ABSTRACT
Several studies have reported that epidermal growth factor receptor (EGFR)-related molecules may serve as predictors of cetuximab treatment for metastatic colorectal cancer (mCRC), such as EGFR gene copy number (GCN), expression of 2 ligands of EGFR, amphiregulin (AREG) and epiregulin (EREG), and EGFR CA simple sequence repeat 1 (CA-SSR1) polymorphism; however, these biomarkers still remain not useful in clinical practice since they have been evaluated using cohorts with patients treated in various settings of chemotherapy. We therefore analyzed associations of mRNA expression of AREG and EREG, EGFR GCN, and CA-SSR1 polymorphism [short (S;≤ 19) / long (L; ≥ 20)] with clinical outcomes in 77 Japanese patients with KRAS exon 2 wild-type mCRC enrolled in phase II trials of FOLFOX (n = 28/57, UMIN000004197) or SOX (n = 49/67, UMIN000007022) plus cetuximab as first-line therapy. High AREG expression correlated with significantly better progression-free survival (median 11.6 vs. 66 months, HR 0.52, P = 0.037); moreover, it remained statistically significant in multivariate analysis (HR: 0.48, P = 0.027). S/S genotype of CA-SSR1 predicted severe skin toxicity (P = 0.040). Patients with both AREG-low and EGFR low-GCN had significantly shorter overall survival than the others (median 22.2 vs. 42.8 months, HR 2.34, P = 0.042). The multivariate analysis showed that molecular status with both AREG-low and EGFR low-GCN was a predictor of worse survival (P = 0.006). In conclusion, AREG mRNA expression and EGFR CA-SSR1 polymorphism predict survival and skin toxicity, respectively, of initial chemotherapy with cetuximab. Our results also suggest potential prognostic value of the combined assessment of AREG and EGFR GCN for first-line cetuximab treatment.
Abbreviations
| EGFR | = | epidermal growth factor receptor |
| mCRC | = | metastatic colorectal cancer |
| AREG | = | amphiregulin |
| EREG | = | epiregulin |
| CA-SSR1 | = | CA simple sequence repeat 1 |
| GCN | = | gene copy number |
| PFS | = | progression-free survival |
| OS | = | overall survival |
| ECOG | = | Eastern Cooperative Oncology Group |
| CT | = | computed tomography |
| ETS | = | early tumor shrinkage |
| FFPE | = | formalin-fixed paraffin-embedded |
Introduction
Cetuximab, an IgG1 monoclonal antibody, binds to ligand-binding domain of epidermal growth factor receptor (EGFR) and thus exerts an inhibitory effect on tumor proliferative signaling in the downstream of EGFR pathway.Citation1 In metastatic colorectal cancer (mCRC) patients with tumors harboring KRAS exon 2 wild-type, this drug became a standard component of first-line treatment according to several evidences with significant survival benefit from clinical trials.Citation2-7 Then, retrospective analyses of large prospective studies to further understand the molecular determinants of responsiveness to anti-EGFR monoclonal antibodies revealed that an expanded RAS analysis can identify more sensitive responders to the antibodies,Citation8,9 proposing the use of cetuximab in only RAS wild-type patients. However, the selection for the responders to cetuximab still remains insufficient since a subgroup analysis of SWOG/CALGB80405 trial showed that survival time in RAS wild-type patients treated with cetuximab-containing regimen was almost same as that in RAS wild-type patients treated with bevacizumab-containing regimen.Citation10 However, which biologic agent should be used up-front as the best companion for cytotoxic chemotherapy in RAS wild-type patients remains inconclusive. Further predictive markers beyond RAS are needed to refine the selection of patients who benefit from cetuximab-based chemotherapy and to prolong survival time in addition to improve cost effectiveness.
Several studies have reported that EGFR-related molecules may serve as predictors of cetuximab treatment, such as copy number of EGFR gene and the intratumoral mRNA expression of 2 EGFR ligands, amphiregulin (AREG) and epiregulin (EREG).Citation11-18 EGFR CA simple sequence repeat 1 (CA-SSR1) polymorphism has been shown to predict skin toxicity (). However, these biomarkers still remain not useful in clinical practice since they have been evaluated in various settings of chemotherapy or in population not restricted by KRAS status. Thence, it should be critical to accumulate evidences on if the molecules will serve as good predictors of cetuximab. In addition, there are few evidences in Japanese patients with regard to association of EGFR-related molecular markers with clinical outcome of chemotherapy with cetuximab. The aim of the present study was to clarify whether previously-reported EGFR-related biomarkers, AREG, EREG, EGFR gene copy number (GCN), and EGFR CA-SSR1 polymorphism, predict clinical outcomes in patients with mCRC harboring KRAS exon 2 wild-type tumors treated with first-line cetuximab-containing chemotherapy.
Table 1. Previously reported biomarkers and clinical outcome in patients receiving cetuximab.
Results
The baseline characteristics of the patient cohort enrolled in this study are summarized in . In the population with a median age of 63 (range 39–79) years and follow-up time of 31.4 months, response rate, median progression-free survival (PFS), and overall survival (OS) were 73 % (95% CI 61–82%), 10.0 months (95% CI 8.8–11.8), and 33.9 months (95% CI 26.5-Not reached), respectively.
Table 2. Patients' characteristics (N = 77).
