ABSTRACT
Aldehyde dehydrogenase 5 family, member A1 (ALDH5A1) belongs to the superfamily of aldehyde dehydrogenases (ALDHs). However, the prognostic value of ALDH5A1 in ovarian cancer remains unclear. The aim of this study was to explore the relationship between ALDH5A1 and the prognosis of patients with ovarian cancer (OC). We compared the expression of ALDH5A1 in OC to that innormal controls, using GSE40595 profiling data. Tissue microarray analysis was conducted for192 OC patients, 14 adjacent normal ovary tissues, and 2 normal ovary tissues. Using the “Kaplan-Meier plotter” (KM plotter) database, updated gene expression data and survival information of a total of 1583 OC patients were used to evaluate the prognostic value of ALDH5A1 in OC patients. We found that ALDH5A1 mRNA expression was downregulated in OC patients compared with that innormal tissues. In survival analyses, we found that ALDH5A1 was positively linked to prognosis in patients with OC, particularly in those with serous ovarian cancer (SOC). In addition, high Ctranscription activity of ALDH5A1 was correlated with better overall survival in SOC patients expressing mutatedTP53, but not in those expressing wild-type TP53. In pathological grades II/III, a high mRNA level of ALDH5A1 was associated with improved overall survival. The positive association between ALDH5A1 and prognosis was found not only in early stages(I and II), but also in advanced stages (III and IV) of SOC patients. results indicate that ALDH5A1 is an excellent predictive factor of OC and may play crucial roles in OC progression.
Introduction
Ovarian cancer (OC) is the most lethal gynecological malignancy worldwide. The incidence and mortality rates of OC have been steadily increasing in China, with an estimated 22,500 OC-related deaths in 2015.Citation1 Numerous studies over the past 2 decades have evaluated OC. Despite advances in diagnostic techniques,Citation2 the long-term prognosis remains poor. It is important to identify reliable prognostic and predictive biomarkers in OC that may help in treatment decisions and eventually lead to the development of more effective therapies.
Aldehyde dehydrogenase 5 family member A1(ALDH5A1), which encodes for succinic semialdehyde dehydrogenase(SSDH), is an enzyme involved in mitochondrial glutamate metabolism. ALDH5A1 belongs to the superfamily of aldehyde dehydrogenases (ALDHs). ALDH activity is essential for the synthesis of various molecules such as retinoic acid, βine, and γ-aminobutyric acid. These molecules are important in cell proliferation, differentiation, and survival.Citation3 Activation of the ALDH5A1 gene has been detected in various cancers. Preliminary characterization of ALDH 5 enzyme activity has been reported in a human hepatoma cell line.Citation4 Higher expression of ALDH5A1 was detected in certain cancers in a meta-analysis of normalized gene expression profiles in the Gene Sapiens “body-wide” microarray database, particularly in glioma, leukemia, and lymphoma. However, breast and reproductive cancers showed moderate expression of ALDH5A1.Citation5 Few studies have examined ALDH5A1 in OC; one clinical outcome study of miRNA-related single-nucleotide polymorphismsandtreatment outcomes revealed that ALDH5A1 single-nucleotide polymorphisms were significantly associated with OC.Citation6 Additionally, SSDH(encoded by ALDH5A1), which is involved in degrading 4-aminobutyric acid (GABA) by catalyzing the oxidation of succinic semialdehyde, was reported to be strongly expressed not only in the brain, but also in the liver, pituitary, heart, and ovary.Citation7
In the present study, we examined the downregulated expression levels of ALDH5A1 in OC by analyzing the GSE40595 database and via immunohistochemistry experiments in a tissue microarray containing 192 OC tissues and 16 normal ovarian tissues. We evaluated the prognostic role of ALDH5A1 in OC patients, using the Kaplan-Meier (KM) plotter database. Our results provide important insights into ALDH5A1 as a promising biomarker of OC, particularly in SOC or OC with mutant TP53.
