ABSTRACT
Curcumin, the primary bioactive component isolated from turmeric, has been shown to possess variety of biologic functions including anti-cancer activity. However, molecular mechanisms in different cancer cells are various. In the present study, we demonstrated that curcumin induced G2/M cell cycle arrest and apoptosis by increasing the expression levels of cleaved caspase-3, cleaved PARP and decreasing the expression of BCL−2 in U937 human leukemic cells but not in K562 cells. We found some interferon induced genes, especially interferon-induced protein with tetratricopeptide repeats 2 (IFIT2), were significantly upregulated when treated with curcumin in U937 cells by gene expression chip array, and further confirmed that the expression of IFIT2 was obviously higher in U937 than that in K562 cells by Western blot assay. In addition, inhibiting the expression of IFIT2 by shRNA in U937 rescued curcumin-induced apoptosis and exogenous overexpression of IFIT2 by lentiviral transduction or treating with IFNγ in K562 cells enhanced anti-cancer activity of curcumin. These results indicated for the first time that curcumin induced leukemic cell apoptosis via an IFIT2-dependent signaling pathways. The present study identified a novel mechanism underlying the antitumor effects of curcumin, and may provide a theoretical basis for curcumin combined with interferon in the cancer therapeutics.
KEYWORDS:
Abbreviations
| IFNs | = | Interferons |
| ISGs | = | IFN-stimulated genes |
| IFIT2 | = | IFN-induced protein with tetratricopeptide repeats 2 |
| TPR | = | tetratricopeptide repeats |
| IRES | = | internal ribosome entry site |
| ECMV | = | encephalomyocarditis virus |
Introduction
Leukemia, cancer of the haematopoietic cells, is the leading cause of cancer death in adults and children.Citation1 In 2012, leukemia developed in 352,000 people globally and caused 265,000 deaths.Citation2 Chemotherapy is the major form of treatment of leukemia, and the survival rates have improved remarkably over the past decades. However, the potential side effects of cytotoxic chemotherapy significantly impact the effect of drugs.Citation3 Thus, new therapeutic weapons, or methods of prevention are required.
Curcumin, an active ingredient derived from turmeric, has been recognized for its medicinal properties.Citation4 Multiple studies in both animals and human indicated that curcumin possesses a variety of potent properties, including antioxidant, anti‐inflammation, radical‐scavenging, anticancer, and so on.Citation5,6 Several studies indicated that curcumin can suppress the proliferation of leukemia and induce cell death.Citation7-9 Besides, it shows no significant side effects compared with other chemotherapeutic drugs.Citation10 Therefore, curcumin may represent a new therapeutic strategy for treatment of leukemia.Citation11
Considerable work has been conducted to explore how curcumin works.Citation7-9 Extensive research over the last decades has revealed that curcumin exerts anticancer effect through affecting numerous molecular and biochemical cascades. The molecular targets of curcumin include growth factors, growth factor receptors, transcription factors, cytokines, enzymes, and genes regulating apoptosis and proliferation.Citation12 However, the molecular mechanisms in different tumor cells are different.
Interferon-induced protein with tetratricopeptide repeats 2 (IFIT2), also known as ISG54, GARG39, and MuP54, is originally discovered as a direct response to type I IFN. Further studies proved that it was also induced as a primary stress response to infection which independent of IFNs, and might play a role in anticancer.Citation13-16 IFIT2 regulates the functions of cell cycle, apoptosis, tumor colonization and viral replication, which confer cellular resistance to viral infections and regulates proliferation, apoptosis and migration of cancer cells.Citation17-20
In this report, we found that curcumin induced expression of interferon regulatory genes (especially IFIT2) in luekemia cell U937. upregulation of IFIT2 by exogenous expressing or treating with IFNγ in K562 increased cells apoptosis and enhanced anticancer effect of curcumin. And shRNA-mediated IFIT2 knockdown inhibited curcumin-induced apoptosis in U937 cells. Our results demonstrated for the first time that there is a cross-talk between curcumin and interferon signaling pathways, which provides the basis for curcumin combined with interferon in cancer therapeutics.
Results
Sensitivity of leukemia cell lines to curcumin
To assess anticancer effect of curcumin on leukemia, 5 leukemia cell lines (NB4, NB4-R1, K562, HL60 and U937) were treated with different concentration of curcumin for 24 h and growth viability were analyzed by MTT assay. As shown in , The concentration of curcumin inhibiting 50% cell proliferation (IC50) in leukemia cell lines were: 15.77 ± 2.17μM for NB4, 16.67 ± 1.63 μM for NB4-R1, 27.86 ± 2.45 μM for HL60, 27.67 ± 2.72 μM for K562 and 8.63 ± 2.34 μM for U937 respectively.