AREG and EREG mRNA expression and clinical outcomes
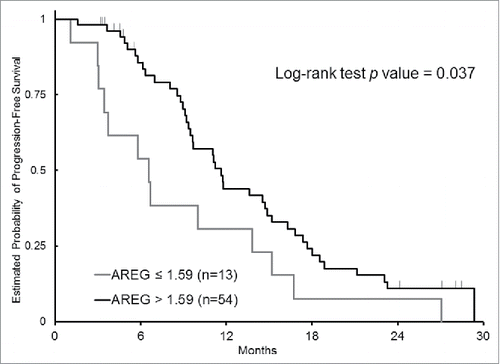
Measuring mRNA expression of AREG and EREG were successful in 84 % and 81 %, respectively. In failed cases, the measurement was not successful because of limited quantity and/or poor quality of isolated total RNA. Patients with AREG high-expression (AREG mRNA level > 1.59) had a significantly longer PFS than those with AREG low-expression (median 11.6 months vs. 6.6 months, HR 0.52, 95% CI 0.28–0.98, P = 0.037) (); furthermore, OS was longer in patients with overexpression of AREG although there was no statistically significance (median 42.8 months vs. 26.5 months, HR 0.50, 95% CI 0.22–1.13, P = 0.084). No association of EREG mRNA expression with outcomes was observed ().
Table 3. Candidate predictors and clinical outcomes.
EGFR GCN and CA-SSR1 polymorphism and clinical outcomes
All enrolled patients were assessable for EGFR GCN and CA-SSR1 polymorphism. An increased GCN of EGFR was observed in 30 (39%) of 77 patients. EGFR GCN was not statistically significantly associated with clinical outcomes although EGFR high-GCN had trends toward longer PFS and OS (P = 0.073 and P = 0.068, respectively). Frequency of L/L, L/S, and S/S genotypes of EGFR CA-SSR1 were 47%, 38%, and 15%, respectively. The CA-SSR1 polymorphism did not predict survival of cetuximab-containing chemotherapy in all patients enrolled in this study. Then, we addressed at investigating association of the CA-SSR1 polymorphism with cetuximab-induced skin toxicity. We found that prevalence of grade 2 or 3 skin toxicity at 8 weeks after administration of treatment was significantly higher in patients harboring S/S genotype of the polymorphism than the other ones (P = 0.040) ().
Combined assessment of both AREG expression and EGFR GCN to predict clinical outcomes of cetuximab-containing chemotherapy
Additionally, we performed an exploratory sub-group analysis according to molecular status of both AREG mRNA expression and EGFR GCN, which are variables regarding EGFR ligand and receptor, respectively. PFS was significantly shorter in patients with either tumors harboring AREG low-expression or EGFR low-GCN compared to those with both AREG-high and EGFR high-GCN (median 9.4 months vs. 11.8 months, HR 1.75, 95% CI 1.00–3.08, P = 0.038). Patients with both AREG-low and EGFR low-GCN had significantly worse response rate and OS than the others (50% vs. 82%, P = 0.043; median 22.2 months vs. 42.8 months, HR 2.34, 95% CI 0.99–5.50, P = 0.042, respectively) ( and ). No difference was observed in AREG expression level by EGFR GCN (P = 0.61, Wilcoxon 2-sample test).
Figure 2. Probability of overall survival by combined assessment with AREG gene expression and EGFR gene copy number (GCN), (A) Kaplan-Meier curves of 3 groups according to AREG gene expression and EGFR GCN, (B) Kaplan-Meier curves of groups with either AREG-high or EGFR GCN-high and both AREG-low and EGFR GCN-low.
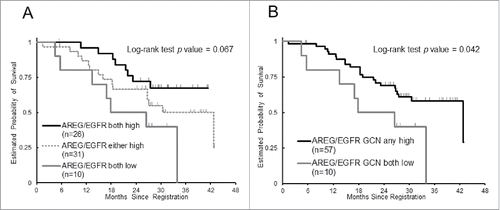
Table 4. Association between AREG mRNA levels and EGFR gene copy number and clinical outcomes.
Factors with prognostic and/or predictive significance at multivariate analysis
A multivariate analysis adjusted for Eastern Cooperative Oncology Group (ECOG) performance status, regimen, and primary tumor site was performed for AREG, EREG, EGFR GCN, and CA-SSR1 polymorphism in all patients. High AREG mRNA levels was independently associated with better PFS and OS compared to patients with AREG-low levels (P = 0.027, P = 0.040, respectively). Molecular status with both AREG-low and EGFR low-GCN was a predictor of worse survival in patients treated with first-line cetuximab and oxaliplatin-based chemotherapy, irrespective of other factors ().
Table 5. Prognostic/predictive factors at multivariate analysis.
Discussion
Our study demonstrates that AREG mRNA expression and EGFR CA-SSR1 polymorphism predict survival and skin toxicity, respectively, in Japanese patients with KRAS exon 2 wild-type mCRC treated with initial chemotherapy with cetuximab, potentially supporting previous data on the expression of AREG mRNA as a predictor for cetuximab. These results also suggest potential prognostic value of the combined assessment of AREG expression and EGFR GCN for cetuximab treatment.
