ABSTRACT
The transcriptional regulator Krüppel-like factor 4 (KLF4) is decreased in human esophageal squamous cell cancer (ESCC), and Klf4 deletion in mice produces squamous cell dysplasia. Nonetheless the mechanisms of KLF4 downregulation in ESCC and the functions of KLF4 during ESCC development and progression are not well understood. Here, we sought to define the regulation of KLF4 and delineate the stage-specific effects of KLF4 in ESCC. We found that KLF4 expression was decreased in human ESCC and in 8 of 9 human ESCC cell lines. However, by genomic sequencing, we observed no KLF4 mutations or copy number changes in any of 52 human ESCC, suggesting other mechanisms for KLF4 silencing. In fact, KLF4 expression in human ESCC cell lines was increased by the DNA methylation inhibitor 5-azacytidine, suggesting an epigenetic mechanism for KLF4 silencing. Surprisingly, while KLF4 decreased in high-grade dysplasia and early stage tumors, KLF4 increased with advanced cancer stage, and KLF4 expression in ESCC was inversely correlated with survival. Interestingly, KLF4 promoted invasion of human ESCC cells, providing a functional link to the stage-specific expression of KLF4. Taken together, these findings suggest that KLF4 loss is necessary for esophageal tumorigenesis but that restored KLF4 expression in ESCC promotes tumor spread. Thus, the use of KLF4 as a diagnostic and therapeutic target in cancer requires careful consideration of context.
Abbreviations
| KLF4 | = | Krüppel-like factor 4 |
| ESCC | = | esophageal squamous cell cancer |
| SNP | = | single nucleotide polymorphism |
| HDAC | = | histone deacetylase |
Introduction
Esophageal cancer affects nearly half a million people and is the 6th most common cause of cancer death in the world.Citation1,2 Worldwide, more than 80% of esophageal cancers are esophageal squamous cell carcinoma (ESCC).Citation2,3 ESCC arises from a precursor lesion, esophageal squamous cell dysplasia,Citation3 but most patients with ESCC present at an advanced stage, leading to a poor overall 5-year survival of 17.9%.Citation4 Not surprisingly, survival rates for ESCC are highly dependent on the stage at diagnosis with 5-year survival of 40.4% for localized disease but only 4.2% for distant spread. Thus, understanding the mechanisms of ESCC development and progression are critical for disease prevention and therapy.
Molecular targets hold promise for the diagnosis, prognosis, and therapy of ESCC, and a number of genetic and protein alterations have been linked to human ESCC.Citation5-8 Understanding the functions and consequences of these alterations requires careful attention to context, and to date, molecular biomarkers have not been integrated into clinical practice for ESCC.Citation7 In particular, the factors that predispose patients with squamous cell dysplasia to develop ESCC and the changes that lead to metastases from the primary tumor are not known. Thus, unfortunately, little progress has been made to improve the prognosis of patients with ESCC.Citation2
The zinc finger transcription factor Krüppel-like factor 4 (KLF4) is implicated in the pathogenesis of numerous cancers, and KLF4 has important context-dependent functions in cancer,Citation9 including in ESCC cell differentiation and survival.Citation10-12 In mice, Klf4 loss leads to the development of esophageal squamous cell dysplasia, and KLF4 is downregulated in human ESCC suggesting that KLF4 has tumor suppressive functions in ESCC.Citation13-15 Moreover, while Klf4 overexpression in murine esophagus leads to the development of inflammation-mediated ESCC, KLF4 is absent from these tumors,Citation16 suggesting that Klf4 silencing may be required for tumor formation. As such, KLF4 appears to be important for the development of ESCC, although the precise functions of KLF4 during ESCC development and progression and the mechanisms of KLF4 downregulation in ESCC are not well understood.
Here, we sought to define the expression of KLF4 during ESCC progression and delineate the mechanisms of KLF4 silencing during ESCC. To this end, we demonstrate that KLF4 is silenced epigenetically but not genetically in human ESCC. Further, while KLF4 decreases early in the process of esophageal carcinogenesis, KLF4 increases with advanced cancer stage, and KLF4 expression is inversely correlated with ESCC survival. Moreover, using a 3-dimensional spheroid model, we find that KLF4 promotes invasion of human ESCC cells, providing a functional link to the stage-specific expression of KLF4. Taken together, these findings suggest that KLF4 may be a “gatekeeper” that prevents the development of ESCC but that in patients with ESCC, reactivation of KLF4 may have negative consequences.