Materials and methods
Patients and samples
Tissue microarray samples were obtained from the University of Michigan, Biomax (OV208a) and included formalin-fixed paraffin-embedded serous papillary adenocarcinoma (n = 130), mucinous papillary adenocarcinoma(n = 24), adenocarcinoma (n = 7), serous adenocarcinoma (n = 2), disgerminoma (n = 5), endodermal sinus carcinoma(n = 7), endometrioid carcinoma(n = 3), embryonal carcinoma (n = 1), immature teratoma(n = 2), mature teratoma(n = 1), clear cell carcinoma (n = 1), transitional cell carcinoma(n = 1), strumal carcinoid(n = 1), squamous cell carcinoma from teratoma with malignant transformation(n = 3), granular cell tumor(n = 4), normal ovarian epithelial tissue (n = 2), and adjacent normal ovary tissue (n = 14). Clinical data (age, histological type, differentiation, FIGO stage, and other information) were obtained from the medical records of each patient.
GEO data sets analysis
Gene expression data (GSE40595 profiling data) was downloaded as raw signals from Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo), interpreted, normalized, and log2-scaled using the online analysis tool GCBI website (https://www.gcbi.com.cn). Identification of differentially expressed gene sets between normal and cancer profiles in GSE40595 was also performed using the GCBI online tool.
Immunohistochemistry
Immunohistochemistry was performed on paraffin blocks. The sections were mounted on poly- L-lysine-coated slides, dried at 60°C overnight, dewaxed, and gradually hydrated. Antigen retrieval was achieved by pressure cooking in 0.01 M citrate buffer for 10 min. The sections were incubated with 20% normal goat serum for 20 min at 60°C Next, the slides were incubated overnight at 4°C with primary polyclonal antibodies against ALDH5A1 (rabbit anti-human ALDH5A1; Proteintech, Rosemont, IL, USA) at a dilution of 1: 100, and then incubated with biotin-labeled anti-rabbit (mouse) IgG antibody for 20 min. The avidin-biotin-peroxidase technique was applied using diaminobenzidine for visualization and hematoxylin for counterstaining. Negative controls were preparedby replacing the primary antibody with phosphate-buffered saline. Immunostaining for ALDH5A1 was defined as the presence of brown or yellowish brown granules in the cytoplasm/membrane, and was evaluated by 2 independent pathologists who were unaware of the patients' outcome using a semiquantitative scoring system.Citation8 Briefly, the intensity of immunostaining (1 = negative; 2 = weak; 3 = moderate; 4 = strong) and proportion of immunopositive cells (1 = ≤ 10%; 2 = >10 to ≤ 50%; 3 = > 50 to ≤ 75%; 4 = >75%) were calculated, and the 2 scores of the corresponding sample were multiplied to obtain a staining index ranging from 1 to 16 points. Points ≤ 4 were marked as −, 5–8 points as +, 9–12 points as ++, and 13–16 points as +++. For statistical analysis, − and + were considered as low expression of ALDH5A, whereas ++ and +++ were considered as high (intensive) expression.
KM plotter
An online databaseCitation9 was used to assess the correlation of individual ALDH5A1 mRNA expression to progression-free survival. OC patients in the database were identified from the Cancer Biomedical Informatics Grid (http://cabig.cancer.gov/), Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/), and The Cancer Genome Atlas (http://cancergenome.nih.gov) ovarian cancer data sets.Citation9 Clinical data including gender, age, histology, stage, grade, TP53 mutation status, and applied chemotherapy were collected for all patients in WinStat 2013. The database was established using gene expression data and survival information of 1307 OC patients downloaded from Gene Expression Omnibus. ALDH5A1 was entered in the database (http://kmplot.com/analysis/index.php?p=serviceandcancer=ovar) to acquire Kaplan-Meier survival plots. Hazard ratio (HR), 95% confidence intervals, and log-rank P were determined and presented on the main plots.