Figure 1. Sensitivity of leukemia cell lines to curcumin (A) Leukemia cells(NB4,NB4-R1, HL60, K562 and U937) were treated with curcumin, IC50 were determined by MTT assay. (B-C) K562 and U937 were exposed to curcumin (0, 2.5, 5 and 10 μM), cell viability were measured by MTT on 24 h and 48 h; Each value were represents the mean ± SD.
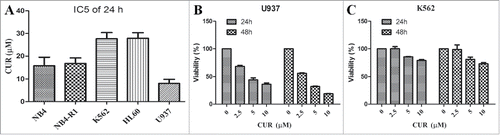
Given the intriguing results, we chose K562 and U937 as research objects to study the mechanism under this condition. We treated K562 and U937 cells with different concentration of curcumin (0–10 μM) for 24 h and 48 h. MTT results showed that curcumin significantly inhibited U937 cell growth in time- and dose-dependent manner (), while K562 cells were not sensitive to curcumin (). These results indicated that sensitivity of leukemic cells to curcumin was different.
Curcumin induces G2/M phase arrest in leukemia cells
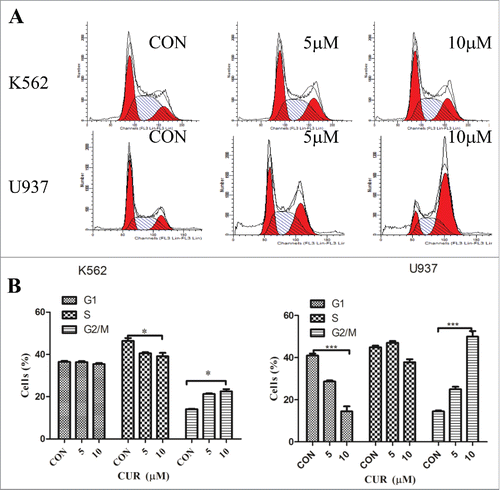
To further define anti-tumor effect of curcumin in leukemia cells, we analyzed cell cycle progression in K562 and U937 cells treated with curcumin (0, 5 and 10 μM) for 24 hours by FCM assay. As the results shown in , although curcumin blocked the cells in the G2/M phase, there were some differences in K562 and U937 cells: an Significantly increased number of cells were detected in G2/M phase and corresponding reduction in G0/G1 phase of U937 cells were observed in a dose-dependent manner. However, in K562 cells, the proportion of G2/M phase slightly increased and accompanied by a decrease in the S phase.
Curcumin induces apoptosis in U937 but not in K562 cells
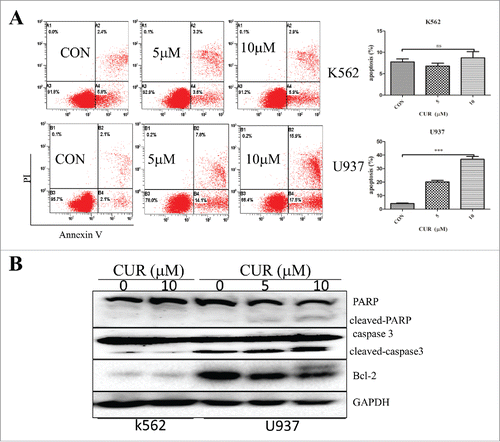
To determine apoptosis induction capacity of curcumin in leukemia cells, we treated U937 and K562 cells with curcumin (0, 5, and 10 μM) for 24 h, and detected the percent of apoptotic cells by Annexin-V/PI staining assay. The results showed that curcumin induced apoptosis in U937 with a dose-depend manner but not in K562 (). Furthermore, in U937 cells, 2 apoptosic proteins (cleaved caspase3 and PARP) were markedly increased and the apoptosis inhibitor protein Bcl−2 was decreased. However, the proteins in the K562 changed slightly ().
Figure 3. Effect of curcumin on inducing apoptosis in leukemia cell lines. (A)The percentage of early apoptosis and late apoptosis were detected by Annexin-V/PtdIns double-staining assay. The percentage of apoptotic cell in different concentration of curcumin. Each value were represents the mean ± SD ns means not significant, *p < 0.05,***p < 0.001. (B) K562 and U937 cells treated with curcumin, apoptosic proteins (caspase3 and PAPR) and the apoptosis inhibitor protein Bcl−2 were dectected by western blot.
Curcumin induces the expression of interferon regulatory genes in U937 but not in K562 cells
To explore the molecular mechanisms of different impact of curcumin on K562 and U937 cells, we analyzed change of gene expression in U937 after treated with curcumin (10 μM) for 24 h by gene expression chip. Chip results showed that over 1000 genes were changed and most genes have been reported previously, such as skp2, myc, and GADD45A. We found that some of the interferon regulatory genes, especially IFIT2 gene expression was significantly upregulated when treated with curcumin ().