Results
KLF4 functions as a tumor suppressor in certain contexts,Citation9 and KLF4 loss is associated with a worse prognosis for several cancers including colorectal and gastric carcinoma.Citation17,18 However, in other contexts, KLF4 exhibits behavior more consistent with an oncogene and is negatively associated with patient survival.Citation19-21 To explore the role of KLF4 in ESCC, we initially examined KLF4 expression in human ESCC using Oncomine. We identified 2 datasetsCitation22,23 containing paired samples of ESCC and adjacent normal tissue; in both of these data sets, KLF4 expression was reduced in ESCC compared to adjacent normal (Fig. S1). Interestingly, while KLF4 expression was reduced overall, there was a large distribution of KLF4 levels among the cancers. Next, we examined KLF4 expression in ESCC cell lines and found that KLF4 mRNA was significantly reduced in 8 of 9 ESCC cell lines (). KLF4 was also reduced in human head and neck squamous cell carcinoma cells (Fig. S2), suggesting parallels between KLF4 in ESCC and in other squamous cell cancers.
Figure 1. KLF4 expression was decreased by DNA methylation is ESCC cell lines. (A) Compared to non-transformed EPC1 primary esophageal squamous epithelial cells, 8 of 9 human ESCC cell lines had decreased KLF4 mRNA expression by quantitative real-time PCR. (B) By quantitative real-time PCR, KLF4 increased in the ESCC cell lines TE8 and HCE4 following treatment with the DNA demethylation DNA methylation inhibitor 5-azacytidine (5-AC) but not the HDAC inhibitor trichostatin A (TSA). In TE2 cells, KLF4 was not upregulated by 5-azacytidine or trichostatin A, suggesting another mechanism exists for KLF4 silencing in this cell line.
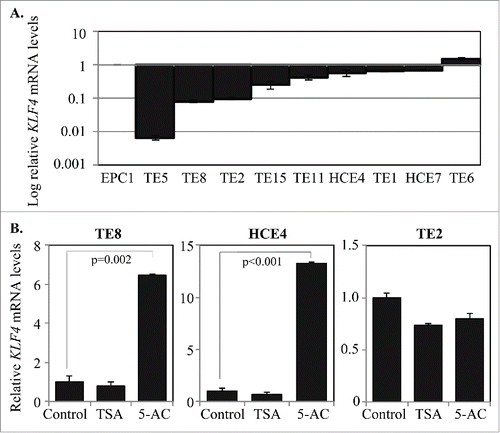
Figure 2. KLF4 is lost in esophageal squamous cell dysplasia and early ESCC but not in advanced ESCC. (A) In normal esophageal squamous epithelium, immunohistochemistry demonstrated brown KLF4 staining predominantly within the suprabasal layer. (B-C) KLF4 staining by immunohistochemistry was nearly absent from esophageal squamous cell dysplasia (B) and early stage ESCC (C). (D) In more advanced ESCC, KLF4 staining was prevalent. Magnification = 100x.
To determine the mechanism of KLF4 reduction in ESCC, we first performed targeted exonic sequencing of the KLF4 gene in 52 patients from China with ESCC. We did not identify KLF4 mutations or copy number changes in any of the cancers, although we did discern several single nucleotide polymorphisms (SNPs) in KLF4 Fig. S3). One of these SNPs (rs2236599), resulting in a G to A change, was identified in 31 of 52 patients with ESCC (0.596), an allele frequency much higher than in the general population (0.140)Citation24; of note, allelic frequency for this SNP is particularly high in East Asians (0.293), in whom ESCC incidence is elevated.Citation2 The KLF4 rs2236599 SNP has been associated with radiotherapy toxicity during breast cancer treatment,Citation25 suggesting that this SNP may have functional consequences. We next investigated whether KLF4 is silenced epigenetically in human ESCC. KLF4 levels were unchanged in the ESCC cell lines TE8, HCE4, and TE2 by treatment with the HDAC inhibitor trichostatin A.Citation26 However, treatment with the DNA methylation inhibitor 5-azacytidineCitation27 increased KLF4 mRNA levels in TE8 and HCE4 cells, suggesting that KLF4 is silenced by hypermethylation in these cell lines (). Interestingly, KLF4 expression in TE2 cells was not altered by either trichostatin A or 5-azacytidine, suggesting that other mechanisms exist for KLF4 downregulation in ESCC.
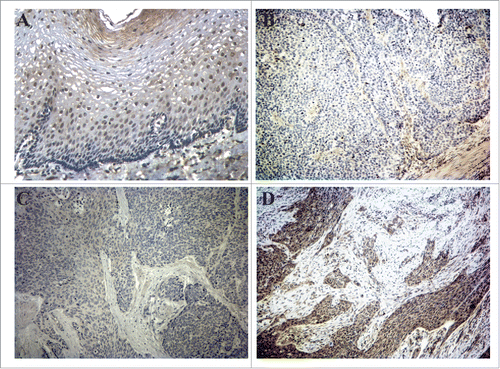
Given the variations of KLF4 expression in human ESCC, we sought to determine whether KLF4 levels in ESCC varied with tumor stage. Esophageal-specific deletion of Klf4 produces squamous cell dysplasia in mice,Citation13 and consistent with these mechanistic findings, KLF4 was markedly decreased in human esophageal dysplasia and in early stage ESCC compared to adjacent normal tissues (). However, KLF4 increased progressively with larger tumor sizes and nodal metastasis such that for more advanced ESCC, KLF4 approached the levels seen in normal tissues (). Moreover, for tumors of similar sizes, KLF4 was significantly higher in those with nodal metastasis than in those without nodal spread. Additionally, levels of KLF4 in ESCC correlated inversely with patient survival (), with a median survival of 46 months for patients with low levels of KLF4 and only 16 months in those with high KLF4 levels.