Statistical analysis
Statistical analysis was performed using SPSS version 17.0 (SPSS Inc., Chicago, IL, USA). Fisher's exact test and logistic regression analysis were performed to evaluate the association of ALDH5A1 expression with clinical factors. Statistical significance was determined by the log-rank test. The results were considered statistically significant at the level of P < 0.05.
Results
ALDH5A1 expression was downregulated in OC Samples
The mRNA level of the ALDH5A1 genes in different ovarian tissue components was analyzed under the GSE40595 expression profile data set in which epithelial cells were laser-micro dissected from normal ovaries and OCs.Citation10 The transcription level of ALDH5A1 in OC was lower than that in normal ovarian tissues (P = 0.03) ().
Figure 1. ALDH5A1 was downregulated in patients with ovarian tumor. (A) Comparison of ALDH5A1 expression in microdissected tumor epithelial tissues with that of microdissected normal epithelial tissues using the GSE40595 database. (B) Immunohistochemical analysis of ALDH5A1 expression in ovarian tissues. IHC score of each patient was plotted as an individual dot in the chart. (C) Ovarian tumor samples weakly positively stained with a staining index marked as +. (D) Ovarian tumor samples moderately positively stained with a staining index marked as ++. (E) Ovarian tumor samples strongly positively stained with a staining index marked as +++. (F) Normal ovarian samples strongly positively stained with a staining index marked as +++. Others. Mucinous papillary adenocarcinoma(n = 24), adenocarcinoma (n = 7), Disgerminoma (n = 5), Endodermal sinus carcinoma(n = 7), Endometrioid carcinoma(n = 3), Embryonal carcinoma (n = 1), Immature teratoma(n = 2), Mature teratoma(n = 1), Clear cell carcinoma (n = 1), Transitional cell carcinoma(n = 1), Strumal carcinoid(n = 1), Squamous cell carcinoma from teratoma with malignant transformation(n = 3), Granular cell tumor(n = 4).
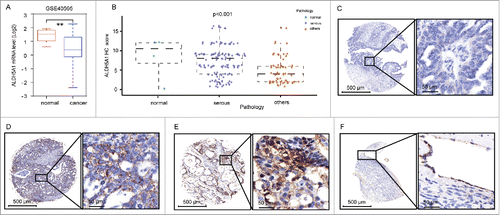
To further investigate the protein expression status of ALDH5A1 in OC tissues, immunohistochemical analysis was conducted in208 tissues containing 192 OC tissues and 16 normal ovarian tissues. As shown in , ALDH5A1 expression was obviously decreased both in SOC tissues and other pathological types. For details (), immunohistochemical analysis showed positive granular staining for ALDH5A1 distributed in the cytoplasm/membra of cells. ALDH5A1 showed strongly positive staining in 3 of 4 (75%) normal ovarian tissues (only normal 4 cases with epithelial cells), 191 OC tissues(epithelial cells were detected in 1 case) were positively stained for ALDH5A1, 123 cases showed low expression of ALDH5A1, and 68 cases showed high expressions of ALDH5A1 (P = 0.000725).
Correlation between ALDH5A1 expression and clinicopathological features
The most common type of epithelial OC is SOC, which accounts for 80–85% of all epithelial OC cases, and thus we focused on SOC. To further investigate the correlation of ALDH5A1 with SOC progress, we analyzed the relationship between ALDH5A1 expression and clinicopathological features of 132 cases of SOC. As shown in , this study consisted of 48 patients aged ≤ 50 y and 88 patients aged >50 y (P = 0.363). Carcinomas with a serous histology were the most frequent (68.8%), most patients (86.3%) presented as FIGO stage I, and a high percentage of carcinomas were GOG grade 3 (69.1%). As shown in , later stages of SOC were detected with downregulated ALDH5A1 (P< 0.05). Notably, ALDH5A1 expression was negatively correlated with the pathologic grades of SOC cancer (P< 0.005). These results indicate that a negative correlation exists between ALDH 5A1 and SOC progress.
Table 1. Association of ALDH5A1 expression with the clinicopathologic variables of primary SOC patients (132Footnoteb cases).