Eleven ligands have been identified in the ErbB family in humans, 2 of which are AREG and EREG. Citation19,20 Ligands binding to receptors induce the formation of receptor homodimers and heterodimers and activation of the intrinsic kinase domain, resulting in phosphorylation of specific tyrosine residues, which triggers intracellular signaling through the RAS/RAF/MAPK and PI3K/AKT pathways that subsequently modulates cell proliferation, adhesion, angiogenesis, migration, and survival. Citation20,21 Several studies have shown that increased intratumoral mRNA expression of AREG or EREG gene is associated with clinical outcome in patients with KRAS exon 2 wild-type mCRC treated with cetuximab-containing therapy.Citation13,15,16,22 These studies enrolled cohorts with heterogeneous patient characteristics, type of therapy (single or combination), treatment in various lines and mixed KRAS status, while our study included a more homogeneous patient cohort that consisted of only KRAS exon 2 wild-type patients and treated with combination therapy with oxaliplatin as first-line treatment, probably indicating our findings become more reliable evidence. Recently, a sub-analysis of a big randomized clinical trial comparing irinotecan plus panitumumab with irinotecan as second-line treatment has been reported. Citation23 The translational study results indicated that high expression of either AREG or EREG is a predictive marker for panitumumab therapy benefit on PFS in RAS wild-type patients, supporting results of our study using a homogeneous patient cohort, which could show significantly univariate and multivariate associations of AREG overexpression with better PFS as well as OS. Our finding supports that AREG expression may serve as a predictor of favorable outcomes of cetuximab treatment. On the other hand, we failed to find significant associations of EREG with clinical outcomes in the current study. Stahler A, et al has revealed the positive prognostic effect of high EREG expression but not AREG expression in patients treated with first-line irinotecan-based chemotherapy. Citation24 EGFR ligands may have different prognostic effects between types of chemotherapy; therefore, there would be of interest to investigate the difference in future translational researches.
Cetuximab inhibited proliferation of colorectal-cancer cells with increased EGFR copy number, but that the same dose did not affect cells with unamplified EGFR.Citation25,26 EGFR GCN has been shown to correlate with better response to anti-EGFR antibodies; Citation11,27-31 however, some studies have reported inconsistent results.Citation32-34 We failed to indicate statistically significant association of EGFR GCN with outcomes of cetuximab treatment. One of possible reasons for these divergent results is heterogeneity of copy number changes in colorectal cancer,Citation35,36 resulting in not only that cut-off value is different among previous studies but also that this molecular marker would be difficult to be included in clinical practice. In our study the increased GCN of EGFR was defined by the cut-off value, 2.92, based on a classification by receiver operating characteristic (ROC) analysis as previously described.Citation29 In addition, the patient number included in previous studies was relatively small. The association of EGFR GCN warrants to be validated in larger-size studies; however, EGFR GCN is less likely to become a clinically useful biomarker due to the difficulties in determining copy number by polymerase chain reaction in samples containing a mixture of somatic and tumor DNA and in standardizing the cut-off value.Citation31
We found that short allele of EGFR CA-SSR1 predicts severe cetuximab-induced skin toxicity in Japanese patients with mCRC. No association with survival time was observed in our study. Some studies have previously reported significant association of long length of the EGFR CA repeat with worse outcomes of cetuximab treatment,Citation37,38 while the other studies have indicated no association,Citation39-41 leading to that it still remains controversial. Our results were consistent with previous reports that the length of the EGFR CA repeat inversely correlates with severe cetuximab-induced skin toxicity.Citation37 There has been shown to be interethnic differences in the repeat number of EGFR CA-SSR1 between Caucasians and Asians.Citation42 In our study, frequency of the short allele was lower than that of the long allele, which is minor allele for Caucasians. To the best of our knowledge, this is the first report regarding distribution of the allele in Japanese mCRC patients.
Our exploratory sub-analysis revealed that patients harboring tumors with both AREG low-expression and EGFR low-GCN had statistically significantly worse survival than the other patients when treated with first-line cetuximab-containing chemotherapy. The association was observed in both univariate and multivariate analyses, suggesting potential prognostic value of the combined assessment of AREG mRNA and EGFR GCN for cetuximab treatment in mCRC. In the current study, overexpression of AREG affected outcome in term of OS, with a median OS of 42.8 months for AREG-high compared to 26.5 months for AREG-low (adjusted P = 0.040). The adjusted p-value of combined variables with AREG and EGFR GCN for OS was 0.006. Moreover, no association between AREG expression and EGFR GCN was observed (P = 0.61). In a previous study evaluating EGFR-related biomarkers, the combined assessment of EGFR pathway activations, which included receptor and downstream pathways such as RAS/MAPK, was not associated with clinical outcome.Citation43 In our study the exploratory analysis involved receptor-related marker and ligand-related marker, suggesting that combined factors of both the receptor and ligand may strongly influence survival in patients treated with cetuximab. Our findings also suggest that although each factor does not play a key role in decision-making for treatment the combined assessment may become a novel tool to distinguish patients who can receive more benefit from cetuximab.
Our study has some limitations. We demonstrated significant results for EGFR-related biomarkers as previously reported, and appeared to support their predictive value; however, sample size of our study was small, leading to some possibility that the patient number has no adequate ability to assess the association between the biomarkers and clinical outcomes. Although all patients were enrolled in a prospective trial, a selection bias cannot be excluded because the samples were collected retrospectively. The cut-off value of EGFR GCN and CA repeat still remains controversial, and the used cut-off value in our study is not validated. Additionally, we did not approach an analysis when restricted to RAS wild-type patients. Cetuximab should be administrated to RAS wild-type patients rather than KRAS exon 2 wild-type patients according several sub-group analyses of randomized clinical trials. Citation9,44 Therefore, our findings should be confirmed in studies using larger cohorts with RAS wild-type patients.
