ABSTRACT
Vascular Endothelial Growth Factor (VEGF) mediates angiogenesis, which is crucial for tumor development and progression. The present study aimed to evaluate the impact of VEGFA gene polymorphisms rs699947, rs833061, rs1570360, rs2010963 and rs3025039 on breast cancer features and prognosis. A cohort of Brazilian women (N = 1038) with unilateral non-metastatic breast cancer was evaluated. The association between VEGFA polymorphisms and histopathological features or pathological complete response (pCR) to neoadjuvant chemotherapy was evaluated by the Chi-square test, with calculation of the respective odds ratio (OR) and 95% confidence intervals (95% CI). The impact of individual categories on disease-free survival was evaluated using Kaplan-Meier curves and multivariate Cox proportional hazards regression models for calculation of adjusted hazard ratios (HRadjusted). Variant genotypes of rs699947 (CA + AA) were significantly associated with high-grade (G2 + G3) tumors (OR = 1.82; 95% CI = 1.15 – 2.89), and with shorter disease-free survival among patients treated with neoadjuvant chemotherapy followed by mastectomy (HRadjusted = 1.82; 95% CI = 1.16 – 2.86). Variant genotypes of rs833061 (TC + CC) were significantly associated with high-grade (G2 + G3) tumors (OR = 1.79; 95% CI = 1.12 – 2.84) and with positive lymph node status (OR = 1.34; 95% CI = 1.01 – 1.77), but showed no independent effect on disease-free survival. Variant haplotypes (*2 to *5) appear to favor pCR (OR = 7.1; 95% CI = 1.7 – 30.1). VEGFA genotyping may add to prognostic evaluation of breast cancer, with rs699947 being the most likely to contribute.
Abbreviations
| CAF-T | = | Cyclophosphamide, Doxorubicin And 5-Fluorouracyl- Docetaxel |
| DNA | = | DNA |
| ER | = | Estrogen Receptor |
| HER2 | = | Human Epidermal Growth Factor Receptor 2 |
| HR | = | Hazard Ratio |
| INCA | = | Brazilian National Cancer Institute |
| OR | = | Odds Ratio |
| PR | = | Progesterone Receptor |
| SNP | = | Single Nucleotide Polymorphism |
| TNM | = | Tumor, Nodes, Metastasis |
| TTP | = | Time To Progression |
| VEGFA | = | Vascular Endothelial Growth Factor |
Introduction
Breast cancer is the leading location of cancer among women,Citation1-3 and its incidence tends to increase all over the world.Citation3 Although breast cancer mortality decreased recently in more developed countries, it remains the first cause of death by cancer among women worldwide.Citation2
The most important prognostic factors in breast cancer are the classical morphological characteristics of the tumor, such as histological type and grade, and the presence of metastasis in lymph nodes or in other sites.Citation4 In addition, the therapeutic approach of breast cancer is based on the tumor status for hormone receptors (estrogen and progesterone), and for the epidermal growth factor receptor 2 (HER2),Citation5 which are predictive of treatment responses to hormonal therapy Citation6 or to trastuzumab,Citation7 respectively. Despite such advances, there is still great interpatient variability on clinical outcomes after standard treatment protocols, and new biomarkers and stage models are being searched, in attempt to improve prognostic evaluation and guide therapy selection.Citation8 Genes involved in carcinogenesis-related processes are natural candidates for exploring the potential of interindividual variations as new prognostic factors.Citation9,10
Angiogenesis is a crucial step in the development of cancer, being necessary to the growth of the primary tumor, invasion and metastasis,Citation11,12 and the Vascular Endothelial Growth Factor A (VEGFA) is one of its key mediators.Citation13 In breast cancer, high tumoral levels of VEGFA have been associated with increased recurrenceCitation14-17 and poor survival.Citation14,18,15-17
The expression of VEGFA gene is highly modulated,Citation19-21 and single nucleotide polymorphisms (SNPs) occur in the promoter region (rs699947, rs1570360 and rs833061), in the 5'-untranslated region (rs2010963, rs25648) and in the 3'-untranslated region (rs3025039, rs10434), next to many potential regulatory elements.Citation22,21,12 VEGFA SNPs were shown to affect VEGFA levels in different cell models,Citation21,23-25 including breast cancer.Citation26 VEGFA SNPs have also been associated with breast cancer susceptibility,Citation11,13,23,26-39 increased risk of progression Citation13,40-44 or poor survival.Citation13,43 However, most of these works focused on the effects of SNPs evaluated separately, and the results from different authors present great disparity.
The present study aimed to explore VEGFA SNPs and haplotypes as potential biomarkers for prognostic evaluation of breast cancer. Newly diagnosed patients from a cohort of Brazilian women with non-metastatic breast cancer were evaluated for the association between VEGFA SNPs or haplotypes and histophatological features of breast cancer and for their impact on the pathological resposnse to neoadjuvant chemotherapy and on the risk of early-onset disease progression.
Results
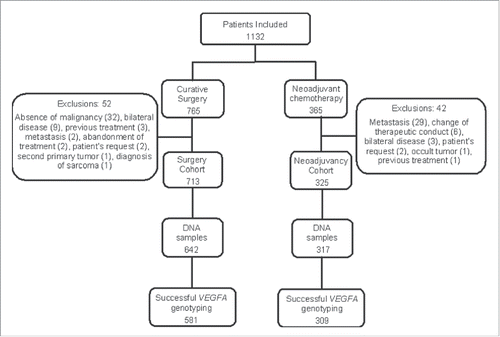
depicts the flowchart of the study, describing the formation of the study subcohorts (surgery or neoadjuvant chemotherapy), the availability of DNA samples, and the number of samples successfully genotyped for at least one SNP. The minimum rate of successful genotyping was 88% for rs1570360. Patients submitted to curative surgery as their first therapeutic approach were further treated by adjuvant chemotherapy (61.4%), hormonal therapy (12.9%), radiotherapy plus hormonal therapy (10.8% each), radiotherapy alone (5.8%) or clinically followed with no secondary intervention (9.1%). The standard chemotherapeutic protocol for both adjuvant and neoadjuvant chemotherapy was CAF-T (3 cycles of cyclophosphamide, doxorubicin and 5-fluorouracyl, followed by 3 cycles of docetaxel), which accounted for 89% of adjuvant chemotherapy and 96% of neoadjuvant chemotherapy.