Prognostic value of different ALDH5A1 in OC
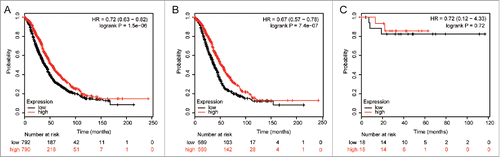
We then determined the prognostic value of different ALDH5A1 in OC, using an online tool (http://kmplot.com/analysis/index.php?p=serviceandcancer=ovar). Survival curves were plotted in www.kmplot.com for all OC patients (n = 1583), serous cancer patients (n = 1144), endometrioid cancer patients (n = 36), TP53 wild-type ovarian cancer patients (n = 87), and TP53 mutation-type ovarian cancer patients (n = 441). We assessed the prognostic effect of the mRNA expression of ALDH5A1 (Affymetrix ID is 203608_at) usingwww.kmplot.com.
High ALDH5A1 mRNA expression was found to be correlated with OS for all OC patients followed for 20 years [hazard ratio (HR) = 0.72 (0.63−0.82), P = 1.5e−06 ()], and significantly correlated with better OS for SOC patients [HR = 0.67 (0.57−0.78), P = 7.4e−07 ()], but not in endometrioid cancer patients [HR = 0.72 (0.12−4.33), P = 0.72()].
Figure 2. The prognostic effect of the ALDH5A1 mRNA level in OC was got from KM plotter (www.kmplot.com). The desired Affymetrix IDs is valid: 203608_at (ALDH5A1). Survival curves are plotted for all patients (n = 1582) (A), serous cancer patients (n = 1138) (B), endometrioid cancer patients (n = 36) (C).
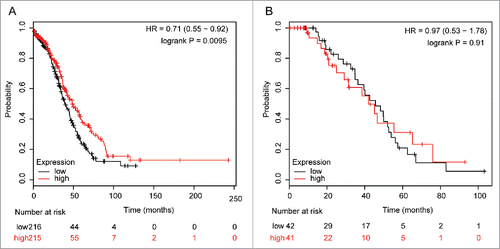
OC is characterized by ubiquitous TP53 mutation.Citation11,12 Thus, the survival curves were stratified by TP53 status to identify which patients may benefit from targeted detection (). Upregulated ALDH5A1 was significantly associated with better OS in TP53 mutation-type SOC patients, with HR = 0.71 (0.55−0.92) and P = 0.0095, but not correlated with OS in TP53 wild-type SOC patients, with HR = 0.97 (0.53−1.78) and P = 0.91.
Figure 3. The prognostic effect of the expression of ALDH5A1 mRNA in www.kmplot.com. The desired Affymetrix IDs is valid: 203608_at (ALDH5A1). Survival curves are plotted for TP53 mutated cases (n = 431) (A), for TP53 wild type cases (n = 83) (B).
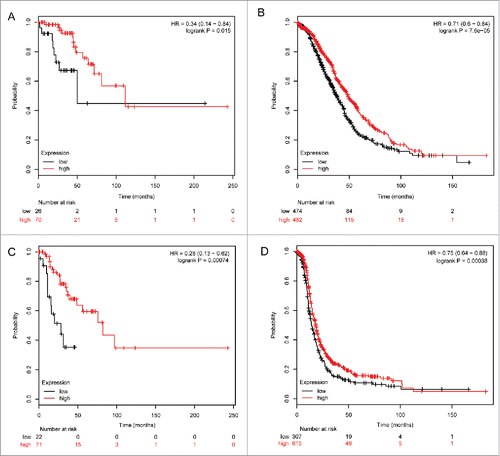
To determine the correlation of ALDH5A1 mRNA expression and other clinicopathological features, we examined the correlation with pathological grades and clinical stages of SOC patients. As the sample size was too small in patients with pathological grade IV (n = 2), we did not analyze the survival curves in this group. As shown in , ALDH5A1 mRNA was not significantly associated with pathological grade I of SOC patients, but upregulation of ALDH5A1 in pathological grade I of SOC patients was linked with better PFS, with HR = 0.24 (0.07−0.81) and P = 0.013(). In pathological grade II and III patients, high expression of ALDH5A1 mRNA was associated with improved OS/PFS [HR = 0.64 (0.44−0.92), P = 0.016 () / HR = 0.62 (0.45−0.84), P = 0.002 ()and HR = 0.64 (0.54−0.77), P = 2.3e−06 () / HR = 0.69 (0.56−0.84), P = 0.00019 ()]. High transcriptional expression of ALDH5A1 was associated with longer OS/PFS in SOC patients with clinical stages I–II [HR = 0.34 (0.14−0.84), P = 0.015 ()/ HR = 0.28 (0.13−0.62), P = 0.00074 ()] and in clinical stages III–IV [HR = 0.71 (0.6−0.844), P = 7.6e−05 () / HR = 0.75 (0.64−0.88), P = 0.00038 ()].