In conclusion, our study demonstrates that AREG mRNA expression and EGFR CA-SSR1 polymorphism predict survival and skin toxicity, respectively, in KRAS exon 2 wild-type mCRC patients treated with cetuximab-containing chemotherapy as first-line treatment. Furthermore, our results suggest that the combined assessment of AREG expression and EGFR GCN may be a predictor of survival in mCRC patients treated with cetuximab.
Materials and methods
Patient population
We investigated mCRC patients with tumor expressing EGFR and harboring KRAS exon 2 wild-type, who participated in either Japanese phase II trial evaluating efficacy of first-line cetuximab in combination with oxaliplatin-based regimen, modified FOLFOX6 (JACCRO CC-05; n = 57, UMIN000004197) or SOX (S-1 plus oxaliplatin) (JACCRO CC-06; n = 67, UMIN000007022). A total of 77 patients with tumor tissue available from the 2 phase II trials were enrolled in this study. Tumor response was evaluated by computed tomography (CT) scan every 8 weeks until disease progression. External reviewers classified objective tumor response according to Response Evaluation Criteria in Solid Tumors (RECIST) v1.1. Patients with complete or partial response were categorized as responders, while those with stable or progressive disease were non-responders. Early tumor shrinkage (ETS) that was evaluated at the first CT scan at 8 weeks after starting treatment was recorded as yes when 20% and more decrease in the sum of diameters of target lesions, otherwise no. PFS was calculated from entry date to either progression of disease or death from any cause. OS was defined as the period from entry date until death. If events were not observed, the endpoints were censored at the last time of contact or follow-up. This study was conducted adhering to the REporting recommendations for tumor MARKer prognostic studies (REMARK).Citation45,46 The tissue analyses presented in this study were carried out at SRL, Inc. (Tokyo, Japan) and Response Genetics, Inc. (RGI; Los Angeles, CA, United States), following approval by the Institutional Review Board in each institute which participated in JACCRO CC-05 or CC-06 trial. Informed consent was obtained from all individual participants included in the study.
DNA and RNA isolation
Formalin-fixed paraffin-embedded (FFPE) tumor specimens were cut into sections with a thickness of 3 or 10 μm. In a preparation for macrodissection, one 3μm slide was stained with H&E and then evaluated for tumor content and marked for areas with dominant tumor foci by a pathologist. Macrodissection by scratching the marked areas was carried out using a blade to ensure that tumor cells as many as possible were dissected. The dissected particles of tissue were transferred to reaction tubes for isolation of genomic DNA and RNA. Genomic DNA was extracted from FFPE tissue derived from tumor samples using QIAamp DNA FFPE Tissue Kit (QIAGEN KK) according to the manufacturer's protocol. RNA isolation from macrodissected FFPE samples was performed using miRNeasy FFPE Kit (QIAGEN KK) according to the manufacturer's instructions. From the total RNA yielded, cDNA was converted using miScript II RT Kit (QIAGEN KK).
Quantitative reverse transcriptase polymerase chain reaction analysis for gene expression
Quantitation of gene mRNA expression levels of AREG, EREG, and an internal reference (β-actin) cDNA was done using a fluorescence-based Real Time PCR. Briefly, isolated RNA was reverse-transcribed to cDNA using random hexamers, followed by Real Time-PCR using specific primers and probes. For each sample, parallel reactions were carried out for each gene of interest and the β-actin reference gene to normalize for input cDNA. Real Time PCR was performed using the ABI PRISM 7900HT Sequence detection System (TaqMan; Perkin-Elmer Applied Biosystems). The obtained ratio between the values provided relative gene expression levels for the gene expression tested.
EGFR gene copy number and CA simple sequence repeat 1 polymorphism
One section with a thickness of 3 μm from FFPE tissue was used for a FISH assay to analyze EGFR GCN. The EGFR FISH assay was carried out with Vysis® LSI® EGFR SpectrumOrange / CEP® 7 SpectrumGreen Probe (Abbott Japan Co., Ltd.) according to methods of described PathVysion® HER-2 DNA Probe Kit (Abbott Japan Co., Ltd.). EGFR gene, which is located on the short arm of chromosome 7 was visualized as orange and green signal is α satellite of chromosome 7 and blue signal is nuclei respectively, with each filters through Monochrome CCD Camera Excel M (Dage-MTI, Inc.) using an automated fluorescence microscopy scanning system: Accord/SOLO (BioView Ltd.). The largest possible area of distant tumor areas were selected guided by the H&E-stained slide and the EGFR signal was counted in at least 30 nuclei per tumor area at X1000 magnification. Mean of ≥ 2.92 gene signals per nucleus was scored as EGFR FISH positive based on a classification by receiver operating characteristic analysis as previously described by Cappuzzo et al.Citation29 EGFR CA-repeat number was determined using PCR, followed by separation with capillary electrophoresis on ABI 3130xl Genetic Analyzer (Thermo Fisher Scientific K.K.). The assay with forward and reverse primers and sequences for analyzing the genetic variant were performed as described previously.Citation47 Based on the trimodal distribution of the EGFR CA repeat alleles in Asians,Citation42 EGFR less than 20 CA repeats and ≥ 20 CA repeats were defined short (S) or long (L) alleles, respectively.