presents the main clinical and histopathological characteristics of the combined study cohort (N = 1038), with the distribution of VEGFA genotypes for patients with available DNA (N = 959). Haplotypes (N= 1780) were inferred from samples with successful VEGFA genotyping (N = 890). The clinical data indicate a wide age range (20–99 years), with a median of 56 y and a high predominance of postmenopausal status. Most tumors had ductal origin, presented with invasive status and showed poorly differentiated histological grades (G2 or G3). With regards to the immunohistochemical profile, most tumors were positive to hormone receptors, Luminal A being the most frequent subtype.
Table 1. Description of the study cohort (N = 1038).
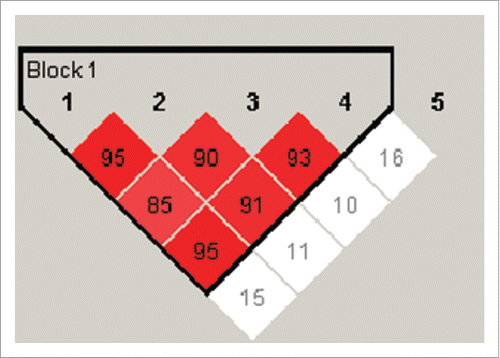
All SNPs were in Hardy-Weinberg equilibrium. The results revealed that rs699947, rs833061, rs1570360, rs2010963 were in strong linkage disequilibrium, forming a single haploblock, whereas and rs3025039 was not linked to the other SNPs (). A total of 11 haplotypes were inferred, but only 5 are presented in , since they summed 95.7% of all haplotypes occurring in the cohort.
Figure 2. Haplotype association analysis for the five VEGFA SNPs in the study cohort. Numbers in boxes indicate decimal places of D’. There was strong linkage disequilibrium across rs699947 (lane 1), rs833061 (lane 2), rs1570360 (lane 3) and 2010963 (lane 4), whereas rs3025039 (lane 5) was not linked to the other SNPs.
presents significant associations between VEGFA genotypes or haplotypes and histopathological features of breast tumors. Significant differences were found for rs699947 and rs833061 according to histological grade and for rs833061 according to the lymph node status. Variant genotypes of rs699947 (CA + AA) were shown to be significantly more prevalent among high-grade (G2 + G3) tumors (OR = 1.82; 95%CI = 1.15 – 2.89). Variant genotypes of rs833061 (TC + CC) were significantly associated with high-grade (G2 + G3) tumors (OR = 1.79; 95%CI = 1.12 – 2.84) and with positive lymph node status (OR = 1.34; 95%CI = 1.01 – 1.77). Haplotypes *3 and *4, which combine the variant alleles of rs699947 and rs833061, were also significantly associated with high-grade tumors, when compared to haplotypes *1 and *2, both containing the major alleles of rs699947 and rs833061 (OR = 1.46; 95% CI = 1.01 – 2.12). The combined haplotypes *3 and *4, however, showed no statistically significant association with positive lymph node status.
Table 2. Distribution of VEGFA genotypes and haplotypes according to histophatological features of breast tumors (N = 890).
shows the influence of clinical and histopathological characteristics of breast cancer, as well as of VEGFA genotypes and haplotypes on the proportion of tumor complete response or pathological complete response to neoadjuvant chemotherapy. The only histopathological feature significantly affecting the sensitivity to neoadjuvant chemotherapy was the status for hormone receptors, with tumors negative for ER/PR presenting higher proportion of tCR (OR = 5.2; 95% CI = 2.2 – 12.1) and pCR (OR = 5.8; 95% CI = 2.3 – 14.7). As a consequence, HER2-like and Triple-Negative tumors also exhibit higher proportion of tCR (OR = 6.7; 95% CI = 2.7 – 16.8) and pCR (OR = 8.1; 95% CI = 2.9 – 23.2), when compared to Luminal tumors. With regards to VEGFA genotypes, the results indicate no significant independent effects on the pathological response to neoadjuvant chemotherapy. However, variant haplotypes (*2 to *5) appear to be increased among patients with tCR (OR = 3.5; 95% CI = 1.3 – 8.9) or pCR (OR = 7.1; 95% CI = 1.7 – 30.1).
Table 3. Influence of histopathological features, VEGFA genotypes or haplotypes on the distribution of pathological response of breast tumors to neoadjuvant chemotherapy (N = 325).
shows the influence of clinical and histopathological characteristics, as well as of VEGFA genotypes and haplotypes, in the rates of breast cancer progression, as estimated by the median times to progression (TTP) and 95% CI. Total patient follow-up was 43,130 person-months, with a median follow-up time per person of 41.1 months. Loco-regional recurrence was observed in 33 patients, whereas distant metastasis occurred in 155 patients. Patients submitted to curative surgery or to neoadjuvant chemotherapy as first treatment were evaluated separately. Large tumor size (T2 or T3), positive lymph node status, high histological grade (G2 + G3) and negative status for hormone receptors were significantly associated with shorter TTP in both subcohorts. As expected, tumor stage, defined by TNM, was also a good predictor of breast cancer progression. Regarding VEGFA SNPs, only rs699947 showed a significant effect on disease-free survival, with variant genotypes significantly increasing the rate of breast cancer progression among patients treated with neoadjuvant chemotherapy (). No significant effects on breast cancer disease-free survival were observed for any of the VEGFA haplotypes, when individual diplotypes were considered.
Figure 3. Disease-free survival curve in breast cancer neoadjuvancy subcohort according to VEGFA genotypes rs699947 (CC vs CA/AA). Multivariate Cox proportional hazards regression was used to calculate the adjusted hazard ratio (HRadjusted), including tumor size, lymph nodes status, tumor grade and status for hormone receptors (estrogen or progesterone) as other independent predictors of breast cancer progression.