Figure 3. KLF4 expression in ESCC is associated with advanced cancer stage and decreased patient survival. (A) Compared to adjacent normal tissue, KLF4 expression was decreased in high-grade dysplasia and early-stage ESCC but not in more advanced ESCC. Note that for tumors of the same size, KLF4 levels were higher for those with nodal metastases than those without. (B) Kaplan-Meier survival curves demonstrated poorer survival for patients with high levels of KLF4 compared to those with low or intermediate levels (p < 0.0001).
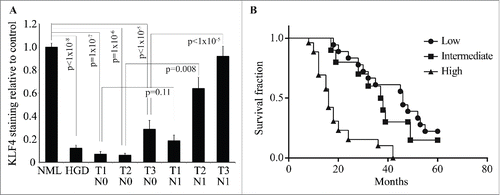
Since KLF4 was elevated in tumors with nodal spread, we hypothesized that KLF4 expression in ESCC cells might contribute to tumor invasion and metastasis. To test this hypothesis, we utilized a 3-dimensional spheroid model which simulates collective cancer cell invasion in vivo.Citation28 Both HCE4 and TE8 ESCC cells had reduced KLF4 expression relative to controls, and when we ectopically expressed KLF4 in these cells, we significantly enhanced tumor cell invasion, with enlarged spheroids relative to controls ( and Fig. S4). Since KLF4 may target p21Waf1/Cip1 and cyclin D1,Citation9 2 cell cycle regulators with effects on tumor invasion,Citation29,30 we examined whether p21Waf1/Cip1 and cyclin D1 were regulated by KLF4 in ESCC. In HCE4 and TE8 ESCC cells, transgenic expression of KLF4 did not significantly alter levels of p21Waf1/Cip1 or cyclin D1 (Fig. S5), suggesting that the effects of KLF4 on tumor cell invasion were not mediated by either p21Waf1/Cip1 or cyclin D1. Overall, KLF4 promotes invasion of ESCC cells and appears to have stage-specific functions in ESCC.
Figure 4. KLF4 promotes ESCC invasion. (A) In 3-dimensional spheroid culture, HCE4 ESCC cells expressing KLF4 invaded more into surrounding matrix that control cells. (B) Indicative of increased cancer cell invasion, HCE4 spheroids that expressed KLF4 were significantly larger at 3 and 7 days, compared to control spheroids (n = 4 for each experimental condition at each time point).
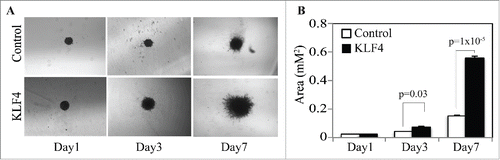
Figure 5. A model for the role of KLF4 during esophageal squamous cell carcinogenesis. KLF4 is silenced, typically by hypermethylation, permitting normal esophageal squamous epithelial cells to become dysplastic and transform into ESCC. In ESCC cells, which are now transformed, reactivation of KLF4 enhances invasion and metastasis.
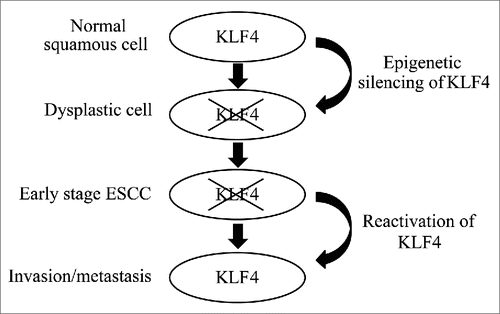
Discussion
During human esophageal squamous cell carcinogenesis, normal squamous epithelial cells become dysplastic and develop into cancer cells.Citation3 These tumor cells then undergo additional molecular changes leading to local invasion and distant metastasis, although the precise nature of these molecular alterations is not clear. Since distant metastases reduce 5-year survival in patients with ESCC by a factor of 10,Citation4 understanding the mechanisms of progression are essential for management of this deadly disease.Citation4 Here, we demonstrate that KLF4 is downregulated early in esophageal squamous cell carcinogenesis, with decreased KLF4 expression already evident in squamous cell dysplasia, and that KLF is reactivated in invasive tumors. In addition, we show that KLF4 increases invasiveness of ESCC cells, providing a potential functional link between KLF4 and patient outcomes.