Statistical analysis
PFS was the primary endpoint of the current study. Tumor response (responder, non-responder), ETS (yes, no), and OS were the secondary endpoints. The associations between each categorical marker and tumor response or ETS were examined using Fisher's exact test. The maximal chi-square approachCitation48,49 was used to test associations between gene expression level of AREG or EREG and tumor response. A cut-off value was identified to separate patients into 2 groups in terms of likelihood of tumor response and P value was adjusted for multiple testing using 2000 bootstrap-like simulations.Citation50 The same cut-off value was used for assessing the associations with other endpoints. The associations between each marker and PFS or OS were assessed using Kaplan-Meir curves and log-rank test in the univariate analyses. Multivariable Cox regression model was performed to evaluate the independent effect of a marker on PFS or OS adjusting for regimen (modified FOLFOX6 vs. SOX), ECOG performance status (0 vs. 1), and primary tumor site (right vs. left).
Sixty-two of 77 patients had progressed or died when receiving the cetuximab-based first-line therapy. The minimum detectable hazard ratios ranged from 2.06 to 2.52 using a 2-sided log-rank test at the significance level of 0.05 with 80% power.
All tests were conducted using the SAS 9.4. (SAS Institute) at a significance level of 0.05. All P values were 2-sided and not adjusted for multiple hypothesis testing due to the nature of the current study.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1975 Helsinki declaration and its later amendments or comparable ethical standards.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank the patients, their families, and the investigators who participated in the JACCRO CC-05/06AR trial. We also thank Atsushi Kakimoto and Nahoko Hirabayashi (SRL, Inc., Tokyo, Japan) for genetic testing and Sachika Koyama for editorial assistance.
Funding
This study was partly funded by Dhont Family Foundation, San Pedro Peninsula Cancer Guild, and the Japan Clinical Cancer Research Organization (JACCRO).
References
- Galizia G, Lieto E, De Vita F, Orditura M, Castellano P, Troiani T, Imperatore V, Ciardiello F. Cetuximab, a chimeric human mouse anti-epidermal growth factor receptor monoclonal antibody, in the treatment of human colorectal cancer. Oncogene 2007; 26:3654-60; PMID:17530019; http://dx.doi.org/10.1038/sj.onc.1210381
- Van Cutsem E, Kohne CH, Hitre E, Zaluski J, Chang Chien CR, Makhson A, D'Haens G, Pinter T, Lim R, Bodoky G, et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Eng J Med 2009; 360:1408-17; PMID:19339720; http://dx.doi.org/10.1056/NEJMoa0805019
- Bokemeyer C, Bondarenko I, Hartmann JT, de Braud F, Schuch G, Zubel A, Celik I, Schlichting M, Koralewski P. Efficacy according to biomarker status of cetuximab plus FOLFOX-4 as first-line treatment for metastatic colorectal cancer: the OPUS study. Ann Oncol 2011; 22:1535-46; PMID:21228335; http://dx.doi.org/10.1093/annonc/mdq632
- Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol 2008; 26:1626-34; PMID:18316791; http://dx.doi.org/10.1200/JCO.2007.14.7116
- Karapetis CS, Khambata-Ford S, Jonker DJ, O'Callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD, Robitaille S, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Eng J Med 2008; 359:1757-65; PMID:18946061; http://dx.doi.org/10.1056/NEJMoa0804385
- Allegra CJ, Jessup JM, Somerfield MR, Hamilton SR, Hammond EH, Hayes DF, McAllister PK, Morton RF, Schilsky RL. American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol 2009; 27:2091-6; PMID:19188670; http://dx.doi.org/10.1200/JCO.2009.21.9170
- Dahabreh IJ, Terasawa T, Castaldi PJ, Trikalinos TA. Systematic review: Anti-epidermal growth factor receptor treatment effect modification by KRAS mutations in advanced colorectal cancer. Ann Intern Med 2011; 154:37-49; PMID:21200037; http://dx.doi.org/10.7326/0003-4819-154-1-201101040-00006
- Douillard JY, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M, Humblet Y, Bodoky G, Cunningham D, Jassem J, et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Eng J Med 2013; 369:1023-34; PMID:24024839; http://dx.doi.org/10.1056/NEJMoa1305275