Figure 4. (A) Curcumin induces the expression of interferon regulatory genes in U937 but not in K562 cells Change of partial genes expression in U937 treated with curcumin (10μmol/L) for 24 h analyzed by gene chip; (B) Validation of genes expression in U937 and K562 when treated with curcumin for 24 h by RT-PCR. Each value were represents the mean ± SD; (D) Validation of genes (SKP2, IFIT22, IFIT3) expression in U937 and K562 when treated with curcumin for 24 h by western blot array.
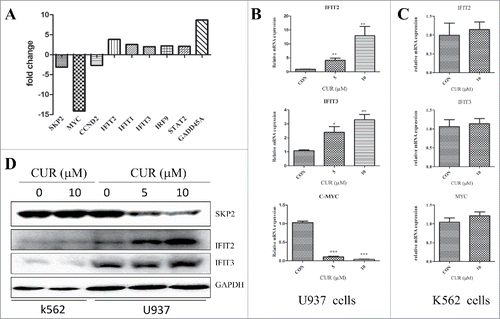
Subsequently, RT-qPCR and western-blot were performed to confirm the change of gene expression in U937 observed by gene chip. SKP2, MYC, IFIT2, IFIT3 were selected owning to their biologic function. U937 cells were treated with different concentration of curcumin (0, 5 and 10 μM), and genes expression of MYC, IFIT2, IFIT3 and proteins level of SKP2, IFIT2, and IFIT3 were determined by RT-qPCR and western blot respectively. As shown in and , the results were consistent with the previous results of gene chip assay. However, there is no difference in K562 cells with or without curcumin (), which suggesting the gene expression induced by curcumin in U937 cells might associated with sensitivity of leukemic cells to curcumin.
Downregulation of IFIT2 by RNA interference rescues curcumin-induced apoptosis in U937 cells
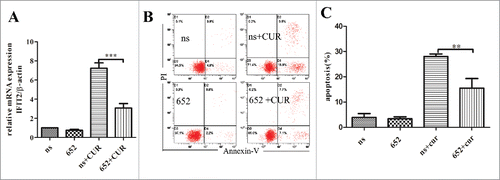
We knocked down IFIT2 in U937 so as to confirm the hypothesis that IFIT2 plays vital role in anticancer process of curcumin. After infected with IFIT2-shRNA or nonspecific shRNA lentiviral for 3 days, U937 cells were exposed with 10 μM curcumin for 24 h, and the knockdown efficiency of IFIT2 were analyzed by using RT-qPCR to examine the expression of IFIT2 in cells. As seen in , compared with nonspecific shRNA, the mRNA of IFIT2 decreased more than 65% in IFIT2-knocked down cells when treated with curcumin. The percentage of apoptotic cells was also reduced significantly in IFIT2-shRNA infected cells after treating with curcumin ( and ). These data add to the growing evidence that IFIT2 plays vital role in the apoptosis-induction effect of curcumin.
IFIT2 enhances sensitivity of K562 cells to curcumin
Based on effect of IFIT2 on cell cycle, apoptosis, tumor colonization and viral replication, we hypothesized that IFIT2 might make contributions to anti-tumor effect of curcumin. To test the hypothesis, IFIT2 gene-containing viruses were packaged in HEK-293T cells, and then infected K562 cells for 5 d. Expression of IFIT2 was analyzed by both RT-qPCR and western blot. As shown in , mRNA and protein levels of IFIT2 were significantly increased, and expression of SKP2 and myc genes was decreased at the same time (), which implying IFIT2 might play a role in inhibiting proliferation in K562 cells.
Figure 5. Knockdown of IFIT2 by shRNA decreases curcumin-induced apoptosis (A) IFIT2 specific shRNA or nonspecific (ns) shRNA lentivirals infected U937 cells for 72 hours, and cells stimulated with curcumin (10μM) for 24 hours. The knockdown efficiency of endogenous IFIT2 were analyzed by RT-qPCR. (B and C) Cells expressing IFIT2 specific shRNA or nonspecific (ns) shRNA were evaluated for their apoptotic response to curcumin by Annexin-V/PtdIns double-staining assay and flow cytometry. **p < 0.01,***p < 0.001.
To investigate the roles of IFIT2 apoptosis-induction activity of curcumin, Annexin-V/PtdIns double-staining assay was performed to analyze apoptosis of the cells exposed to curcumin (10 μM) for 24 h. As the results seen in , compared with vector group, the percentage of apoptotic cells was found to be obviously increased in the IFIT2 overexpressed cells, and curcumin further potentiated apoptosis of IFIT2 overexpressed cells. These results support the hypothesis that IFIT2 plays an important role in the apoptosis-induction activity of curcumin.