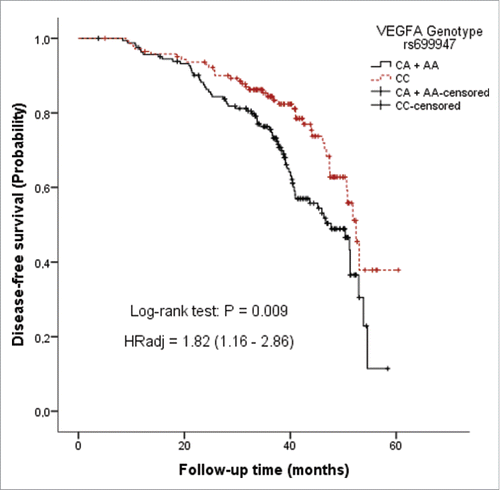
Table 4. Influence of individual features on the 5-year disease-free survival of breast cancer patients
Discussion
The present study aimed to evaluate the prognostic contribution of five common VEGFA SNPs (rs699947, rs833061, rs1570360, rs2010963, rs3025039), and its haplotypes, in breast cancer. The study was conducted in a prospective manner, based on a single-institution cohort of Brazilian women with first diagnosis of unilateral non-metastatic breast cancer. INCA is a reference institution for treatment of cancer in Brazil, and patients were mostly residents from different cities of the state of Rio de Janeiro (16.5 million inhabitants). Although relatively limited in the sample size, the study design allowed full verification of the patients' clinical history, as well as minimization of diversity in therapeutic conducts and in follow-up routines. Such design aspects contribute to reduce uncontrolled confounding factors that may affect association analyses. However, as a main limitation, the study was based on a recently established cohort, with most patients being diagnosed in less than five years. Therefore, the available follow-up time was too short to evaluate overall survival, and even the results on disease-free survival must be regarded as suggestive, rather than conclusive.
Our finding of a positive association between variant rs699947 genotypes and high-grade tumors is in opposite direction of that reported by Jin et al,Citation23 who found an association between variant homozygous AA and low-grade tumors, whereas other authors reported no association between rs699947 and tumor grade.Citation37,45,46 Likewise, the results indicating positive association of variant genotypes of rs833061 with high histological grade and lymph node metastases at diagnosis also appear to contradict those of Sa-Nguanraksa et al,Citation44 who reported lower lymphovascular invasion of breast tumors with the variant homozygous genotype CC. The impact of rs833061 in breast cancer has also been studied by Rahoui et al.,Citation37 who found no significant associations with histopathological features. Nevertheless, the authors only included 70 patients, which limit the statistical power for association analyses.
Because rs699947 and rs833061 are in strong linkage disequilibrium, it is likely that they might have similar effects. Accordingly, our results indicate that the combined presence of alleles A of rs699947 and C of rs833061, in haplotypes *3 and *4, maintains the magnitude of the association with high-grade tumors.
The positive association of both rs699947 and rs833061 with high-grade tumors, and of rs833061 with higher lymph node invasion, suggests that these two SNPs, and possibly their haplotypes, either segregate with more aggressive breast cancer phenotypes or may lead to higher VEGF levels, which is known to be associated with worse outcomes of breast cancer.Citation14,15-18 In agreement with the latter notion, Radovich et al.,Citation47 who characterized the impact of VEGFA haplotypes in gene reporter systems, showed that the two haplotypes containing the alleles A of rs699947 and C of rs833061 resulted in higher luciferase expression in most cell lines, both in normoxia and in hypoxia conditions. Similarly, Koukourakis et al. Citation25 reported increased VEGF expression for the A allele of rs699947.
With regards to rs1570360 or rs3025039, our results indicate no significant associations with breast cancer characteristics, which appear to corroborate previous reports either for rs1570360 Citation23,42 or rs3025039.Citation23,26,30,42,45
In relation to rs2010963, however, our lack of significant associations with histopathological features of breast cancer contradicts previous studies, which have indicated that patients with the variant CC Citation23,35,42 or GC genotypes Citation44 have larger tumor size (> 20 mm),if compared to those with the major GG genotype. Luo et al.Citation35 also found positive association between the variant homozygous CC genotype of rs2010963 and high histological grade, whereas Sa-Nguanraksa et al.Citation44 found positive associations for the GC genotype with perineural invasion and advanced tumor stage (II-IV). In accordance, Balasubramanian et al.Citation48 reported an association between the C allele and maximum size of invasive component. In contrast, Langsenlehner et al.Citation45 found that the C allele was associated with lower tumor size.
Such disparities of results among studies might be due to differences in study design, as well as to different haploblocks among populations. Our results indicate a single haploblock, formed by rs699947, rs833061, rs1570360, rs2010963, whereas rs3025039 is not linked to the others. Among Brazilians, the variant rs2010963 C appears to occur alone, being a tag SNP for haplotype *2, which accounts for approximately one third of breast cancer patients. This pattern of haploblock corroborates previous reports, which point the haplotype containing the variant C allele of rs2010963 as the most frequent in the USA,Citation47 Thailand Citation44 or Morroco,Citation37 as well as in Brazil, among endometriosis patients or controls.Citation49 Although Kidd et al. Citation43 studied only rs699947, rs1570360 and rs2010963, their haplotype analysis also indicated rs2010963 C allele occurring alone, and forming the major haplotype among Americans, with a frequency of 0.35. Jin et al.Citation23 also reported linkage disequilibrium involving rs699947, rs1570360 and rs2010963, but not rs3025039, among Europeans. In contrast, Langsenlehner et al.Citation45 suggested two separate blocks of linkage disequilibrium for VEGFA in Austrians, one formed by five SNPs upstream of the coding sequence and another with the two SNPs downstream of the coding sequence.