- Heinemann V, von Weikersthal LF, Decker T, Kiani A, Vehling-Kaiser U, Al-Batran SE, Heintges T, Lerchenmuller C, Kahl C, Seipelt G, et al. FOLFIRI plus cetuximab versus FOLFIRI plus bevacizumab as first-line treatment for patients with metastatic colorectal cancer (FIRE-3): a randomised, open-label, phase 3 trial. Lancet Oncol 2014; 15:1065-75; PMID:25088940; http://dx.doi.org/10.1016/S1470-2045(14)70330-4
- Lenz H, Niedzwiecki D, Innocenti F, Blanke C, Mahony MR, O'Neil BH, Shaw JE, Polite B, Hochster H, Atkins J, et al. 501OCALGB/SWOG 80405: PHASE III TRIAL OF IRINOTECAN/5-FU/LEUCOVORIN (FOLFIRI) OR OXALIPLATIN/5-FU/LEUCOVORIN (MFOLFOX6) WITH BEVACIZUMAB (BV) OR CETUXIMAB (CET) FOR PATIENTS (PTS) WITH EXPANDED RAS ANALYSES UNTREATED METASTATIC ADENOCARCINOMA OF THE COLON OR RECTUM (MCRC). Ann Oncol 2014; 25; PMID:25411415; http://dx.doi.org/10.1093/annonc/mdu438.13
- Moroni M, Veronese S, Benvenuti S, Marrapese G, Sartore-Bianchi A, Di Nicolantonio F, Gambacorta M, Siena S, Bardelli A. Gene copy number for epidermal growth factor receptor (EGFR) and clinical response to antiEGFR treatment in colorectal cancer: a cohort study. Lancet Oncol 2005; 6:279-86; PMID:15863375; http://dx.doi.org/10.1016/S1470-2045(05)70102-9
- Frattini M, Saletti P, Romagnani E, Martin V, Molinari F, Ghisletta M, Camponovo A, Etienne LL, Cavalli F, Mazzucchelli L. PTEN loss of expression predicts cetuximab efficacy in metastatic colorectal cancer patients. Br J Cancer 2007; 97:1139-45; PMID:17940504; http://dx.doi.org/10.1038/sj.bjc.6604009
- Khambata-Ford S, Garrett CR, Meropol NJ, Basik M, Harbison CT, Wu S, Wong TW, Huang X, Takimoto CH, Godwin AK, et al. Expression of epiregulin and amphiregulin and K-ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. J Clin Oncol 2007; 25:3230-7; PMID:17664471; http://dx.doi.org/10.1200/JCO.2006.10.5437
- Saridaki Z, Tzardi M, Papadaki C, Sfakianaki M, Pega F, Kalikaki A, Tsakalaki E, Trypaki M, Messaritakis I, Stathopoulos E, et al. Impact of KRAS, BRAF, PIK3CA mutations, PTEN, AREG, EREG expression and skin rash in >/= 2 line cetuximab-based therapy of colorectal cancer patients. PloS One 2011; 6:e15980; PMID:21283802; http://dx.doi.org/10.1371/journal.pone.0015980
- Pentheroudakis G, Kotoula V, De Roock W, Kouvatseas G, Papakostas P, Makatsoris T, Papamichael D, Xanthakis I, Sgouros J, Televantou D, et al. Biomarkers of benefit from cetuximab-based therapy in metastatic colorectal cancer: interaction of EGFR ligand expression with RAS/RAF, PIK3CA genotypes. BMC cancer 2013; 13:49; PMID:23374602; http://dx.doi.org/10.1186/1471-2407-13-49
- Jacobs B, De Roock W, Piessevaux H, Van Oirbeek R, Biesmans B, De Schutter J, Fieuws S, Vandesompele J, Peeters M, Van Laethem JL, et al. Amphiregulin and epiregulin mRNA expression in primary tumors predicts outcome in metastatic colorectal cancer treated with cetuximab. J Clin Oncol 2009; 27:5068-74; PMID:19738126; http://dx.doi.org/10.1200/JCO.2008.21.3744
- Loupakis F, Cremolini C, Fioravanti A, Orlandi P, Salvatore L, Masi G, Schirripa M, Di Desidero T, Antoniotti C, Canu B, et al. EGFR ligands as pharmacodynamic biomarkers in metastatic colorectal cancer patients treated with cetuximab and irinotecan. Targeted Oncol 2014; 9:205-14; PMID:23821377; http://dx.doi.org/10.1007/s11523-013-0284-7
- Di Fiore F, Sesboue R, Michel P, Sabourin JC, Frebourg T. Molecular determinants of anti-EGFR sensitivity and resistance in metastatic colorectal cancer. Br J Cancer 2010; 103:1765-72; PMID:21139621; http://dx.doi.org/10.1038/sj.bjc.6606008
- Yarden Y. The EGFR family and its ligands in human cancer. signalling mechanisms and therapeutic opportunities. Eur J Cancer 2001; 37 Suppl 4:S3-8; PMID:11597398; http://dx.doi.org/10.1016/S0959-8049(01)00230-1
- Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer 2005; 5:341-54; PMID:15864276; http://dx.doi.org/10.1038/nrc1609
- Scaltriti M, Baselga J. The epidermal growth factor receptor pathway: a model for targeted therapy. Clin Cancer Res 2006; 12:5268-72; PMID:17000658; http://dx.doi.org/10.1158/1078-0432.CCR-05-1554
- Stintzing S, Jung A, Kapaun C, Reiche J, Modest DP, Giessen CA, Vehling-Kaiser U, Stauch M, Hass H, Fischer von Weikersthal L, et al. Ligand expression of the EGFR ligands amphiregulin, epiregulin, and amplification of the EGFR gene to predict for treatment efficacy in KRAS wild-type mCRC patients treated with cetuximab plus CAPIRI and CAPOX: Analysis of the randomized AIO CRC-0104 trial. ASCO Meeting Abstracts 2012; 30:3519.