In contrast to our findings, KLF4 has also been reported to decrease ESCC cell migration and invasion.Citation11,31 However, the context of these studies must be considered carefully, and these prior studies were performed in a 2-dimensional context using Transwell assays. Here, we have utilized a multicellular, 3-dimensional spheroid invasion assay, which better recapitulates tumor invasion in vivo.Citation28 Nonetheless, additional studies are necessary to better define the context-dependent effects of KLF4 on tumor cell invasion in vivo. Moreover, it will be important to determine whether KLF4 expression is increased specifically at the leading edge of the tumor during invasion and in distant metastases. Of note, while KLF4 was reported in one study to portend improved survival in ESCC,Citation32 the established patterns of KLF4 expression for normal squamous epitheliaCitation13,33-35 raise questions about the specificity of KLF4 staining in that study. Nonetheless, additional studies of KLF4 expression and ESCC survival are warranted
Importantly, while we show here that KLF4 promotes ESCC invasion, KLF4 also regulates other processes that are key for tumor development and progression,Citation9,36,37 including in ESCC.Citation10-12 Interestingly, KLF4 maintains putative cancer stem cell populations,Citation9 and the maintenance of ESCC cancer stem cells could be critical for ESCC growth and spread and for the inverse association between KLF4 expression and survival. In addition, tumor-promoting inflammation is an emerging hallmark of cancer,Citation38 and Klf4 overexpression in murine esophageal epithelia in vivo promotes inflammation and causes inflammation-mediated ESCC.Citation16 As such, it will be important to assess the consequences of KLF4 on cancer stem cells and other hallmarks of cancer during ESCC progression.
A number of genetic modifications have been identified in human ESCC,Citation5,39 and epigenetic changes, including hypermethylation of known tumor suppressor genes,Citation40 occur frequently in ESCC. Since KLF4 is consistently silenced early in esophageal squamous cell carcinogenesis but is not genetically modified, reactivation of KLF4 in squamous cell dysplasia might seem at first to provide a potential strategy for the prevention of ESCC. Yet, KLF4 expression and function appear to change during malignant progression, suggesting functional switching of KLF4 as described for other KLFs.Citation9 As such, dissecting the pathways required for the tumor suppressive properties of KLF4 from those that contribute to ESCC invasion and metastasis may permit the identification of novel targets in ESCC downstream of KLF4. Moreover, KLF4, as well as specific KLF4 SNPs, may serve as useful biomarkers for ESCC.
In sum, our findings suggest that KLF4 downregulation or loss is necessary for the development of ESCC but KLF4 reactivation in ESCC promotes tumor cell invasion, leading to metastases, advanced disease, and decreased survival (). Thus, KLF4 may function as a “gatekeeper” for human ESCC and has distinct, context-dependent functions during tumor progression.
Materials and methods
ESCC tissue and cell lines
Specimens of ESCC and adjacent normal tissue were collected from 52 Chinese patients who had no prior therapy and underwent esophagectomy for treatment of their ESCC at Sun Yat-Sen Memorial Hospital of Sun Yat-Sen University in Guangzhou, China. Pathology was reviewed by a surgical pathologist at Sun Yat-Sen University, and cancer stages were recorded prior to resection. For outcomes data, patients were followed for up to 60 months post-surgery. IRB approvals were obtained from both Sun Yat-Sen University and the University of Pennsylvania. KLF4 expression was also compared in human ESCC and normal tissue using the Oncomine Platform (Thermo Fisher Scientific). For these analyses, we utilized 2 sets of microarray data from 34Citation22 and 106Citation23 paired samples of ESCC and adjacent normal tissue from patients undergoing surgical resection of their tumor who had received no prior therapy.
Non-transformed primary human esophageal keratinocytes (EPC1 cells), were a gift of Dr. Anil Rustgi (University of Pennsylvania).Citation41 EPC1 cells were cultured in keratinocyte serum-free medium (Thermo Fisher Scientific) supplemented with 1.0 ng/mL human recombinant Epidermal Growth Factor 1-53 and 40 μg/mL Bovine Pituitary Extract. Human ESCC cell linesCitation42,43 were cultured in 1:1 Dulbecco's Modified Eagle's Medium (DMEM)/Ham's F12 medium (Thermo Fisher Scientific) with 10% FBS (Sigma-Aldrich), 100 U/mL penicillin, and 100 μg/mL streptomycin (Thermo Fisher Scientific). For demethylation and acetylation treatment, cells were grown to 60% confluence in 100 mm dishes, and 5 μM 5-azacytidine (Sigma-Aldrich) or 1 μM trichostatin A (Sigma-Aldrich) dissolved in DMSO was added into the growth medium for 24 hours.