With regards to disease-free survival of non-metastatic breast cancer patients, seven previous studies have explored the role of VEGFA SNPs.Citation13,38,40,42-44,50 However, the studies differed about design, population characteristics and SNPs selected, and the results must be compared with caution. In relation to rs699947, Maae et al. Citation42 and Sa-Nguanraksa et al. Citation44 found no significant results on disease-free survival, whereas Kidd et al. Citation43 reported an increased risk of breast cancer recurrence (HR = 1.58, 95% CI = 1.06–2.35; p = 0.03) for the haplotype formed by rs699947 C, rs1570360 G and rs2010963 G. The authors claimed that such effect might be related to rs699947 C allele, which was not independently predictive of breast cancer outcomes, but improved the 5- and 8-year predictive accuracy of standard prognostic indicators. It is not clear, however, how patients were treated or how the tumor samples were selected. In our study, unlike the results by Kidd et al.Citation43, there was no significant effect of any VEGFA haplotype on the risk of breast cancer recurrence, but the rs699947 A allele (present in variant genotypes CA or AA) was associated with lower disease-free survival for patients initially treated with neoadjuvant chemotherapy.
Our data regarding rs699947 might be explained by increased VEGF expression for the A allele,Citation25 and hence the increase of angiogenesis, leading to worse clinical outcomes.Citation14,15-18 The lack of significant effect of variant rs699947 genotypes on the disease-free survival after curative surgery appears to be related to a lower risk of progression in comparison with the neoadjuvant chemotherapy subcohort (see ). In fact, patients were referred to neoadjuvant chemotherapy when curative surgery was not recommended due to large tumor size or local neoplastic invasion of non-mammary tissues (e.g. skin). These morphological differences suggest a more pronounced role of angiogenesis among patients from the neoadjuvant chemotherapy subcohort. Accordingly, our results indicate that VEGFA variant haplotypes favor tCR and pCR to neoadjuvant chemotherapy, possibly because the increased angiogenesis might improve tumor access of cytotoxic antineoplastic chemotherapy. In a recent paper, Hein et al.,Citation51 exploring 125 SNPs in 15 genes involved in the VEGFA pathway, showed that rs833058 and rs699947 may also favor pCR in patients treated with bevacizumab. With regards to rs1570360, one study reported lower disease-free survival for the heterozygous genotype,Citation44 whereas two other studies failed to confirm such risk association.Citation42,43 In relation to rs2010963, three studies described increased breast cancer recurrence for the homozygous variant CC genotype,Citation13,42,44 which was not observed by Lu et al.Citation13 In addition, despite the reported strong linkage disequilibrium between rs699947, rs833061 and rs1570360, the studies failed to confirm risk associations for their variant haplotypes.Citation13,42,44
Finally, with respect to rs3025039, which is not linked to the other VEGFA SNPs, Etienne-Grimaldi et al. Citation41 described a significantly beneficial effect for the T allele (P = 0.022), with an increased time to progression (11.5 months) when compared to CC genotype (9.7 months) among metastatic breast cancer patients treated with bevacizumab. The beneficial effect of rs3025039 T allele could be due to a reduction on the stability of mRNA transcripts of VEGFA,Citation19 leading to lower VEGF plasma levels compared with subjects homozygous for the C allele.Citation26,41,52 However, such potentially protective effect of rs3025039 T allele against breast cancer recurrence was not observed with non-metastatic cases.Citation13,42 Moreover, Absenger et al.Citation40 reported an increased risk of breast cancer recurrence for the T allele among non-metastatic patients (HR 1.880; 95% CI 1.020–3.465; P = 0.043).
Although there is no clear consensus regarding the association between VEGFA SNPs or haplotypes and breast cancer characteristics, our results support the notion that they might contribute for further classification of tumor subtypes, and should be evaluated for their potential role as prognostic biomarkers, especially among HER2-like or Triple-Negative tumors, which have less therapeutic options, and are more likely to benefit from new therapies, including bevacizumab.Citation53-55
Materials and methods
Subjects and study design
The study population consisted of an on-going prospective hospital-based cohort of Brazilian women with first diagnosis of unilateral breast carcinoma and no identification of distant metastases. Patients were recruited when assigned for curative surgery or neoadjuvant chemotherapy as their first therapeutic approach at the Brazilian National Cancer Institute (INCA), during the period from February 2009 to April 2013. The study protocol was approved by the Ethics Committees of the Brazilian National Cancer Institute (INCA #129/08) and of the National School of Public Health (FIOCRUZ/CAAE36047514.5.0000.5240), and all patients gave written consent to participate. The REMARK guidelines (REporting recommendations for tumor MARKer prognostic studies) were followed.Citation56
Histopathological characterization
The histopathological evaluation was performed with tumor biopsies obtained for diagnostic purposes, as part of institutional routine procedures. All individual data were obtained from electronic medical records, and the data on hormone receptors and HER2 status were used for biological classification of tumors,Citation5 with definition of four subtypes: Luminal A, positive for both estrogen receptor (ER) and progesterone receptor (PR), but negative for HER2; Luminal B, positive for either ER or PR, regardless of HER2 status, or positive for the three receptors; HER2-like, negative for both ER and PR, but positive for HER2; and Triple-Negative, when negative for the three receptors.
Pathological response to neoadjuvant chemotherapy
The pathological response was evaluated based on previously described regression grading systems.Citation5,57 Briefly, pathological complete response (pCR) was defined by no microscopic evidence of residual viable tumor cells (invasive or noninvasive) in all resected specimens of the breast and lymph nodes, whereas tumor complete response (tCR) was characterized by no residual tumor or only residual noninvasive (in situ) in breast tissue irrespective of lymph node involvement.
Survival outcomes
Disease-free survival was defined as the primary clinical endpoint of the study. Disease progression was characterized by the occurrence of loco-regional or contra-lateral recurrence of breast cancer, or by the detection of any distant metastasis. The time to progression (TTP) was calculated as the period of time between the first therapeutic approach (surgery or neoadjuvant chemotherapy) and the date of the first exam indicating disease progression. Patients were considered disease-free if they had no suggestive clinical symptoms or imaging diagnosis of disease progression until their last medical consult. New primary cancer lesions or deaths by causes unrelated to disease progression were censored in survival analyses. Patients achieving five y of follow-up were also censored.