Figure 4. Correlation of ALDH5A1 mRNA high expression with pathological grades of ovarian cancer patients, the prognostic effect of the expression of ALDH5A1 in www.kmplot.com. The desired Affymetrix IDs is valid: 203608_at (ALDH5A1). OS (A) and PFS (D) for cases in grade I, OS (B) and PFS (E) for cases in grade II, OS (C) and PFS (F) for cases in grade III.
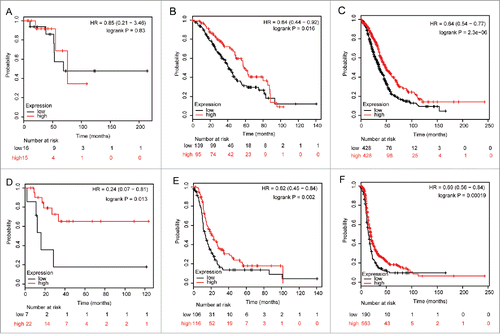
Figure 5. Correlation of ALDH5A1 mRNA high expression with clinical stages of ovarian cancer patients. The prognostic effect of the expression of ALDH5A1 in www.kmplot.com. The desired Affymetrix IDs is valid: 203608_at (ALDH5A1). OS (A) and PFS (C) for cases in stage I+II, OS (B) and PFS (D) for cases in stage III+IV.
Discussion
ALDH5A1 belongs to the ALDH family, and the gene product of ALDH5A1, also known as SSDH, is involved in catabolism of the neurotransmitter GABA, which is highly expressed in the brain.Citation5 ALDH5A1 is a novel transcript that has not previously been associated with OC. Although there is limited information regarding the role of ALDH5A1 in OC, the present study found that GABA was elevated in invasive OC tissues compared with that in in borderline ovarian tumors.Citation13 The elevated concentration of GABA in the urine of OC patients supported deregulation of the SSADH pathway.Citation14 Moreover, we found that the expression level of ALDH5A1 was significantly associated with tumor stage and grade of OC patients, and low ALDH5A1 expression was a significant indicator of poor clinical prognosis for patients with SOC. Thus, ALDH5A1 represents a novel predicative marker for the prognosis of patients with SOC.
In this study, we found significantly higher expression of ALDH5A1 in normal ovary tissues than in OC tissues, based on the GSE40595 expression profile data sets. Next, we measured and compared the expression levels of ALDH5A1 in human 192 OC tissues by immunohistochemistry. Statistical analysis revealed that the expression of ALDH5A1 was negatively correlated with advanced tumor FIGO stage and GOG grade, suggesting a crucial role forALDH5A1 in OC progression.
To further explore the potential prognostic value of ALDH5A1 in SOC, the correlation between ALDH5A1 expression and OS in OC patients was analyzed by KM plotter analysis. KM plotter isa widely used database that includes gene expression data, clinicopathological information, and survival information fora total of 1307 OC patients.Citation9 It can be used to analyze individual genes with clinical results and total patient survival. Several genes have been explored and/or recognized by KM plotter in OC.Citation15,16 We examined the prognostic values of ALDH5A1 levels in OC, using KM plotter. In this study, we found that low ALDH5A1 may predict poorer survival in OC. Additional studies are needed to explore the mechanism of ALDH5A1 in OC.
