lymphoma. Cancer 2006; 107:108-15; PMID:16708354; http://dx.doi.org/10.1002/cncr.21971
- Teta MJ, Lau E, Sceurman BK, Wagner ME. Therapeutic radiation for lymphoma: risk of malignant mesothelioma. Cancer 2007; 109:1432-8; PMID:17315168; http://dx.doi.org/10.1002/cncr.22526
- Travis LB, Fossa SD, Schonfeld SJ, McMaster ML, Lynch CF, Storm H, Hall P, Holowaty E, Andersen A, Pukkala E, et al. Second cancers among 40,576 testicular cancer patients: focus on long-term survivors. J Natl Cancer Inst 2005; 97:1354-65; PMID:16174857; http://dx.doi.org/10.1093/jnci/dji278
- Brown LM, Chen BE, Pfeiffer RM, Schairer C, Hall P, Storm H, Pukkala E, Langmark F, Kaijser M, Andersson M, et al. Risk of second non-hematological malignancies among 376,825 breast cancer survivors. Breast Cancer Res Treat 2007; 106:439-51; PMID:17277968; http://dx.doi.org/10.1007/s10549-007-9509-8
- Bott M, Brevet M, Taylor BS, Shimizu S, Ito T, Wang L, Creaney J, Lake RA, Zakowski MF, Reva B, et al. The nuclear deubiquitinase BAP1 is commonly inactivated by somatic mutations and 3p21.1 losses in malignant pleural mesothelioma. Nat Genet 2011; 43:668-72; PMID:21642991; http://dx.doi.org/10.1038/ng.855
- Bredel M, Scholtens DM, Yadav AK, Alvarez AA, Renfrow JJ, Chandler JP, Yu IL, Carro MS, Dai F, Tagge MJ, et al. NFKBIA deletion in glioblastomas. N Engl J Med 2011; 364:627-37; PMID:21175304; http://dx.doi.org/10.1056/NEJMoa1006312
- Baratti D, Kusamura S, Deraco M. Diffuse malignant peritoneal mesothelioma: systematic review of clinical management and biological research. J Surg Oncol 2011; 103:822-31; PMID:21283990; http://dx.doi.org/10.1002/jso.21787
- Larson T, Melnikova N, Davis SI, Jamison P. Incidence and descriptive epidemiology of mesothelioma in the United States, 1999-2002. Int J Occup Environ Health 2007; 13:398-403; PMID:18085053; http://dx.doi.org/10.1179/oeh.2007.13.4.398
- McDonald AD, McDonald JC. Malignant mesothelioma in North America. Cancer 1980; 46:1650-6; PMID:7417959; http://dx.doi.org/10.1002/1097-0142(19801001)46:7%3c1650::AID-CNCR2820460726%3e3.0.CO;2-Y
- Robinson BM. Malignant pleural mesothelioma: an epidemiological perspective. Ann Cardiothorac Surg 2012; 1:491-6; PMID:23977542; http://dx.doi.org/10.3978/j.issn.2225-319X.2012.11.04
- McDonald JC, McDonald AD. The epidemiology of mesothelioma in historical context. Eur Respir J 1996; 9:1932-42; PMID:8880114; http://dx.doi.org/10.1183/09031936.96.09091932
- Howlader NNA, Krapcho M, Garshell J, Miller D, Altekruse SF, Kosary CL, Yu M, Ruhl J, Tatalovich Z, Mariotto A, et al, Cronin KA SEER Cancer Statistics Review, 1975-2012. Bethesda, MD: National Cancer Institute., 2014.