Genotyping analyses
Peripheral blood samples (3 mL) were collected from all subjects, and DNA was extracted using the Blood Genomic Prep Mini Spin Kit (GE Heathcare, Buckinghamshire, UK), following the procedures recommended by the manufacturer.
VEGFA SNPs were selected based on their frequency and on previous reports of functional effects on the regulation of VEGFA expression.Citation21,24,25,58
Patients were genotyped for rs699947 (−2578C>A), rs833061(-1498T>C or −460T>C), rs1570360 (−1154G>A), rs2010963 (−634G>C or 405G>C) and rs3025039 (936C>T) by allelic discrimination using TaqMan→ SNP Genotyping Assays (Applied Biosystems, Warrington, UK), as previously described.Citation49
Statistical analyses
A descriptive study of the cohort was conducted, presenting relative frequencies for each categorical variable. Allelic and genotypic frequencies were derived by gene counting, and the adherence to the Hardy–Weinberg principle was evaluated by the Chi-square test for goodness of-fit. Haplotype patterns were inferred using Haploview 4.2 [Haploview internet version], based on the algorithm of expectation and maximization.Citation59 Individual diplotypes were inferred using the Haplo Stats software, version 1.3.Citation60
Individual features were dichotomized for better and worse prognostic values, and their associations with VEGFA genotypes or alleles were evaluated by the Chi-square or Fisher's exact tests. Significant associations (p < 0.05) were further tested in multiple regression analyses, with calculation of adjusted odds ratios (ORadjusted), and respective 95% confidence intervals (95% CI).
Disease-free survival curves were estimated using the Kaplan–Meier product-limit method, with the influence of individual variables on the mean time to disease progression being evaluated with the two-sided log-rank test. The impact of individual variables on disease-free survival rates was estimated by calculation of their hazard ratios (HR), and 95% confidence intervals (95%CI). The significant covariates were included in multivariate Cox proportional hazards regression models to calculate the adjusted HR (HRadjusted) and respective 95%CI of new identified independent prognostic factors of breast cancer progression.
All statistical analyses were conducted using SPSS 13.0 for Windows (SPSS Inc., Chicago, Illinois).
Compliance with ethical standards
The study was conducted following the international precepts of ethics in research, including the 1975 Helsinki declaration and its later amendments, and of good clinical practice. The authors complied with the Brazilian regulation of clinical research. The protocol was approved by the Ethics Committee of the Brazilian National Cancer Institute (INCA #129/08) and of the National School of Public Health (FIOCRUZ/CAAE36047514.5.0000.5240), and all patients gave written consent to participate.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors thank Dr. Guilherme Suarez-Kurtz for the use of laboratory facilities, and the personnel from the Breast Cancer Hospital (HC3-INCA), and from the National Bank of Tumors in the Brazilian National Cancer Institute (BNT-INCA), for logistic support in sample and data collection. This study was supported by Conselho Nacional de Pesquisa e Desenvolvimento (CNPq 474522/2010–5), Fundação Carlos Chagas Filho de Amparo à Pesquisa no Rio de Janeiro (FAPERJ E-26/110.356/2010 and E-26/010.002644/2014), and INCT para Controle do Câncer (CNPq 573806/2008–0; FAPERJ E26/170.026/2008). HAVM, DRFA and TNC were awarded by Fundação Carlos Chagas Filho de Amparo à Pesquisa no Rio de Janeiro; JMAD and MSL were awarded by Conselho Nacional de Pesquisa e Desenvolvimento. CTN, JAP and SMTG-C were awarded by Fundação do Câncer.
References
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin 2011; 61:69-90; PMID:21296855; http://dx.doi.org/10.3322/caac.20107
- Ferlay J, Shin H-R, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 2010; 127:2893-917; PMID:21351269; http://dx.doi.org/10.1002/ijc.25516
- Ly D, Forman D, Ferlay J, Brinton LA, Cook MB. An international comparison of male and female breast cancer incidence rates. Int J Cancer 2013; 132:1918-26; PMID:22987302; http://dx.doi.org/10.1002/ijc.27841
- Soerjomataram I, Louwman MWJ, Ribot JG, Roukema JA, Coebergh JWW. An overview of prognostic factors for long-term survivors of breast cancer. Breast Cancer Res Treat 2008; 107:309-30; PMID:17377838; http://dx.doi.org/10.1007/s10549-;007-9556-1
- Huober J, Minckwitz G, Denkert C, Tesch H, Weiss E, Zahm DM, Belau A, Khandan F, Hauschild M, Thomssen C, et al. Effect of neoadjuvant anthracycline–taxane-based chemotherapy in different biological breast cancer phenotypes: overall results from the GeparTrio study. Breast Cancer Res Treat 2010; 124:133-40; PMID:20697801; http://dx.doi.org/10.1007/s10549-010-1103-9
- Early Breast Cancer Trialists' Collaborative Group. Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet 2005; 365:1687-717; PMID:15894097; http://dx.doi.org/10.1016/S0140-6736(05)66544-0