IFNγ enhances curcumin-induced apoptosis in K562 cells
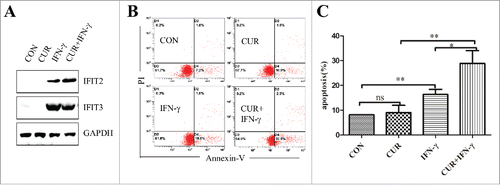
Since IFIT2 is an IFN induced protein, we supposed that IFNγ could enhance the anticancer effect of curcumin by inducing the expression of IFIT2 protein. K562 cells were treated with IFNγ (5000 u/ml) alone or combined with curcumin (10 μM) for 24 h. We found that IFNγ induced the expression of IFIT2 and IFIT3 proteins (), and then we detected the apoptosis of cells treated with regents by FCM analysis. As shown in and , although curcumin had little effect on the apoptosis of K562 cell, 5000u/ml IFNγ induced apoptosis of K562 cells, and IFNγ combined with curcumin even further strengthen apoptosis-induction effect. Depending on these results, we concluded that IFN induced IFIT2 expression and enhanced the efficacy of curcumin for the treatment of leukemia.
Figure 6. Exogenous overexpression of IFIT2 enhances sensitivity of K562 cells to curcumin (A) FIT2 gene-containing lentiviral infected K562 cells and detected the expression of IFIT2 by RT-PCR and western blot; (B) the expression of c-myc and skp2 were analyzed by RT-PCR after overexpression of IFIT2; (C) K562, K562-Vector and K562-IFIT2 were treated with or without 10μmol/L for 24 h, Annexin-V/PtdIns double-staining assay was used to detect percentage of apoptotic cells. *P < 0.05; ***P < 0.001.
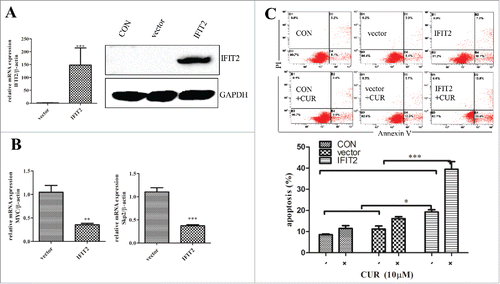
Figure 7. Effect of curcumin combined with IFN on inducing apoptosis of K562. (A) Expression of IFIT2 and IFIT3 in K562 cells was measured by western blot after treating with curcumin, IFNγ, or both IFNγ and curcumin; (B) The apoptosis were analyzed in K562 cells following treatment of curcumin, FNγ, or FNγ combined with curcmin by FMC assay; (C) The percentage of apoptotic cells in K562 treated with curcumin, IFNγ, or both curcumin and IFNγ. Each value were represents the mean ± SD(n = 3). ns means not significant; *P < 0.05; **P < 0.01.
Discussion
In our present study, we investigated the role of IFIT2 in anti-leukemia process of curcumin, and explored whether Interferens (IFNs) enhanced the apoptosis-induction effect of curcumin. Initially, we studied a panel of leukemia cell lines and found that sensitivity of these cell lines to curcumin were different. As curcumin is a promised anticancer drug, exploration of mechanism of curcumin can widen its clinical application and bring more benefit to patients. To clarify it, one sensitive cell line U937 and one resistant cell line K562 were chosen in the present study. We found that gene expression of IFIT2 was decreased with curcumin in both dose- and time-dependent manner in U937 cells but not in K562. To further investigate the role of IFIT2, IFIT2 was overexpressed in K562 and knocked down in U937. The results showed that IFIT2 could increase the anticancer effect of curcumin, and when combined with IFNγ, curcumin has a better effect on leukemia cells. These results not only demonstrate the efficacy of IFIT2, but also suggest the synergistic effects of curcumin and IFNγ.
IFIT2, which is a member of ISRE-containing genes, have been shown could be a biomarker of cancer.Citation19,21,22 Lee TC et al. reported that IFIT2 could inhibit migration and metastasis of oral squamous cell carcinoma (OSCC) cells by activation of atypical PKC signaling.Citation19,23,24 In addition, IFIT2 also be proved to suppress development of human adencarcinoma of gastric esophageal junction.Citation25 These results indicated that IFIT2 was related to anti-cancer effect. IFIT2 has been demonstrated involved in anticancer process of other agents, but its role in the treatment of leukemia by curcumin still unknown.Citation26,27 Several researchers have shown that IFIT2 can induce apoptosis via a mitochondrial pathway dependent on Bcl−2Citation28,29, and curcumin has been reported inducing apoptosis via BCL−2 signaling pathway as well.Citation30-32 The previous study suggested that BCR-ABL kinase domain inhibited the expression of ISRE-containing genes,Citation33 which could in part associated with resistant of K562. All the data are consistent with our findings.