OC is characterized by ubiquitous TP53 mutation, and in SOC, TP53 somatic mutation was rather common, detected in more than 50% of tumors.Citation17 Our results have potential clinical utility, as low expression of ALDH5A1 was associated with a worse OS inTP53 mutation-type SOC patients. Thus, the significance of the correlation between p53 and individual ALDH5A1 in OC should be further analyzed.
In this study, in early stage (I and II) tumors compared with advanced stage (III and IV) tumors, ALDH5A1 had the same effect on clinical outcome. Notably, low expression of ALDH5A1 was associated with worse OS/PFS in early stage diseases(HR = 0.34/0.28), and thus identification of tumors with worse prognosis may help in the determination of optimal individual therapy. Upregulation of ALDH5A1 in stage III and IV OC was linked with better OS/PFS, but with HR = 0.71/0.75, which is likely explained by the fact that in stage III and IV, some patients have a worse outcome. A negative correlation between ALDH5A1 and OS was found in pathological grades II and III, but not in grade I. Considering that only 31 patients in the analysis were grade I, this result must be further confirmed.
In conclusion, this is the first report evaluating the differential expression of ALDH5A1 in OC. The prognostic signature observed in our study is linked to downregulation of ALDH5A1. Our findings revealed the expression of ALDH5A1 in OC specimens and indicated that a high percentage of cells expressing ALDH5A1 was associated with a longer OS and that ALDH5A1 was a favorable prognostic factor in OC, but the mechanism remains unknown. Further studies are needed to clarify this issue. Our results confirmed that ALDH5A1 can be used as a biomarker, with low expression predicting worse outcomes in patients with OC.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This research was supported by funds from the National Basic Research Program of China (973 Program, No. 2013CB911304 to Hui Wang and No. 2015CB553903 to Ding Ma), Nature and Science Fundation of China (81472783, 81230038), Clinical Medical Research Projects of Municipal Health Bureau, Wuhan City (WX11B05). Nature and Science Fundation of China (No. 81402158 to Zheng Hu).
References
- Chen W, Zheng R, Baade PD et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016; 66:115-32; PMID:26808342; http://dx.doi.org/10.3322/caac.21338
- Klangsin S, Suntharasaj T, Suwanrath C, et al. Comparison of the five sonographic morphology scoring systems for the diagnosis of malignant ovarian tumors. Gynecol Obstet Invest 2012; 76:248-53; PMID:24192793; http://dx.doi.org/10.1159/000355563
- Jackson B, Brocker C, Thompson DC, Black W, Vasiliou K, Nebert DW, Vasiliou V. Update on the aldehyde dehydrogenase gene (ALDH) superfamily. Hum Genomics 2011; 5:283-303; PMID:7779080; http://dx.doi.org/10.1186/1479-7364-5-4-283
- Stewart MJ, Malek K, Xiao Q, Dipple KM, Crabb DW. The novel aldehyde dehydrogenase gene, ALDH5, encodes an active aldehyde dehydrogenase enzyme. Biochem Biophys Res Commun 1955; 211:144-51; PMID:7779080; http://dx.doi.org/10.1006/bbrc.1995.1789
- Kaur H, Mao S, Li Q, Sameni M, Krawetz SA, Sloane BF, Mattingly RR. RNA-Seq of human breast ductal carcinoma in situ models reveals aldehyde dehydrogenase isoform 5A1 as a novel potential target. PLoS ONE 2012; 7:e50249; PMID:23236365; http://dx.doi.org/10.1371/journal.pone.0050249