- Pritchard KI, Shepherd LE, O'Malley FP, Andrulis IL, Tu D, Bramwell VH, Levine MN. HER2 and Responsiveness of Breast Cancer to Adjuvant Chemotherapy. N Engl J Med 2006; 354:2103-11; PMID:16707747; http://dx.doi.org/10.1056/NEJMoa054504
- Danova M, Delfanti S, Manzoni M, Mariucci S. Tissue and Soluble Biomarkers in Breast Cancer and Their Applications: Ready to Use?. JNCI Monogr 2011; 2011:75-8; PMID:22043046; http://dx.doi.org/10.1093/jncimonographs/lgr023
- Ruiz C, Tolnay M, Bubendorf L. Application of personalized medicine to solid tumors: opportunities and challenges. Swiss Med Wkly 2012; 142:w13587; PMID:22714262; http://dx.doi.org/10.4414/smw.2012.13587
- Chung CC, Chanock SJ. Current status of genome-wide association studies in cancer. Hum Genet 2011; 130:59-78; PMID:21678065; http://dx.doi.org/10.1007/s00439-011-1030-9
- Jakubowska A, Gronwald J, Menkiszak J, Górski B, Huzarski T, Byrski T, Edler L, Lubiński J, Scott RJ, Hamann U. The VEGF_936_C&T 3′UTR polymorphism reduces BRCA1-associated breast cancer risk in Polish women. Cancer Lett 2008; 262:71-6; PMID:18171601; http://dx.doi.org/10.1016/j.canlet.2007.11.029
- Sa-nguanraksa D, O-charoenrat P. The Role of vascular endothelial growth factor a polymorphisms in breast cancer. Int J Mol Sci 2012; 13:14845-64; PMID:23203097; http://dx.doi.org/10.3390/ijms131114845
- Lu H. Association of genetic polymorphisms in the vegf gene with breast cancer survival. Cancer Res 2005; 65:5015-9; PMID:15958542; http://dx.doi.org/10.1158/0008-5472.CAN-04-2786
- Gasparini G, Bonoldi E, Gatti C, Vinante O, Toi M, Tominaga T, Gion M, Dittadi R, Verderio P, Boracchi P, et al. Prognostic Significance of Vascular Endothelial Growth Factor Protein in Node-Negative Breast Carcinoma. JNCI J Natl Cancer Inst 1997; 89:139-47; PMID:8998183; http://dx.doi.org/10.1093/jnci/89.2.139
- Gasparini G. Prognostic value of vascular endothelial growth factor in breast cancer. Oncologist 2000; 5:37-44; PMID:10804090; http://dx.doi.org/10.1634/theoncologist.5-suppl_1-37
- Linderholm BK, Lindahl T, Holmberg L, Klaar S, Lennerstrand J, Henriksson R, Bergh J. The expression of vascular endothelial growth factor correlates with mutant p53 and poor prognosis in human breast cancer. Cancer Res 2001; 61:2256-60; PMID:11280795
- Bando H, Weich HA, Brokelmann M, Horiguchi S, Funata N, Ogawa T, Toi M. Association between intratumoral free and total VEGF, soluble VEGFR-1, VEGFR-2 and prognosis in breast cancer. Br J Cancer [Internet] 2005; 92(3):553-61; PMID:15668703; http://dx.doi.org/10.1038/sj.bjc.6602374
- Linderholm B, Tavelin B, Grankvist K, Henriksson R. Vascular endothelial growth factor is of high prognostic value in node-negative breast carcinoma. J Clin Oncol Off J Am Soc Clin Oncol 1998; 16:3121-8; PMID:9738584
- Shih S-C, Claffey KP. Regulation of human vascular endothelial growth factor mrna stability in hypoxia by heterogeneous nuclear ribonucleoprotein L. J Biol Chem 1999; 274:1359-65; PMID:9880507; http://dx.doi.org/10.1074/jbc.274.3.1359
- Schultz A, Lavie L, Hochberg I, Beyar R, Stone T, Skorecki K, Lavie P, Roguin A, Levy AP. Interindividual heterogeneity in the hypoxic regulation of VEGF : significance for the development of the coronary artery collateral circulation. Circulation 1999; 100:547-52; PMID:10430770; http://dx.doi.org/10.1161/01.CIR.100.5.547
- Watson CJ, Webb NJ., Bottomley MJ, Brenchley PE. Identification of polymorphisms within the vascular endothelial growth factor (VEGF) gene: correlation with variation in VEGF protein production. Cytokine 2000; 12:1232-5; PMID:10930302; http://dx.doi.org/10.1006/cyto.2000.0692
- Brogan IJ, Khan N, Isaac K, Hutchinson JA, Pravica V, Hutchinson IV. Novel polymorphisms in the promoter and 5′ UTR regions of the human vascular endothelial growth factor gene. Hum Immunol 1999; 60:1245-9; PMID:10626738; http://dx.doi.org/10.1016/S0198-8859(99)00132-9
- Jin Q. Vascular endothelial growth factor polymorphisms in relation to breast cancer development and prognosis. Clin Cancer Res 2005; 11:3647-53; PMID:15897560; http://dx.doi.org/10.1158/1078-0432.CCR-04-1803
- Awata T, Inoue K, Kurihara S, Ohkubo T, Watanabe M, Inukai K, Inoue I, Katayama S. A common polymorphism in the 5’-untranslated region of the VEGF gene is associated with diabetic retinopathy in Type 2 diabetes. Diabetes 2002; 51:1635-9; PMID:11978667; http://dx.doi.org/10.2337/diabetes.51.5.1635
- Koukourakis MI, Papazoglou D, Giatromanolaki A, Bougioukas G, Maltezos E, Siviridis E. VEGF gene sequence variation defines VEGF gene expression status and angiogenic activity in non-small cell lung cancer. Lung Cancer Amst Neth 2004; 46:293-8; PMID:15541813; http://dx.doi.org/10.1016/j.lungcan.2004.04.037
- Krippl P, Langsenlehner U, Renner W, Yazdani-Biuki B, Wolf G, Wascher TC, Paulweber B, Haas J, Samonigg H. A common 936 C/T gene polymorphism of vascular endothelial growth factor is associated with decreased breast cancer risk. Int J Cancer 2003; 106:468-71; PMID:12845639; http://dx.doi.org/10.1002/ijc.11238