IFNγ used to be a therapeutic agent against chronic myeloid leukemia with a moderate efficiency.Citation34 In this study, we found that combination of curcumin and IFNγ enhanced curcumin-mediated anti-cancer activity, and showed for the first time that curcumin-induced leukemic cell apoptosis is an IFIT2-dependent signaling pathway.
Taken up, our results suggested for the first time that IFIT2 enhance the effect of curcumin, and supported that combinational of curcumin and IFNγ would shed a new light on leukemia therapy. Because of great strides could be made in the use of curcumin, further mechanistic exploration of it is warranted.
Materials and methods
Cells and reagents
U937, K562, NB4, NB4-R1, HL60 cells were maintained in RPMI-1640 containing 10% fetal bovine serum under 37 °C with 5% CO2. Curcumin (Sigma, St. Louis, MO) was dissolved in DMSO as a stock solution at 5 mM. IFNγ was available from R&D systems and dissolved in PBS buffer at 106 u/ml. The antibodies against GAPDH, PARP, caspase 3, Bcl−2, Skp2, IFIT2 and IFIT3 were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Annexin-V-FITC/PtdIns kit was purchased from Bestbio Biotechnology (Bestbio, China). Cell cycle detection kit (COULTER DNA PREP reagent kit) was from Beckman coulter, Inc. Reverse transcription reagent and SuperReal qPCR PreMix (SYBR Green) reagent kit were purchased from TIANGEN Biotech (Beijing) Co., Ltd. The lentiviral expression vector pLVX-Puro, pLVX-shRNA1 vector and the lentiviral packaging plasmids were provided by Clontech Laboratories, Inc. The pseudoviral particle producer cell line 293T was cultured with DMEM supplemented with 10% fetal bovine serum.
MTT assays
Two × 104 cells per well were seeded into 96-well plates in 100 μl volume and cultivated in RPMI1640 medium with 10% fetal bovine serum at 37 °C. Cells were grown for 24 h and 48 h, respectively. 10 μl of MTT reagent was added at indicated time. Then the cells were incubated in 37 °C after covered in aluminum foil. 2 hours later, centrifuged and removed the supernatant. 100 μl DMSO was added to each well and read the results by Elisa reader after an additional 15-minutes incubation.
Cell cycle detection by PtdIns single-staining assay
To detect effect of curcumin on cell cycle arrest, PtdIns single-staining assay was conducted. Cells were treated with the drug and cultivated under 37°C saturated humidity and 5% CO2 for 24 h. 106 cells were harvested in the appropriate manner (centrifuged at 2,000 rpm for 5 min) and removed the supernatant. Add 50 µl DNA PREP LPR reagent for 1 min according to the instructions, and then added 300 µl DNA PREPStain reagent and place it at room temperature for 30 min. and then detected by CYTOMICS FC500 ( Beckman coulter, inc ) and analyzed cell cycle by MODFIT2 software.
Apoptosis analysis by FMC
Cells were treated with or without drug and cultured under 37°C for 24 h. 106 cells were harvested in the appropriate manner( centrifuged at 2,000 rpm for 5 min), 5 µl Annexin-V were added after adding 300µl Annexin-V binding solution and placed the mixture at 4°C for 15min. Added 10 µl PtdIns at indicated time, then analyzed the results by cytometer. Annexin V+ and/or PtdIns+ cells are apoptosis cells.
RT-qPCR
Cells ( 2 × 106 ) were grown in cell culture bottles and then treated with the indicated concentrations of drug for 24 h. Total RNA were extracted by TRIzol reagent (Invitrogen) and reverse-transcribed to the cDNA synthesis and the following qPCR with the specific primers were as follows: IFIT2: 5′-ctgcaaccatgagtgagaac-3′ and antisense 5′-caggtgaccagacttctgat-3′; IFTIT3: sence 5′-tcatgagtgaggtcaccaag-3′, antisense 5′- cctcgttgttaccatctagg-3′; MYC: sence 5′-gctgcttagacgcgattt-3′ and antisence 5′-caccgagtcgtagtcgaggt-3′; Skp2: sence 5′-atgccccaatcttgtccatct-3′and antisence 5′-caccgactgagtgataggtgt-3′; β-actin: sense 5′-catgtacgttgctatccaggc-3′ and antisense 5′- ctccttaatgtcacgcacgat-3′.
The relative quantity of target gene expression was analyzed using the comparative CT (2-ΔΔCT) method.