Sequencing and data analysis
DNA was extracted from tissues and cell lines with QIAamp DNA Mini Kit (Qiagen) following the manufacturer's instruction. The TruSeq Custom Amplicon kit (Illumina) was used for library preparation for deep exonic sequencing of the human KLF4 gene. Briefly, oligonucleotides probes were designed with Illumina DesignStudio, to generate multiple amplicons of 250 bp in size that crossed the exon-intron boundary by 50 bp. Input DNA (100 ng) was hybridized with multiple probes, followed by extension and ligation, with ligation products serving as templates that were PCR-amplified with 2 unique primers containing index sequences specific to each sample. The products of this PCR reaction were converted to a single-stranded, adapter-ligated, normalized library using a bead-based protocol as per the manufacturer's instructions. Sequencing was performed on an Illumina MiSeq and de-multiplexed using standard Illumina software. Sequencing reads were aligned to the UCSC hg19 human genome reference with the Burrows-Wheeler Aligner (BWA),Citation44 and the Genome Analysis Toolkit (GATK)Citation45 was applied for base quality score recalibration, local insertion/deletion (indel) realignment, substitution, and SNP and INDEL discovery across all samples using standard hard filtering parameters or variant quality score recalibration according to GATK Best Practices recommendations.Citation46,47 Exonic sequence data was visualized using the Integrative Genomics Viewer.Citation48,49
Quantitative polymerase chain reaction
To test KLF4 expression in ESCC cell lines, total RNA was isolated with the RNeasy Micro Kit (Qiagen), and cDNA was synthesized with a High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). Quantitative real-time PCR was performed in triplicate on 3 samples for each experimental condition using an ABI StepOne Plus Real-Time PCR System (Thermo Fisher Scientific) and SYBR Green PCR Master Mix (Thermo Fisher Scientific). TATA box-binding protein served as the internal control. Primer sequences are listed in Supplemental Table 1.
Immunohistochemistry
Sections containing adjacent normal tissue, squamous cell dysplasia, and ESCC were stained with anti-KLF4 antibodies as described previously.Citation13 Briefly, after wax removal and rehydration, slides were immersed in 10 mM citric acid buffer (pH 6.0) and microwaved for 15 minutes for antigen retrieval. Nonspecific binding of avidin-biotin was blocked with the Avidin/Biotin Blocking Kit (Vector Laboratories), and nonspecific antibody binding was blocked with Starting Block T20 Blocking Buffer (Thermo Fisher Scientific). Sections were incubated overnight at 4°C with 1:1000 rabbit anti-KLF4.Citation11 Biotinylated species-specific secondary antibodies (Vector Laboratories) were added, and antibody binding was detected with the Vectastain Elite ABC Kit (Vector Laboratories) and DAB Peroxidase Substrate Kit (Vector Laboratories), followed by counterstaining with hematoxylin. KLF4 expression was scored using the following scale, as previously describedCitation50: 0, no staining; 1, very weak staining; 2, weak staining; 3, moderate staining; 4, strong staining; and 5, very strong staining. For each normal tissue, high-grade dysplasia, or invasive ESCC, 100 cells were scored. For survival analyses, tumors were grouped by KLF4 levels as low (mean staining 0-1), intermediate (mean staining 2-3), and high (mean staining 4-5), and data were plotted using GraphPad Prism (GraphPad Software). Survival curves were compared using the log-rank test.
Spheroid invasion assay
CELLSTAR 96 well suspension culture plates with U-bottoms (Greiner Bio-One) were coated with 50 μl 2% Poly(2-hydroxyethyl methacrylate) (pHEMA) (Sigma-Aldrich) in 95% Ethanol to resist cell adhesion. HCE4 and TE8 ESCC cells were infected with the neomycin-selectable retrovirus, pFB-mCherry-hKLF4, which was constructed by cloning the mCherry-KLF4 cassette from pLM-mCherry-KLF4 (gift of Michel Sadelain, Addgene plasmid #23243)Citation51 into the multiple cloning site of pFB-neo (Agilent Technologies) or pFB-neo empty vector control. To permit spheroid formation, we plated 1000 cells in 100 μl medium into each well and incubated at 37°C for 48 hours, then gently added 100 ul of 10% Matrigel (BD, Franklin Lakes, NJ) to embed the cell spheroid. Spheroids were examined after 1, 3, and 7 days, and cell invasion was visualized with a Leica DM IRB microscope (Leica Microsystems) and quantitated by measurement of spheroid diameter with QCapture software (QImaging).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by NIH NIDDK R01 DK069984 to JPK, and by the University of Pennsylvania Center for Molecular Studies in Digestive and Liver Diseases (NIH NIDDK P30 DK050306) through the Molecular Pathology and Imaging Core, the Molecular Biology/Gene Expression Core, and the Cell Culture Core and by NIH NCI P01 CA098101 (“Mechanisms of Esophageal Carcinogenesis”).