- Jacobs E, Feigelson H, Bain E, Brady K, Rodriguez C, Stevens V, Patel A, Thun M, Eugenia E. Polymorphisms in the vascular endothelial growth factor gene and breast cancer in the Cancer Prevention Study II cohort. Breast Cancer Res 2006; 8:1-6; PMID:16613616; http://dx.doi.org/10.1186/bcr1400
- Kataoka N. Population-Based case-control study of VEGF gene polymorphisms and breast cancer risk among chinese women. Cancer Epidemiol Biomarkers Prev 2006; 15:1148-52; PMID:16775174; http://dx.doi.org/10.1158/1055-9965.EPI-05-0871
- Gerger A, Langsenlehner U, Renner W, Weitzer W, Eder T, Yazdani-Biuki B, Hofmann G, Samonigg H, Krippl P. A multigenic approach to predict breast cancer risk. Breast Cancer Res Treat 2007; 104:159-64; PMID:17058024; http://dx.doi.org/10.1007/s10549-006-9408-4
- Eroğlu A, Öztürk A, Çam R, Akar N. Vascular endothelial growth factor gene 936 C/T polymorphism in breast cancer patients. Med Oncol 2008; 25:54-5; PMID:18188715; http://dx.doi.org/10.1007/s12032-007-0046-4
- Schneider BP, Radovich M, Sledge GW, Robarge JD, Li L, Storniolo AM, Lemler S, Nguyen AT, Hancock BA, Stout M, et al. Association of polymorphisms of angiogenesis genes with breast cancer. Breast Cancer Res Treat 2008; 111:157-63; PMID:17891484; http://dx.doi.org/10.1007/s10549-007-9755-9
- Wehrschuetz M, Schöllnast H, Wehrschuetz E, Renner W, Luschin G. VEGF 936C > T Polymorphism and Association of BI-RADS score in women with suspected breast cancer. Breast Cancer Basic Clin Res 2009; 3:77-81; PMID:21556250
- Oliveira C, Lourenço GJ, Silva PMR, Cardoso-Filho C, Favarelli MHC, Gonçales NSL, Gurgel MSC, Lima CSP. Polymorphisms in the 5′- and 3′-untranslated region of the VEGF gene and sporadic breast cancer risk and clinicopathologic characteristics. Tumor Biol 2011; 32:295-300; PMID:20981515; http://dx.doi.org/10.1007/s13277-010-0121-x
- Rodrigues P, Furriol J, Tormo E, Ballester S, Lluch A, Eroles P. The single-nucleotide polymorphisms +936 C/T VEGF and −710 C/T VEGFR1 are associated with breast cancer protection in a Spanish population. Breast Cancer Res Treat 2012; 133:769-78; PMID:22315135; http://dx.doi.org/10.1007/s10549-012-1980-1
- Luo T, Chen L, He P, Hu Q-C, Zhong X-R, Sun Y, Yang Y-F, Tian T-L, Zheng H. Vascular endothelial growth factor (vegf) gene polymorphisms and breast cancer risk in a chinese population. Asian Pac J Cancer Prev 2013; 14:2433-7; PMID:23725153; http://dx.doi.org/10.7314/APJCP.2013.14.4.2433
- Kapahi R, Guleria K, Sambyal V, Manjari M, Sudan M, Uppal MS, Singh NR. Vascular endothelial growth factor (VEGF) gene polymorphisms and breast cancer risk in Punjabi population from North West India. Tumour Biol J Int Soc Oncodevelopmental Biol Med 2014; 35:11171-81; PMID:25106408; http://dx.doi.org/10.1007/s13277-014-2404-0
- Rahoui J, Laraqui A, Sbitti Y, Touil N, Ibrahimi A, Ghrab B, Al Bouzidi A, Moussaoui Rahali D, Dehayni M, Ichou M, et al. Investigating the association of vascular endothelial growth factor polymorphisms with breast cancer: a Moroccan case–control study. Med Oncol 2014; 31:193. PMID:25148899; http://dx.doi.org/10.1007/s12032-014-0193-3
- Slattery ML, John EM, Torres-Mejia G, Lundgreen A, Lewinger JP, Stern MC, Hines L, Baumgartner KB, Giuliano AR, Wolff RK. Angiogenesis genes, dietary oxidative balance and breast cancer risk and progression: The breast cancer health disparities study: angiogenesis and breast cancer risk and progression. Int J Cancer 2014; 134:629-44; PMID:23832257; http://dx.doi.org/10.1002/ijc.28377
- Yan Y, Liang H, Li T, Guo S, Li M, Li S, Qin X. Vascular endothelial growth factor +936C/T polymorphism and breast cancer risk: a meta-analysis of 13 case–control studies. Tumor Biol 2014; 35:2687-92; PMID:24390659; http://dx.doi.org/10.1007/s13277-013-1354-2
- Absenger G, Szkandera J, Stotz M, Pichler M, Winder T, Langsenlehner T, Langsenlehner U, Samonigg H, Renner W, Gerger A. A common and functional gene variant in the vascular endothelial growth factor a predicts clinical outcome in early-stage breast cancer: VEGF-A predicts outcome in breast cancer. Mol Carcinog 2013; 52:96-102; PMID:23625573; http://dx.doi.org/10.1002/mc.22028
- Etienne-Grimaldi M-C, Formento P, Degeorges A, Pierga JY, Delva R, Pivot X, Dalenc F, Espié M, Veyret C, Formento J-L, et al. Prospective analysis of the impact of VEGF-A gene polymorphisms on the pharmacodynamics of bevacizumab-based therapy in metastatic breast cancer patients: VEGF-A gene polymorphisms and bevacizumab-based treatment. Br J Clin Pharmacol 2011; 71:921-8; PMID:21204912; http://dx.doi.org/10.1111/j.1365-2125.2010.03896.x
- Maae E, Andersen RF, Steffensen KD, Jakobsen EH, Brandslund I, Sørensen FB, JAKOBSEN A. Prognostic Impact of VEGFA Germline Polymorphisms in patients with HER2-positive primary breast cancer. Anti Cancer Res 2012; 32:3619-28; PMID:22993299
- Kidd LR, Brock GN, VanCleave TT, Benford ML, Lavender NA, Kruer TL, Wittliff JL. Angiogenesis-associated sequence variants relative to breast cancer recurrence and survival. Cancer Causes Control 2010; 21:1545-57; PMID:20571871; http://dx.doi.org/10.1007/s10552-010-9583-9