- Seligmann JF, Elliott F, Richman SD, Jacobs B, Hemmings G, Brown S, Barrett JH, Tejpar S, Quirke P, Seymour MT. Combined Epiregulin and Amphiregulin Expression Levels as a Predictive Biomarker for Panitumumab Therapy Benefit or Lack of Benefit in Patients With RAS Wild-Type Advanced Colorectal Cancer. JAMA Oncol 2016; PMID:26867820; http://dx.doi.org/10.1001/jamaoncol.2015.6065
- Stahler A, Heinemann V, Giessen-Jung C, Crispin A, Schalhorn A, Stintzing S, Fischer von Weikersthal L, Vehling-Kaiser U, Stauch M, Quietzsch D, et al. Influence of mRNA expression of epiregulin and amphiregulin on outcome of patients with metastatic colorectal cancer treated with 5-FU/LV plus irinotecan or irinotecan plus oxaliplatin as first-line treatment (FIRE 1-trial). Int J Cancer 2016; 138:739-46; PMID:26284333; http://dx.doi.org/10.1002/ijc.29807
- Yang JL, Qu XJ, Russell PJ, Goldstein D. Regulation of epidermal growth factor receptor in human colon cancer cell lines by interferon alpha. Gut 2004; 53:123-9; PMID:14684586; http://dx.doi.org/10.1136/gut.53.1.123
- Matar P, Rojo F, Cassia R, Moreno-Bueno G, Di Cosimo S, Tabernero J, Guzman M, Rodriguez S, Arribas J, Palacios J, et al. Combined epidermal growth factor receptor targeting with the tyrosine kinase inhibitor gefitinib (ZD1839) and the monoclonal antibody cetuximab (IMC-C225): superiority over single-agent receptor targeting. Clin Cancer Res 2004; 10:6487-501; PMID:15475436; http://dx.doi.org/10.1158/1078-0432.CCR-04-0870
- Laurent-Puig P, Cayre A, Manceau G, Buc E, Bachet JB, Lecomte T, Rougier P, Lievre A, Landi B, Boige V, et al. Analysis of PTEN, BRAF, and EGFR status in determining benefit from cetuximab therapy in wild-type KRAS metastatic colon cancer. J Clin Oncol 2009; 27:5924-30; PMID:19884556; http://dx.doi.org/10.1200/JCO.2008.21.6796
- Sartore-Bianchi A, Moroni M, Veronese S, Carnaghi C, Bajetta E, Luppi G, Sobrero A, Barone C, Cascinu S, Colucci G, et al. Epidermal growth factor receptor gene copy number and clinical outcome of metastatic colorectal cancer treated with panitumumab. J Clin Oncol 2007; 25:3238-45; PMID:17664472; http://dx.doi.org/10.1200/JCO.2007.11.5956
- Cappuzzo F, Finocchiaro G, Rossi E, Janne PA, Carnaghi C, Calandri C, Bencardino K, Ligorio C, Ciardiello F, Pressiani T, et al. EGFR FISH assay predicts for response to cetuximab in chemotherapy refractory colorectal cancer patients. Ann Oncol 2008; 19:717-23; PMID:17974556; http://dx.doi.org/10.1093/annonc/mdm492
- Scartozzi M, Bearzi I, Mandolesi A, Pierantoni C, Loupakis F, Zaniboni A, Negri F, Quadri A, Zorzi F, Galizia E, et al. Epidermal Growth Factor Receptor (EGFR) gene copy number (GCN) correlates with clinical activity of irinotecan-cetuximab in K-RAS wild-type colorectal cancer: a fluorescence in situ (FISH) and chromogenic in situ hybridization (CISH) analysis. BMC Cancer 2009; 9:303; PMID:19712476; http://dx.doi.org/10.1186/1471-2407-9-303
- Personeni N, Fieuws S, Piessevaux H, De Hertogh G, De Schutter J, Biesmans B, De Roock W, Capoen A, Debiec-Rychter M, Van Laethem JL, et al. Clinical usefulness of EGFR gene copy number as a predictive marker in colorectal cancer patients treated with cetuximab: a fluorescent in situ hybridization study. Clinical Cancer Res 2008; 14:5869-76; PMID:18794099; http://dx.doi.org/10.1158/1078-0432.CCR-08-0449
- Perrone F, Lampis A, Orsenigo M, Di Bartolomeo M, Gevorgyan A, Losa M, Frattini M, Riva C, Andreola S, Bajetta E, et al. PI3KCA/PTEN deregulation contributes to impaired responses to cetuximab in metastatic colorectal cancer patients. Ann Oncol 2009; 20:84-90; PMID:18669866; http://dx.doi.org/10.1093/annonc/mdn541
- Razis E, Briasoulis E, Vrettou E, Skarlos DV, Papamichael D, Kostopoulos I, Samantas E, Xanthakis I, Bobos M, Galanidi E, et al. Potential value of PTEN in predicting cetuximab response in colorectal cancer: an exploratory study. BMC cancer 2008; 8:234; PMID:18700047; http://dx.doi.org/10.1186/1471-2407-8-234
- Stintzing S, Kapaun C, Laubender RP, Jung A, Neumann J, Modest DP, Giessen C, Moosmann N, Wollenberg A, Kirchner T, et al. Prognostic value of cetuximab-related skin toxicity in metastatic colorectal cancer patients and its correlation with parameters of the epidermal growth factor receptor signal transduction pathway: results from a randomized trial of the GERMAN AIO CRC Study Group. Int J Cancer 2013; 132:236-45; PMID:22644776; http://dx.doi.org/10.1002/ijc.27654
- Miyazaki M, Furuya T, Shiraki A, Sato T, Oga A, Sasaki K. The relationship of DNA ploidy to chromosomal instability in primary human colorectal cancers. Cancer Res 1999; 59:5283-5; PMID:10537310