References
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin 2015; 65:87-108; PMID:25651787; http://dx.doi.org/10.3322/caac.21262
- Zhang Y. Epidemiology of esophageal cancer. World J Gastroenterol: WJG 2013; 19:5598-606; PMID:24039351; http://dx.doi.org/10.3748/wjg.v19.i34.5598
- Taylor PR, Abnet CC, Dawsey SM. Squamous dysplasia–the precursor lesion for esophageal squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev 2013; 22:540-52; PMID:23549398; http://dx.doi.org/10.1158/1055-9965.EPI-12-1347
- Howlader N, Noone AM, Krapcho M, Garshell J, Neyman N, Altekruse SF, Kosary CL, Yu M, Ruhl J, Tatalovich Z, et al. SEER Cancer Statistics Review 1975–2012, Bethesda, MD: National Cancer Institute. http://seer.cancer.gov/csr/1975_2012/, based on November 2014 SEER data submission, posted to the SEER web site, April 2015.
- Mandard AM, Hainaut P, Hollstein M. Genetic steps in the development of squamous cell carcinoma of the esophagus. Mutat Res 2000; 462:335-42; PMID:10767643; http://dx.doi.org/10.1016/S1383-5742(00)00019-3
- Lin DC, Du XL, Wang MR. Protein alterations in ESCC and clinical implications: a review. Dis Esophagus 2009; 22:9-20; PMID:18564170; http://dx.doi.org/10.1111/j.1442-2050.2008.00845.x
- Shang L, Wang M. Molecular alterations and clinical relevance in esophageal squamous cell carcinoma. Front Med 2013; 7:401-10; PMID:24002746; http://dx.doi.org/10.1007/s11684-013-0286-y
- Wu C, Li D, Jia W, Hu Z, Zhou Y, Yu D, Tong T, Wang M, Lin D, Qiao Y, et al. Genome-wide association study identifies common variants in SLC39A6 associated with length of survival in esophageal squamous-cell carcinoma. Nat Genet 2013; 45:632-8; PMID:23644492; http://dx.doi.org/10.1038/ng.2638
- Tetreault MP, Yang Y, Katz JP. Kruppel-like factors in cancer. Nat Rev Cancer 2013; 13:701-13; PMID:24060862; http://dx.doi.org/10.1038/nrc3582
- He H, Li S, Hong Y, Zou H, Chen H, Ding F, Wan Y, Liu Z. Kruppel-like factor 4 promotes esophageal squamous cell carcinoma differentiation by up-regulating keratin 13 expression. J Biol Chem 2015; 290:13567-77; PMID:25851906; http://dx.doi.org/10.1074/jbc.M114.629717
- Yang Y, Goldstein BG, Chao HH, Katz JP. KLF4 and KLF5 regulate proliferation, apoptosis and invasion in esophageal cancer cells. Cancer Biol Ther 2005; 4:1216-21; PMID:16357509; http://dx.doi.org/10.4161/cbt.4.11.2090
- Zhang G, Zhu H, Wang Y, Yang S, Liu M, Zhang W, Quan L, Bai J, Liu Z, Xu N. Krüppel-like factor 4 represses transcription of the survivin gene in esophageal cancer cell lines. Biol Chem 2009; 390:463-9; PMID:19361279; http://dx.doi.org/10.1515/BC.2009.060
- Tetreault MP, Yang Y, Travis J, Yu QC, Klein-Szanto A, Tobias JW, Katz JP. Esophageal squamous cell dysplasia and delayed differentiation with deletion of Krüppel-like factor 4 in murine esophagus. Gastroenterology 2010; 139:171-81; PMID:20347813; http://dx.doi.org/10.1053/j.gastro.2010.03.048
- Luo A, Kong J, Hu G, Liew CC, Xiong M, Wang X, Ji J, Wang T, Zhi H, Wu M, et al. Discovery of Ca2+-relevant and differentiation-associated genes downregulated in esophageal squamous cell carcinoma using cDNA microarray. Oncogene 2004; 23:1291-9; PMID:14647409; http://dx.doi.org/10.1038/sj.onc.1207218
- Wang N, Liu ZH, Ding F, Wang XQ, Zhou CN, Wu M. Down-regulation of gut-enriched Krüppel-like factor expression in esophageal cancer. World J gastroenterol: WJG 2002; 8:966-70; PMID:12439907; http://dx.doi.org/10.3748/wjg.v8.i6.966
- Tetreault MP, Wang ML, Yang Y, Travis J, Yu QC, Klein-Szanto AJ, Katz JP. Klf4 overexpression activates epithelial cytokines and inflammation-mediated esophageal squamous cell cancer in mice. Gastroenterology 2010; 139:2124-34; PMID:20816834; http://dx.doi.org/10.1053/j.gastro.2010.08.048
- Wei D, Gong W, Kanai M, Schlunk C, Wang L, Yao JC, Wu TT, Huang S, Xie K. Drastic down-regulation of Krüppel-like factor 4 expression is critical in human gastric cancer development and progression. Cancer Res 2005; 65:2746-54; PMID:15805274; http://dx.doi.org/10.1158/0008-5472.CAN-04-3619