- Liao L, Testa JR, Yang H. The roles of chromatin-remodelers and epigenetic modifiers in kidney cancer. Cancer Genet 2015; 208:206-14; PMID:25873528; http://dx.doi.org/10.1016/j.cancergen.2015.02.008
- Brevet M, Shimizu S, Bott MJ, Shukla N, Zhou Q, Olshen AB, Rusch V, Ladanyi M. Coactivation of receptor tyrosine kinases in malignant mesothelioma as a rationale for combination targeted therapy. J Thorac Oncol 2011; 6:864-74; PMID:21774103; http://dx.doi.org/10.1097/JTO.0b013e318215a07d
- Varghese S, Chen Z, Bartlett DL, Pingpank JF, Libutti SK, Steinberg SM, Wunderlich J, Alexander HR, Jr. Activation of the phosphoinositide-3-kinase and mammalian target of rapamycin signaling pathways are associated with shortened survival in patients with malignant peritoneal mesothelioma. Cancer 2011; 117:361-71; PMID:20839315; http://dx.doi.org/10.1002/cncr.25555
- Enomoto Y, Kasai T, Takeda M, Takano M, Morita K, Kadota E, Iizuka N, Maruyama H, Haratake J, Kojima Y, et al. A comparison of epidermal growth factor receptor expression in malignant peritoneal and pleural mesothelioma. Pathol Int 2012; 62:226-31; PMID:22449226; http://dx.doi.org/10.1111/j.1440-1827.2011.02778.x
- Olut A, Firat P, Ertugrul D, Gungen Y, Emri S. Ras oncoprotein expression in erionite- and asbestos-induced Turkish malignant pleural mesothelioma patients–a pilot study. Respir Med 2001; 95:697-8; PMID:11530960; http://dx.doi.org/10.1053/rmed.2001.1122
- Lin Z, Liu T, Kamp DW, Wang Y, He H, Zhou X, Li D, Yang L, Zhao B, Liu G. AKT/mTOR and c-Jun N-terminal kinase signaling pathways are required for chrysotile asbestos-induced autophagy. Free Radical Biol Med 2014; 72:296-307; PMID:24735948; http://dx.doi.org/10.1016/j.freeradbiomed.2014.04.004
- Riquelme E, Suraokar MB, Rodriguez J, Mino B, Lin HY, Rice DC, Tsao A, Wistuba, II. Frequent coamplification and cooperation between C-MYC and PVT1 oncogenes promote malignant pleural mesothelioma. J Thorac Oncol 2014; 9:998-1007; PMID:24926545; http://dx.doi.org/10.1097/JTO.0000000000000202
- De Rienzo A, Jhanwar SC, Testa JR. Loss of heterozygosity analysis of 13q and 14q in human malignant mesothelioma. Genes Chromosomes Cancer 2000; 28:337-41; PMID:10862040; http://dx.doi.org/10.1002/1098-2264(200007)28:3%3c337::AID-GCC12%3e3.0.CO;2-B
- Beder LB, Gunduz M, Ouchida M, Fukushima K, Gunduz E, Ito S, Sakai A, Nagai N, Nishizaki K, Shimizu K. Genome-wide analyses on loss of heterozygosity in head and neck squamous cell carcinomas. Lab Invest 2003; 83:99-105; PMID:12533690; http://dx.doi.org/10.1097/01.LAB.0000047489.26246.E1
- El-Rifai W, Sarlomo-Rikala M, Andersson LC, Miettinen M, Knuutila S. High-resolution deletion mapping of chromosome 14 in stromal tumors of the gastrointestinal tract suggests two distinct tumor suppressor loci. Genes Chromosomes Cancer 2000; 27:387-91; PMID:10719369; http://dx.doi.org/10.1002/(SICI)1098-2264(200004)27:4%3c387::AID-GCC8%3e3.0.CO;2-C
- Abujiang P, Mori TJ, Takahashi T, Tanaka F, Kasyu I, Hitomi S, Hiai H. Loss of heterozygosity (LOH) at 17q and 14q in human lung cancers. Oncogene 1998; 17:3029-33; PMID:9881705; http://dx.doi.org/10.1038/sj.onc.1202230
- Kroeger N, Klatte T, Chamie K, Rao PN, Birkhauser FD, Sonn GA, Riss J, Kabbinavar FF, Belldegrun AS, Pantuck AJ. Deletions of chromosomes 3p and 14q molecularly subclassify clear cell renal cell carcinoma. Cancer 2013; 119:1547-54; PMID:23335244; http://dx.doi.org/10.1002/cncr.27947