- Sa-Nguanraksa D, Chuangsuwanich T, Pongpruttipan T, Kummalue T, Rojananin S, Ratanawichhitrasin A, Prasarttong-Osoth P, Chuthatisith S, Pisarnturakit P, Aeumrithaicharoenchok W, et al. Vascular endothelial growth factor 634G/C polymorphism is associated with increased breast cancer risk and aggressiveness. Mol Med Rep 2013; 8:1242-50; PMID:23904001; http://dx.doi.org/10.3892/mmr.2013.1607
- Langsenlehner U, Wolf G, Langsenlehner T, Gerger A, Hofmann G, Clar H, Wascher TC, Paulweber B, Samonigg H, Krippl P, et al. Genetic polymorphisms in the vascular endothelial growth factor gene and breast cancer risk. The Austrian “tumor of breast tissue: incidence, genetics, and environmental risk factors” study. Breast Cancer Res Treat 2008; 109:297-304; PMID:17636397; http://dx.doi.org/10.1007/s10549-007-9655-z
- Koutras AK, Kotoula V, Papadimitriou C, Dionysopoulos D, Zagouri F, Kalofonos HP, Kourea HP, Skarlos DV, Samantas E, Papadopoulou K, et al. Vascular endothelial growth factor polymorphisms and clinical outcome in patients with metastatic breast cancer treated with weekly docetaxel. Pharmacogenomics J 2014; 14:248-55; PMID:24061601; http://dx.doi.org/10.1038/tpj.2013.36
- Radovich M, Hancock BA, Kassem N, Mi D, Skaar TC, Schneider BP. Resequencing of the vascular endothelial growth factor promoter reveals haplotype structure and functional diversity. Angiogenesis 2010; 13:211-8; PMID:20552269; http://dx.doi.org/10.1007/s10456-010-9173-1
- Balasubramanian SP, Cox A, Cross SS, Higham SE, Brown NJ, Reed MW. Influence of VEGF-A gene variation and protein levels in breast cancer susceptibility and severity. Int J Cancer 2007; 121:1009-16; PMID:17471570; http://dx.doi.org/10.1002/ijc.22772
- Perini J, Cardoso J, Berardo P, Vianna-Jorge R, Nasciutti L, Bellodi-Privato M, Machado D, Abrão M. Role of vascular endothelial growth factor polymorphisms (−2578C> A, −460 T> C, −1154G> A, +405G > C and +936C> T) in endometriosis: a case–control study with Brazilians. BMC Womens Health 2014; 14:117; PMID:25255852; http://dx.doi.org/10.1186/1472-6874-14-117
- Dorjgochoo T, Zheng Y, Gao Y-T, Ma X, Long J, Bao P, Zhang B, Wen W, Lu W, Zheng W, et al. No association between genetic variants in angiogenesis and inflammation pathway genes and breast cancer survival among Chinese women. Cancer Epidemiol 2013; 37:619-24; PMID:23850146; http://dx.doi.org/10.1016/j.canep.2013.06.005
- Hein A, Lambrechts D, von Minckwitz G, Häberle L, Eidtmann H, Tesch H, Untch M, Hilfrich J, Schem C, Rezai M, et al. Genetic variants in VEGF pathway genes in neoadjuvant breast cancer patients receiving bevacizumab: Results from the randomized phase III GeparQuinto study. Int J Cancer 2015; 137:2981-8; PMID:26100253; http://dx.doi.org/10.1002/ijc.29656
- Renner W, Kotschan S, Hoffmann C, Obermayer-Pietsch B, Pilger E. A Common 936 C/T mutation in the gene for vascular endothelial growth factor is associated with vascular endothelial growth factor plasma levels. J Vasc Res 2000; 37:443-8; PMID:11146397; http://dx.doi.org/10.1159/000054076
- Brufsky AM, Hurvitz S, Perez E, Swamy R, Valero V, O'Neill V, Rugo HS. RIBBON-2: A randomized, double-blind, placebo-controlled, phase III trial evaluating the efficacy and safety of bevacizumab in combination with chemotherapy for second-line treatment of human epidermal growth factor receptor 2-negative metastatic breast cancer. J Clin Oncol 2011; 29:4286-93; PMID:21990397; http://dx.doi.org/10.1200/JCO.2010.34.1255
- Kristensen T, Knutsson M, Wehland M, Laursen B, Grimm D, Warnke E, Magnusson N. Anti-vascular endothelial growth factor therapy in breast cancer. Int J Mol Sci 2014; 15:23024-41; PMID:25514409; http://dx.doi.org/10.3390/ijms151223024
- Fakhrejahani E, Toi M. Antiangiogenesis therapy for breast cancer: an update and perspectives from clinical trials. Jpn J Clin Oncol 2014; 44:197-207; PMID:24474817; http://dx.doi.org/10.1093/jjco/hyt201
- McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. Statistics Subcommittee of the NCI-EORTC Working Group on Cancer Diagnostics. Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK). JNCI J Natl Cancer Inst 2005; 97:1180-4; PMID:16106022http://dx.doi.org/10.1093/jnci/dji237
- Sinn HP, Schmid H, Junkermann H, Huober J, Leppien G, Kaufmann M, Bastert G, Otto HF. Histologic regression of breast cancer after primary (neoadjuvant) chemotherapy. Geburtshilfe Frauenheilkd 1994; 54:552-8; PMID:8001751; http://dx.doi.org/10.1055/s-2007-1022338
- Stevens A, Soden J, Brenchley PE, Ralph S, Ray DW. Haplotype analysis of the polymorphic human vascular endothelial growth factor gene promoter. Cancer Res 2003; 63:812-6; PMID:12591731
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005; 21:263-5; PMID:15297300; http://dx.doi.org/10.1093/bioinformatics/bth457
- Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet 2002; 70:425-34; PMID:11791212; http://dx.doi.org/10.1086/338688