- Patel NV, Ghaleb AM, Nandan MO, Yang VW. Expression of the tumor suppressor Kruppel-like factor 4 as a prognostic predictor for colon cancer. Cancer Epidemiol Biomarkers Prev 2010; 19:2631-8; PMID:20699379; http://dx.doi.org/10.1158/1055-9965.EPI-10-0677
- Chen YJ, Wu CY, Chang CC, Ma CJ, Li MC, Chen CM. Nuclear Kruppel-like factor 4 expression is associated with human skin squamous cell carcinoma progression and metastasis. Cancer Biol Ther 2008; 7:777-82; PMID:18376139; http://dx.doi.org/10.4161/cbt.7.5.5768
- Valencia-Hipomicronlito A, Hernandez-Atenogenes M, Vega GG, Maldonado-Valenzuela A, Ramon G, Mayani H, Pena Alonso Y, Martinez-Maza O, Mendez-Tenorio A, Huerta-Yepez S, et al. Expression of KLF4 is a predictive marker for survival in pediatric Burkitt lymphoma. Leukemia & Lymphoma 2014; 55:1806-14; PMID:24067139; http://dx.doi.org/10.3109/10428194.2013.848437
- Pandya AY, Talley LI, Frost AR, Fitzgerald TJ, Trivedi V, Chakravarthy M, Chhieng DC, Grizzle WE, Engler JA, Krontiras H, et al. Nuclear localization of KLF4 is associated with an aggressive phenotype in early-stage breast cancer. Clin Cancer Res 2004; 10:2709-19; PMID:15102675; http://dx.doi.org/10.1158/1078-0432.CCR-03-0484
- Hu N, Clifford R, Yang H, Wang C, Goldstein A, Ding T, Taylor P, Lee M. Genome wide analysis of DNA copy number neutral loss of heterozygosity (CNNLOH) and its relation to gene expression in esophageal squamous cell carcinoma. BMC Genomics 2010; 11:576; PMID:20955586; http://dx.doi.org/10.1186/1471-2164-11-576
- Su H, Hu N, Yang HH, Wang C, Takikita M, Wang QH, Giffen C, Clifford R, Hewitt SM, Shou JZ, et al. Global gene expression profiling and validation in esophageal squamous cell carcinoma and its association with clinical phenotypes. Clin Cancer Res 2011; 17:2955-66; PMID:21385931; http://dx.doi.org/10.1158/1078-0432.CCR-10-2724
- 1000 Genomes Project Consortium, Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature 2012; 491:56-65; PMID:23128226; http://dx.doi.org/10.1038/nature11632
- Talbot CJ, Tanteles GA, Barnett GC, Burnet NG, Chang-Claude J, Coles CE, Davidson S, Dunning AM, Mills J, Murray RJS, et al. A replicated association between polymorphisms near TNF[alpha] and risk for adverse reactions to radiotherapy. Br J Cancer 2012; 107:748-53; PMID:22767148; http://dx.doi.org/10.1038/bjc.2012.290
- Vigushin DM, Ali S, Pace PE, Mirsaidi N, Ito K, Adcock I, Coombes RC. Trichostatin A is a histone deacetylase inhibitor with potent antitumor activity against breast cancer in vivo. Clin Cancer Res 2001; 7:971-6; PMID:11309348
- Christman JK. 5-Azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene 2002; 21:5483-95; PMID:12154409; http://dx.doi.org/10.1038/sj.onc.1205699
- Kramer N, Walzl A, Unger C, Rosner M, Krupitza G, Hengstschlager M, Dolznig H. In vitro cell migration and invasion assays. Mutat Res 2013; 752:10-24; PMID:22940039; http://dx.doi.org/10.1016/j.mrrev.2012.08.001
- Casimiro MC, Crosariol M, Loro E, Li Z, Pestell RG. Cyclins and cell cycle control in cancer and disease. Genes Cancer 2012; 3:649-57; PMID:23634253; http://dx.doi.org/10.1177/1947601913479022
- Romanov VS, Pospelov VA, Pospelova TV. Cyclin-dependent kinase inhibitor p21(Waf1): contemporary view on its role in senescence and oncogenesis. Biochemistry Biokhimiia 2012; 77:575-84; PMID:22817456; http://dx.doi.org/10.1134/S000629791206003X
- Tian Y, Luo A, Cai Y, Su Q, Ding F, Chen H, Liu Z. MicroRNA-10b promotes migration and invasion through KLF4 in human esophageal cancer cell lines. J Biol Chem 2010; 285:7986-94; PMID:20075075; http://dx.doi.org/10.1074/jbc.M109.062877
- Ma MQ, Zhang HD, Tang P, Jiang HJ, Chen CG. Association of Kruppel-like factor 4 expression with the prognosis of esophageal squamous cell carcinoma patients. Int J Clin Exp Pathol 2014; 7:6679-85; PMID:25400747