Western blot analysis
Proteins were extracted by lysis buffer (1% Triton X-100, 50 mM Tris (pH 8.0), 150 mM NaCl, 1 mM PMSF, 1 mM Na3VO4, and protease inhibitor cocktail). Proteins from cell lysates (20µg) were electrotransferred to nitrocellulose membranes after separated on 10% SDS-PAGE. Before blotted with primary antibody overnight at 4°C, membranes were sealed for 1 h at room temperature in Tris-buffered saline-0.05% Tween-20 (TBST) containing 5% non-fat dry milk. After 3 × 10 min washes in PBS, membranes were incubated with peroxidase-conjugated secondary antibody for 1 hr. Following 3 additional 10 min washes with TBST, and proteins were imaged by enhanced chemi-luminescence detection reagent and detected with Bio-Rad ChemiDoc XRS+ chemiluminescence imaging system (Bio-rad laboratories Inc.).
Gene chip expression
Total RNA were isolated from cells using TRIzol reagent (Invitrogen) and sent to Shanghai Genechem Co.,Ltd where further analyses were conducted. RNA integrity number for all samples should be greater than 7.0 (Agilent 2100 Bioanalyzer, Agilent, Santa Clara, CA). Hybridization and labeling (GeneChip Hybridization Oven 645) was used where aRNA was synthesized by GeneChip 3′IVT Express Kit. Posthybridization and washes were performed by GeneChip Fluidics Station 450(Affymetrix,Santa Clara, CA).The results scanned by GeneChip Scanner 3000 (Affymetrix,Santa Clara, CA) was analyzed by GenePix Pro 4.0 software.
Lentiviral Transduction
The lentiviral expression vector and the lentiviral packaging plasmids were transfected into 293T cells by EndoFectin™ (GeneCopoeia, Inc. US) according to the manufacturer's procedure. Lentivirus were harvested and concentrated at ultrahigh speed for 1 h and stored at −80°C.Cells were incubated 20 hours in 16-well plate at 37°C in a humidified incubator in an atmosphere of 5% CO2. Then 1 ml medium and Hexadimethrine bromide were added after removal of old medium. Before adding 200μl of lentiviral particles, gently swirl the plate to mix. Then the mixture was incubated at 37°C in a humidified incubator in an atmosphere of 5–7% CO2. 24 h later, the media containing lentiviral particles was removed from wells and fresh media was added. Later, the cells were cultivated for another 48 h.
shRNA Knockdown
Nucleotide sequences (IFIT2–652) used for IFIT2 gene silencing was got from document.Citation28 Double-stranded oligonucleotides targeting human IFIT2 cDNA was cloned into pLVX-shRNA1 vector. Virus packaging and infection were seen above.
Statistical analysis
All data were repeated 3 times and presented as mean ± SD. (n = ≥ 3) SPSS13.0 software (SPSS Inc., Chicago, IL) was used for all analyses.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Author contributions
Zhanglin Zhang contributed to the study design., Yonglu Zhang, Yunyuan Kong, Shuyuan Liu, Lingbing Zeng, Lagen Wan, Zhanglin Zhang preformed the research and conducted the data analysis. Zhanglin Zhang and Yonglu Zhang wrote the manuscript.
Acknowledgments
We thank all members of Department of Clinical Laboratory of the First Affiliated Hospital of Nanchang University for their support.
Funding
This work was supported by Natural Science Foundation of Jiangxi province (20122BAB215012 and 20142BAB215068); science and technology support program of Jiangxi province (20121BBG70049).
References
- Jacquel A, Herrant M, Legros L, Belhacene N, Luciano F, Pages G, Hofman P, Auberger P, et al. Imatinib induces mitochondria-dependent apoptosis of the Bcr-Abl-positive K562 cell line and its differentiation toward the erythroid lineage. Faseb J 2003; 17(14):2160-2; PMID:14597677; http://dx.doi.org/10.1096/fj.03-0322fje
- World Cancer Report 2014. World Health Organization 2014. p. 13.