- Rouault A, Banneau G, Macgrogan G, Jones N, Elarouci N, Barouk-Simonet E, Venat L, Coupier I, Letouze E, de Reynies A, et al. Deletion of chromosomes 13q and 14q is a common feature of tumors with BRCA2 mutations. PloS one 2012; 7:e52079; PMID:23284877; http://dx.doi.org/10.1371/journal.pone.0052079
- Chirieac LR, Barletta JA, Yeap BY, Richards WG, Tilleman T, Bueno R, Baldini EH, Godleski J, Sugarbaker DJ. Clinicopathologic characteristics of malignant mesotheliomas arising in patients with a history of radiation for Hodgkin and non-Hodgkin lymphoma. J Clin Oncol 2013; 31:4544-9; PMID:24248693; http://dx.doi.org/10.1200/JCO.2013.49.9616
- Oikawa M, Yoshiura K, Kondo H, Miura S, Nagayasu T, Nakashima M. Significance of genomic instability in breast cancer in atomic bomb survivors: analysis of microarray-comparative genomic hybridization. Radiation oncology 2011; 6:168; PMID:22152285; http://dx.doi.org/10.1186/1748-717X-6-168
- Yang XR, Killian JK, Hammond S, Burke LS, Bennett H, Wang Y, Davis SR, Strong LC, Neglia J, Stovall M, et al. Characterization of genomic alterations in radiation-associated breast cancer among childhood cancer survivors, using comparative genomic hybridization (CGH) arrays. PloS one 2015; 10:e0116078; PMID:25764003; http://dx.doi.org/10.1371/journal.pone.0116078
- Tarkkanen M, Wiklund TA, Virolainen MJ, Larramendy ML, Mandahl N, Mertens F, Blomqvist CP, Tukiainen EJ, Miettinen MM, Elomaa AI, et al. Comparative genomic hybridization of postirradiation sarcomas. Cancer 2001; 92:1992-8; PMID:11745275; http://dx.doi.org/10.1002/1097-0142(20011001)92:7%3c1992::AID-CNCR1719%3e3.0.CO;2-2
- Stein L, Rothschild J, Luce J, Cowell JK, Thomas G, Bogdanova TI, Tronko MD, Hawthorn L. Copy number and gene expression alterations in radiation-induced papillary thyroid carcinoma from chernobyl pediatric patients. Thyroid 2010; 20:475-87; PMID:19725780; http://dx.doi.org/10.1089/thy.2009.0008
- Kimmel RR, Zhao LP, Nguyen D, Lee S, Aronszajn M, Cheng C, Troshin VP, Abrosimov A, Delrow J, Tuttle RM, et al. Microarray comparative genomic hybridization reveals genome-wide patterns of DNA gains and losses in post-Chernobyl thyroid cancer. Radiation Res 2006; 166:519-31; PMID:16953671; http://dx.doi.org/10.1667/RR0547.1
- Richter H, Braselmann H, Hieber L, Thomas G, Bogdanova T, Tronko N, Zitzelsberger H. Chromosomal imbalances in post-chernobyl thyroid tumors. Thyroid 2004; 14:1061-4; PMID:15650359; http://dx.doi.org/10.1089/thy.2004.14.1061
- Arlt MF, Rajendran S, Birkeland SR, Wilson TE, Glover TW. Copy number variants are produced in response to low-dose ionizing radiation in cultured cells. Environ Mol Mutagen 2014; 55:103-13; PMID:24327335; http://dx.doi.org/10.1002/em.21840
- Garte SJ, Burns FJ, Ashkenazi-Kimmel T, Felber M, Sawey MJ. Amplification of the c-myc oncogene during progression of radiation-induced rat skin tumors. Cancer Res 1990; 50:3073-7; PMID:2185880
- Lyng H, Landsverk KS, Kristiansen E, DeAngelis PM, Ree AH, Myklebost O, Hovig E, Stokke T. Response of malignant B lymphocytes to ionizing radiation: gene expression and genotype. Int J Cancer J Int du Cancer 2005; 115:935-42; PMID:15723354; http://dx.doi.org/10.1002/ijc.20962
- Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR. A census of human cancer genes. Nat Rev Cancer 2004; 4:177-83; PMID:14993899; http://dx.doi.org/10.1038/nrc1299