- Liang D, Meyer L, Chang DW, Lin J, Pu X, Ye Y, Gu J, Wu X, Lu K. Genetic variants in microRNA biosynthesis pathways and binding sites modify ovarian cancer risk, survival, and treatment response. Cancer Res 2010; 70(23):9765-76; PMID:21118967; http://dx.doi.org/10.1158/0008-5472.CAN-10-0130
- Chambliss KL, Zhang YA, Rossier E, Vollmer B, Gibson KM. Enzymatic and immunologic identification of succinic semialdehyde dehydrogenase in rat and human neural and nonneural tissues. J Neurochem 1995; 65:851-5; PMID:7616245; http://dx.doi.org/10.1046/j.1471-4159.1995.65020851.x
- EL-Hadaad HA. Immunohistochemical expression and prognostic relevance of Bmi-1, a stem cell factor, in epithelial ovarian cancer. Ann Diagn Pathol 2014; 18:58-62; PMID:24342665; http://dx.doi.org/10.1016/j.anndiagpath.2013.11.004
- Gyorffy B, Lanczky A, Szallasi Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr Relat Cancer 2012; 19:197-208; PMID:22277193; http://dx.doi.org/10.1530/ERC-11-0329
- Yeung TL, Leung CS, Wong KK, Samimi G, Thompson MS, Liu J, Zaid TM, Ghosh S, Birrer MJ, Mok SC. TGF-beta modulates ovarian cancer invasion by upregulating CAF-derived versican in the tumor microenvironment. Cancer Res. 2013; 73:5016-28; PMID:23824740; http://dx.doi.org/10.1158/0008-5472.CAN-13-0023
- Ahmed AA, Etemadmoghadam D, Temple J, Lynch AG, Riad M, Sharma R, Stewart C, Fereday S, Caldas C, Defazio A, et al. Driver mutations in TP53 are ubiquitous in high grade serous carcinoma of the ovary. J Pathol 2010; 221:49-56; PMID:20229506; http://dx.doi.org/10.1002/path.2696
- The Cancer Genome Atlas Research Network. Integrated genomic analysis of ovarian cancer. Nature 2011; 474:609-15; PMID:21720365; http://dx.doi.org/10.1038/nature10166
- Denkert C, Budczies J, Kind T, Weichert W, Tablack P, Sehouli J, Niesporek S, Könsgen D, Dietel M, Fiehn O. Mass spectrometry-based metabolic profiling reveals different metabolite patterns in invasive ovarian carcinomas and ovarian borderline tumors. Cancer Res 2006; 66:10795-804; PMID:17108116; http://dx.doi.org/10.1158/0008-5472.CAN-06-0755
- Nicholson-Guthrie CS, Guthrie GD, Sutton GP, Baenziger JC. Urine GABA levels in ovarian cancer patients: Elevated GABA in malignancy. Cancer Lett 2001; 162:27-30; PMID:11121859; http://dx.doi.org/10.1016/S0304-3835(00)00620-0
- Kamieniak MM, Rico D, Milne RL, Muñoz-Repeto I, Ibáñez K, Grillo MA, Domingo S, Borrego S, Cazorla A, García-Bueno JM. Deletion at 6q24.2-26 predicts longer survival of high-grade serous epithelial ovarian cancer patients. Mol Oncol 2015; 9:422-36; PMID:25454820; http://dx.doi.org/10.1016/j.molonc.2014.09.010
- Gayarre J, Kamieniak MM, Cazorla-Jimenez A, Muñoz-Repeto I, Borrego S, García-Donas J, Hernando S, Robles-Díaz L, García-Bueno JM, Ramón Y Cajal T, et al. The NER related gene GTF2H5 predicts survival in high-grade serous ovarian cancer patients. J Gynecol Oncol 2015; 12:12; PMID:26463438; http://dx.doi.org/10.3802/jgo.2016.27.e7
- Giselly Encinas1, Simone Maistro2, Maria Aparecida Azevedo Koike Folgueira, et al. Somatic mutations in breast and serous ovarian cancer young patients: a systematic review and meta-analysis. Rev Assoc Med Bras 2015; 61(5):474-83; PMID:26603012; http://dx.doi.org/10.1590/1806-9282.61.05.474