- Dilnawaz F, Singh A, Sahoo SK. Transferrin-conjugated curcumin-loaded superparamagnetic iron oxide nanoparticles induce augmented cellular uptake and apoptosis in K562 cells. Acta Biomater 2012; 8(2):704-19; PMID:22051236; http://dx.doi.org/10.1016/j.actbio.2011.10.022
- Grynkiewicz G, Ślifirski P. Curcumin and curcuminoids in quest for medicinal status. Acta Biochim Pol 2012; 59(2):201-12. PMID:22590694
- Maheshwari RK, Singh AK, Gaddipati J, Srimal RC. Multiple biological activities of curcumin: a short review. Life Sci 2006; 78(18):2081-7; PMID:16413584; http://dx.doi.org/10.1016/j.lfs.2005.12.007
- Yao QY, Xu BL, Wang JY, Liu HC, Zhang SC, Tu CT. Inhibition by curcumin of multiple sites of the transforming growth factor-beta1 signalling pathway ameliorates the progression of liver fibrosis induced by carbon tetrachloride in rats. BMC Complement Altern Med 2012; 12:156; PMID:22978413; http://dx.doi.org/10.1186/1472-6882-12-156
- Mapoung S, Pitchakarn P, Yodkeeree S, Ovatlarnporn C, Sakorn N, Limtrakul P. Chemosensitizing Effects of Synthetic Curcumin Analogs on Human Multi-drug Resistance Leukemic Cells. Chem Biol Interact 2015; 244:140-8; PMID:26689174; http://dx.doi.org/10.1016/j.cbi.2015.12.001
- Nagy LI, Fehér LZ, Szebeni GJ, Gyuris M, Sipos P, Alföldi R, Ózsvári B, Hackler L Jr, Balázs A, Batár P, et al. Curcumin and Its Analogue Induce Apoptosis in Leukemia Cells and Have Additive Effects with Bortezomib in Cellular and Xenograft Models. Biomed Res Int 2015; 2015:968981; PMID:26075279; http://dx.doi.org/10.1155/2015/968981
- Taverna S, Giallombardo M, Pucci M, Flugy A, Manno M, Raccosta S, et al. Curcumin inhibits in vitro and in vivo chronic myelogenous leukemia cells growth: a possible role for exosomal disposal of miR-21. Oncotarget 2015; 6:21918-33; PMID:26116834; http://dx.doi.org/10.18632/oncotarget.4204
- Banerjee S, Chakravarty AR. Metal complexes of curcumin for cellular imaging, targeting, and photoinduced anticancer activity. Acc Chem Res 2015; 48(7):2075-83; PMID:26158541; http://dx.doi.org/10.1021/acs.accounts.5b00127
- Hatcher H, Planalp R, Cho J, Torti FM, Torti SV. Curcumin: from ancient medicine to current clinical trials. Cell Mol Life Sci 2008; 65(11):1631-52; PMID:18324353; http://dx.doi.org/10.1007/s00018-008-7452-4
- Goel A, Kunnumakkara AB, Aggarwal BB. Curcumin as “Curecumin:” from kitchen to clinic. Biochem Pharmacol 2008; 75(4):787-809; PMID:17900536; http://dx.doi.org/10.1016/j.bcp.2007.08.016
- Daly C, Reich NC. Double-stranded RNA activates novel factors that bind to the interferon-stimulated response element. Mol Cell Biol 1993; 13(6):3756-64; PMID:8388546; http://dx.doi.org/10.1128/MCB.13.6.3756
- Daly C, Reich NC. Characterization of specific DNA-binding factors activated by double-stranded RNA as positive regulators of interferon alpha/beta-stimulated genes. J Biol Chem 1995; 270(40):23739-46; PMID:7559546; http://dx.doi.org/10.1074/jbc.270.40.23739
- Lai KC, Chang KW, Liu CJ, Kao SY, Lee TC. IFN-induced protein with tetratricopeptide repeats 2 inhibits migration activity and increases survival of oral squamous cell carcinoma. Mol Cancer Res 2008; 6(9):1431-9; PMID:18819931; http://dx.doi.org/10.1158/1541-7786.MCR-08-0141
- Levy D, Larner A, Chaudhuri A, Babiss LE, Darnell JE. Interferon-stimulated transcription: isolation of an inducible gene and identification of its regulatory region. Proc Natl Acad Sci USA 1986; 83(23):8929-33; PMID:3466167; http://dx.doi.org/10.1073/pnas.83.23.8929
- Sen GC, Fensterl V. Crystal structure of IFIT2 (ISG54) predicts functional properties of IFITs. Cell research 2012; 22(10):1407-9; PMID:22964712; http://dx.doi.org/10.1038/cr.2012.130
- Stawowczyk M, Van Scoy S, Kumar KP, Reich NC. The Interferon Stimulated Gene 54 Promotes Apoptosis. J Biol Chem 2011; 286(9):7257-66; PMID:21190939; http://dx.doi.org/10.1074/jbc.M110.207068
- Lai KC, Liu CJ, Chang KW, Lee TC. Depleting IFIT2 mediates atypical PKC signaling to enhance the migration and metastatic activity of oral squamous cell carcinoma cells. Oncogene 2013; 32(32):3686-97; PMID:22986528; http://dx.doi.org/10.1038/onc.2012.384
- Fensterl V, Wetzel JL, Ramachandran S, Ogino T, Stohlman SA, Bergmann CC, Diamond MS, Virgin HW, Sen GC. Interferon-induced Ifit2/ISG54 protects mice from lethal VSV neuropathogenesis. PLoS pathogens 2012; 8(5):e1002712; PMID:22615570; http://dx.doi.org/10.1371/journal.ppat.1002712
- Lai K C CKW, Liu C J, et al. Enhanced expression of ASB6 and IFIT2 in oral squamous cell carcinoma. Cancer Res 2006; 66(8):215.