- Ooi A, Takehana T, Li X, Suzuki S, Kunitomo K, Iino H, Fujii H, Takeda Y, Dobashi Y. Protein overexpression and gene amplification of HER-2 and EGFR in colorectal cancers: an immunohistochemical and fluorescent in situ hybridization study. Mod Pathol 2004; 17:895-904; PMID:15143334; http://dx.doi.org/10.1038/modpathol.3800137
- Graziano F, Ruzzo A, Loupakis F, Canestrari E, Santini D, Catalano V, Bisonni R, Torresi U, Floriani I, Schiavon G, et al. Pharmacogenetic profiling for cetuximab plus irinotecan therapy in patients with refractory advanced colorectal cancer. J Clin Oncol 2008; 26:1427-34; PMID:18349392; http://dx.doi.org/10.1200/JCO.2007.12.4602
- Pander J, Gelderblom H, Antonini NF, Tol J, van Krieken JH, van der Straaten T, Punt CJ, Guchelaar HJ. Correlation of FCGR3A and EGFR germline polymorphisms with the efficacy of cetuximab in KRAS wild-type metastatic colorectal cancer. Eur J Cancer 2010; 46:1829-34; PMID:20418097; http://dx.doi.org/10.1016/j.ejca.2010.03.017
- Lurje G, Nagashima F, Zhang W, Yang D, Chang HM, Gordon MA, El-Khoueiry A, Husain H, Wilson PM, Ladner RD, et al. Polymorphisms in cyclooxygenase-2 and epidermal growth factor receptor are associated with progression-free survival independent of K-ras in metastatic colorectal cancer patients treated with single-agent cetuximab. Clin Cancer Res 2008; 14:7884-95; PMID:19047118; http://dx.doi.org/10.1158/1078-0432.CCR-07-5165
- Park JH, Han SW, Oh DY, Im SA, Jeong SY, Park KJ, Kim TY, Bang YJ, Park JG. Analysis of KRAS, BRAF, PTEN, IGF1R, EGFR intron 1 CA status in both primary tumors and paired metastases in determining benefit from cetuximab therapy in colon cancer. Cancer Chemother Pharmacol 2011; 68:1045-55; PMID:21340604; http://dx.doi.org/10.1007/s00280-011-1586-z
- Dahan L, Norguet E, Etienne-Grimaldi MC, Formento JL, Gasmi M, Nanni I, Gaudart J, Garcia S, Ouafik L, Seitz JF, et al. Pharmacogenetic profiling and cetuximab outcome in patients with advanced colorectal cancer. BMC Cancer 2011; 11:496; PMID:22117530; http://dx.doi.org/10.1186/1471-2407-11-496
- Liu W, Innocenti F, Chen P, Das S, Cook EH, Jr., Ratain MJ. Interethnic difference in the allelic distribution of human epidermal growth factor receptor intron 1 polymorphism. Clin Cancer Res 2003; 9:1009-12; PMID:12631599
- Tol J, Dijkstra JR, Klomp M, Teerenstra S, Dommerholt M, Vink-Borger ME, van Cleef PH, van Krieken JH, Punt CJ, Nagtegaal ID. Markers for EGFR pathway activation as predictor of outcome in metastatic colorectal cancer patients treated with or without cetuximab. Eur J Cancer 2010; 46:1997-2009; PMID:20413299; http://dx.doi.org/10.1016/j.ejca.2010.03.036
- Van Cutsem E, Lenz HJ, Kohne CH, Heinemann V, Tejpar S, Melezinek I, Beier F, Stroh C, Rougier P, van Krieken JH, et al. Fluorouracil, leucovorin, and irinotecan plus cetuximab treatment and RAS mutations in colorectal cancer. J Clin Oncol 2015; 33:692-700; PMID:25605843; http://dx.doi.org/10.1200/JCO.2014.59.4812
- De Vriendt V, De Roock W, Di Narzo AF, Tian S, Biesmans B, Jacobs B, Budinska E, Sagaert X, Rossi S, D'Ario G, et al. DUSP 4 expression identifies a subset of colorectal cancer tumors that differ in MAPK activation, regardless of the genotype. Biomarkers 2013; 18:516-24; PMID:23875912; http://dx.doi.org/10.3109/1354750X.2013.819038
- Oliveras-Ferraros C, Vazquez-Martin A, Cufi S, Queralt B, Baez L, Guardeno R, Hernandez-Yague X, Martin-Castillo B, Brunet J, Menendez JA. Stem cell property epithelial-to-mesenchymal transition is a core transcriptional network for predicting cetuximab (Erbitux) efficacy in KRAS wild-type tumor cells. J Cell Biochem 2011; 112:10-29; PMID:21104905; http://dx.doi.org/10.1002/jcb.22952
- Amador ML, Oppenheimer D, Perea S, Maitra A, Cusatis G, Iacobuzio-Donahue C, Baker SD, Ashfaq R, Takimoto C, Forastiere A, et al. An epidermal growth factor receptor intron 1 polymorphism mediates response to epidermal growth factor receptor inhibitors. Cancer Res 2004; 64:9139-43; PMID:15604284; http://dx.doi.org/10.1158/0008-5472.CAN-04-1036
- Miller R, Siegmund D. Maximally selected chi square statistics. Biometrics 1982; 38:1011-16; http://dx.doi.org/10.2307/2529881
- Halpern J. Maximally selected chi square statistics for small samples. Biometrics 1982; 38:1017-23; http://dx.doi.org/10.2307/2529882
- Vallbohmer D, Zhang W, Gordon M, Yang DY, Yun J, Press OA, Rhodes KE, Sherrod AE, Iqbal S, Danenberg KD, et al. Molecular determinants of cetuximab efficacy. J Clin Oncol 2005; 23:3536-44; PMID:15908664; http://dx.doi.org/10.1200/JCO.2005.09.100