- Goldstein BG, Chao HH, Yang Y, Yermolina YA, Tobias JW, Katz JP. Overexpression of Krüppel-like factor 5 in esophageal epithelia in vivo leads to increased proliferation in basal but not suprabasal cells. Am J Physiol Gastrointest Liver Physiol 2007; 292:G1784-92; PMID:17395897; http://dx.doi.org/10.1152/ajpgi.00541.2006
- Segre JA, Bauer C, Fuchs E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat Genet 1999; 22:356-60; PMID:10431239; http://dx.doi.org/10.1038/11926
- Jaubert J, Cheng J, Segre JA. Ectopic expression of Krüppel-like factor 4 (Klf4) accelerates formation of the epidermal permeability barrier. Development 2003; 130:2767-77; PMID:12736219; http://dx.doi.org/10.1242/dev.00477
- Ghaleb AM, Nandan MO, Chanchevalap S, Dalton WB, Hisamuddin IM, Yang VW. Krüppel-like actors 4 and 5: the yin and yang regulators of cellular proliferation. Cell Res 2005; 15:92-6; PMID:15740636; http://dx.doi.org/10.1038/sj.cr.7290271
- McConnell BB, Ghaleb AM, Nandan MO, Yang VW. The diverse functions of Krüppel-like factors 4 and 5 in epithelial biology and pathobiology. Bioessays 2007; 29:549-57; PMID:17508399; http://dx.doi.org/10.1002/bies.20581
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell; 144:646-74; PMID:21376230; http://dx.doi.org/10.1016/j.cell.2011.02.013
- Lin DC, Hao JJ, Nagata Y, Xu L, Shang L, Meng X, Sato Y, Okuno Y, Varela AM, Ding LW, et al. Genomic and molecular characterization of esophageal squamous cell carcinoma. Nat Genet 2014; 46:467-73; PMID:24686850; http://dx.doi.org/10.1038/ng.2935
- Kaz AM, Grady WM. Epigenetic biomarkers in esophageal cancer. Cancer Lett 2014; 342:193-9; PMID:22406828; http://dx.doi.org/10.1016/j.canlet.2012.02.036
- Andl CD, Mizushima T, Nakagawa H, Oyama K, Harada H, Chruma K, Herlyn M, Rustgi AK. Epidermal growth factor receptor mediates increased cell proliferation, migration, and aggregation in esophageal keratinocytes in vitro and in vivo. J Biol Chem 2003; 278:1824-30; PMID:12435727; http://dx.doi.org/10.1074/jbc.M209148200
- Nishihira T, Hashimoto Y, Katayama M, Mori S, Kuroki T. Molecular and cellular features of esophageal cancer cells. J Cancer Res Clin Oncol 1993; 119:441-9; PMID:8509434; http://dx.doi.org/10.1007/BF01215923
- Banks-Schlegel SP, Quintero J. Growth and differentiation of human esophageal carcinoma cell lines. Cancer Res 1986; 46:250-8; PMID:2415247
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009; 25:1754-60; PMID:19451168; http://dx.doi.org/10.1093/bioinformatics/btp324
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010; 20:1297-303; PMID:20644199; http://dx.doi.org/10.1101/gr.107524.110
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011; 43:491-8; PMID:21478889; http://dx.doi.org/10.1038/ng.806
- Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, et al. From FastQ Data to High-Confidence Variant Calls: The Genome Analysis Toolkit Best Practices Pipeline. Current Protocols in Bioinformatics: John Wiley & Sons, Inc., 2013 Oct 15;11(1110):11.10.1-11.10.33. PMID: 25431634
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotech 2011; 29:24-6; PMID:21221095; http://dx.doi.org/10.1038/nbt.1754
- Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Briefings Bioinformatics 2013; 14:178-92; PMID:22517427; http://dx.doi.org/10.1093/bib/bbs017
- Yang Y, Nakagawa H, Tetreault MP, Billig J, Victor N, Goyal A, Sepulveda AR, Katz JP. Loss of transcription factor KLF5 in the context of p53 ablation drives invasive progression of human squamous cell cancer. Cancer Res 2011; 71:6475-84; PMID:21868761; http://dx.doi.org/10.1158/0008-5472.CAN-11-1702
- Papapetrou EP, Tomishima MJ, Chambers SM, Mica Y, Reed E, Menon J, Tabar V, Mo Q, Studer L, Sadelain M. Stoichiometric and temporal requirements of Oct4, Sox2, Klf4, and c-Myc expression for efficient human iPSC induction and differentiation. Proc Natl Acad Sci 2009; 106:12759-64; PMID:19549847; http://dx.doi.org/10.1073/pnas.0904825106