- Sarma SN, Kim YJ, Ryu JC. Gene expression profiles of human promyelocytic leukemia cell lines exposed to volatile organic compounds. Toxicology 2010; 271(3):122-30; PMID:20359517; http://dx.doi.org/10.1016/j.tox.2010.03.014
- Regmi P LTC. Ectopic expression of truncated IFIT2 at C-terminal tetratricopeptide repeats enhances the migration activity of oral squamous cell carcinoma cells. Cancer Research 2015; 75(15):527-; http://dx.doi.org/10.1158/1538-7445.AM2015-527
- Lai KC, Liu CJ, Lin TJ, Mar AC, Wang HH, Chen CW, Hong ZX, Lee TC. Blocking TNF-α inhibits angiogenesis and growth of IFIT2-depleted metastatic oral squamous cell carcinoma cells. Cancer Lett 2016; 370(2):207-15; PMID:26515391; http://dx.doi.org/10.1016/j.canlet.2015.10.016
- Feng X, Wang Y, Ma Z, Yang R, Liang S, Zhang M, Song S, Li S, Liu G, Fan D, et al. MicroRNA-645, up-regulated in human adencarcinoma of gastric esophageal junction, inhibits apoptosis by targeting tumor suppressor IFIT2. BMC Cancer 2014; 14:633; PMID:25174799; http://dx.doi.org/10.1186/1471-2407-14-633
- Gao F, Zhao ZL, Zhao WT, Fan QR, Wang SC, Li J, Zhang YQ, Shi JW, Lin XL, Yang S, et al. miR-9 modulates the expression of interferon-regulated genes and MHC class I molecules in human nasopharyngeal carcinoma cells. Biochem Biophys Res Commun 2013; 431(3):610-6; PMID:23291181; http://dx.doi.org/10.1016/j.bbrc.2012.12.097
- Motaghed M, Al-Hassan FM, Hamid SS. Thymoquinone regulates gene expression levels in the estrogen metabolic and interferon pathways in MCF7 breast cancer cells. Int J Mol Med 2014; 33(1):8-16; PMID:24270600; http://dx.doi.org/10.3892/ijmm.2013.1563
- Stawowczyk M, Van Scoy S, Kumar KP, Reich NC. The interferon stimulated gene 54 promotes apoptosis. J Biol Chem 2011; 286(9):7257-66; PMID:21190939; http://dx.doi.org/10.1074/jbc.M110.207068
- Reich NC. A death-promoting role for ISG54/IFIT2. J Interferon Cytokine Res 2013; 33(4):199-205; PMID:23570386; http://dx.doi.org/10.1089/jir.2012.0159
- Chen MB, Wu XY, Tao GQ, Liu CY, Chen J, Wang LQ, Lu PH. Perifosine sensitizes curcumin-induced anti-colorectal cancer effects by targeting multiple signaling pathways both in vivo and in vitro. Int J Cancer 2012; 131(11):2487-98; PMID:22438101; http://dx.doi.org/10.1002/ijc.27548
- Zhang Y, Hou Z, Ge Y, Deng K, Liu B, Li X, Li Q, Cheng Z, Ma P, Li C, et al. DNA-Hybrid-Gated Photothermal Mesoporous Silica Nanoparticles for NIR-Responsive and Aptamer-Targeted Drug Delivery. ACS Appl Mater Interfaces 2015; 7:20696-706; PMID:26325285; http://dx.doi.org/10.1021/acsami.5b05522
- Zhou X, Wang W, Li P, Zheng Z, Tu Y, Zhang Y, You T. Curcumin Enhances the Effects of 5-Fluorouracil and Oxaliplatin in Inducing Gastric Cancer Cell Apoptosis Both In Vitro and In Vivo. Oncol Res 2016; 23(1):29-34; PMID:26802648; http://dx.doi.org/10.3727/096504015X14452563486011
- Katsoulidis E, Sassano A, Majchrzak-Kita B, Carayol N, Yoon P, Jordan A, Druker BJ, Fish EN, Platanias LC. Suppression of interferon (IFN)-inducible genes and IFN-mediated functional responses in BCR-ABL-expressing cells. J Biol Chem 2008; 283(16):10793-803; PMID:18287094; http://dx.doi.org/10.1074/jbc.M706816200
- Talpaz M, Kantarjian HM, McCredie KB, Keating MJ, Trujillo J, Gutterman J. Clinical investigation of human alpha interferon in chronic myelogenous leukemia. Blood 1987; 69(5):1280-8. PMID:3471281